Citrobacter tructae sp. nov. Isolated from Kidney of Diseased Rainbow Trout (Oncorhynchus mykiss)
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Characterization of Bacterial Strain SNU WT2
2.2. Phylogenetic Analysis and Genome Sequencing of Strain SNU WT2
2.3. Chemotaxonomic Analysis
2.4. Antibiotic Susceptibility Testing
2.5. Bacterium Challenge Trial
2.6. Histopathological Analysis
2.7. Genome Analysis of Strain SNU WT2
3. Results and Discussion
3.1. Phylogenetic and Genome Analysis of Strain SNU WT2
3.2. Description of Strain SNU WT2 Citrobacter tructae sp. nov.
3.3. Multiple Antibiotic Resistance of Strain SNU WT2
3.4. Pathogenicity of Strain SNU WT2 and Histopathological Findings
3.5. Genome Features of Strain SNU WT2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Chang, S.; Chen, Y.; Luh, K. Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods. J. Microbiol. Immunol. Infect. 2000, 33, 258–262. [Google Scholar] [PubMed]
- Ribeiro, T.G.; Goncalves, B.R.; da Silva, M.S.; Novais, Â.; Machado, E.; Carrico, J.A.; Peixe, L. Citrobacter portucalensis sp. nov., isolated from an aquatic sample. Int. J. Syst. Evol. Microbiol. 2017, 67, 3513–3517. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Wang, J.; Chen, Z.; Xiong, K.; Xu, Q.; Hu, F. Characterization of swarming motility in Citrobacter freundii. FEMS Microbiol. Lett. 2011, 317, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.G.; Clermont, D.; Branquinho, R.; Machado, E.; Peixe, L.; Brisse, S. Citrobacter europaeus sp. nov., isolated from water and human faecal samples. Int. J. Syst. Evol. Microbiol. 2017, 67, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Ganji, L.; Alebouyeh, M.; Shirazi, M.H.; Eshraghi, S.S.; Mirshafiey, A.; Daryani, N.E.; Zali, M.R. Dysbiosis of fecal microbiota and high frequency of Citrobacter, Klebsiella spp., and Actinomycetes in patients with irritable bowel syndrome and gastroenteritis. Gastroenterol. Hepatol. Bed Bench 2016, 9, 325. [Google Scholar]
- Brenner, D.J.; O’Hara, C.M.; Grimont, P.A.; Janda, J.M.; Falsen, E.; Aldova, E.; Steigerwalt, A.G. Biochemical Identification of Citrobacter Species Defined by DNA Hybridization and Description of Citrobacter gillenii sp. nov. (Formerly Citrobacter Genomospecies 10) and Citrobacter murliniae sp. nov. (Formerly Citrobacter Genomospecies 11). J. Clin. Microbiol. 1999, 37, 2619–2624. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.H.; Wang, N.Y.; Wu, A.Y.J.; Lin, C.C.; Lee, C.M.; Liu, C.P. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018, 51, 565–572. [Google Scholar] [CrossRef]
- Joaquin, A.; Khan, S.; Russell, N.; Al Fayez, N. Neonatal meningitis and bilateral cerebellar abscesses due to Citrobacter freundii. Pediatr. Neurosurg. 1991, 17, 23–24. [Google Scholar] [CrossRef]
- Svetlana, J.; Dobrila, J.D.; Lj, V. Citrobacter freundii as a cause of disease in fish. Acta. Vet. 2003, 53, 399–410. [Google Scholar] [CrossRef]
- Capkin, E.; Terzi, E.; Altinok, I. Occurrence of antibiotic resistance genes in culturable bacteria isolated from Turkish trout farms and their local aquatic environment. Dis. Aquat. Organ. 2015, 114, 127–137. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, Y.L.; Cai, J.C.; Zhou, H.W.; Yamaguchi, N.; Ichijo, T.; Chen, G.X. High prevalence of qnr and aac (6’)-Ib-cr genes in both water-borne environmental bacteria and clinical isolates of Citrobacter freundii in China. Microbes Environ. 2009. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Ros, A.; Carricajo, A.; Berthelot, P.; Pozzetto, B.; Bernabeu, S.; Nordmann, P. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemot. 2011, 55, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Sanz, F.; Coll, J. Techniques for diagnosing viral diseases of salmonid fish. Dis. Aquat. Org. 1992, 13, 211–223. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Duman, M.; Saticioglu, I.B.; Buyukekiz, A.G.; Balta, F.; Altun, S. Molecular characterization and antimicrobial resistance profile of atypical Citrobacter gillenii and Citrobacter sp. isolated from diseased rainbow trout (Oncorhynchus mykiss). J. Glob. Antimicrob. Resist. 2017, 10, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.G.; Novais, Â.; Branquinho, R.; Machado, E.; Peixe, L. Phylogeny and comparative genomics unveil independent diversification trajectories of qnrB and genetic platforms within particular Citrobacter species. Antimicrob. Agents Chemot. 2015, 59, 5951–5958. [Google Scholar] [CrossRef]
- Clermont, D.; Motreff, L.; Passet, V.; Fernandez, J.C.; Bizet, C.; Brisse, S. Multilocus sequence analysis of the genus Citrobacter and description of Citrobacter pasteurii sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1486–1490. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Efron, B. Better bootstrap confidence intervals. J. Am. Stat. Assoc. 1987, 82, 171–185. [Google Scholar] [CrossRef]
- EzBioCloud OrthoANIu. Available online: https://www.ezbiocloud.net/tools/ani (accessed on 10 October 2020).
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Genome-to-Genome Distance Calculator 2.1. Available online: http://ggdc.dsmz.de/distcalc2.php (accessed on 10 October 2020).
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; Technical note #101; MIDI: Newark, DE, USA, 1990; pp. 1–6. [Google Scholar]
- Sasser, M. “Tracking” a Strain Using the Sherlock Microbial Identification System (MIS); Technical Note #102; MIDI: Sandy Drive Newark, DE, USA, 2001; p. 14. [Google Scholar]
- Wayne, P.A. Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; pp. 32–39. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Pepperell, C.; Kus, J.V.; Gardam, M.A.; Humar, A.; Burrows, L.L. Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 2002, 46, 3555–3560. [Google Scholar] [CrossRef] [PubMed]
- Toranzo, A.E.; Cutrin, J.M.; Roberson, B.S.; Nunez, S.; Abell, J.M.; Hetrick, F.M.; Baya, A.M. Comparison of the taxonomy, serology, drug resistance transfer, and virulence of Citrobacter freundii strains from mammals and poikilothermic hosts. Appl. Environ. Microbiol. 1994, 60, 1789–1797. [Google Scholar] [CrossRef]
- Mediterranee-Infection. Available online: http://en.mediterranee-infection.com/articlc.php?laref=283&titre=arg-annot- (accessed on 21 September 2020).
- VFDB: Virulence Factor Database. Available online: http://www.mgc.ac.cn/VFs/ (accessed on 15 December 2020).
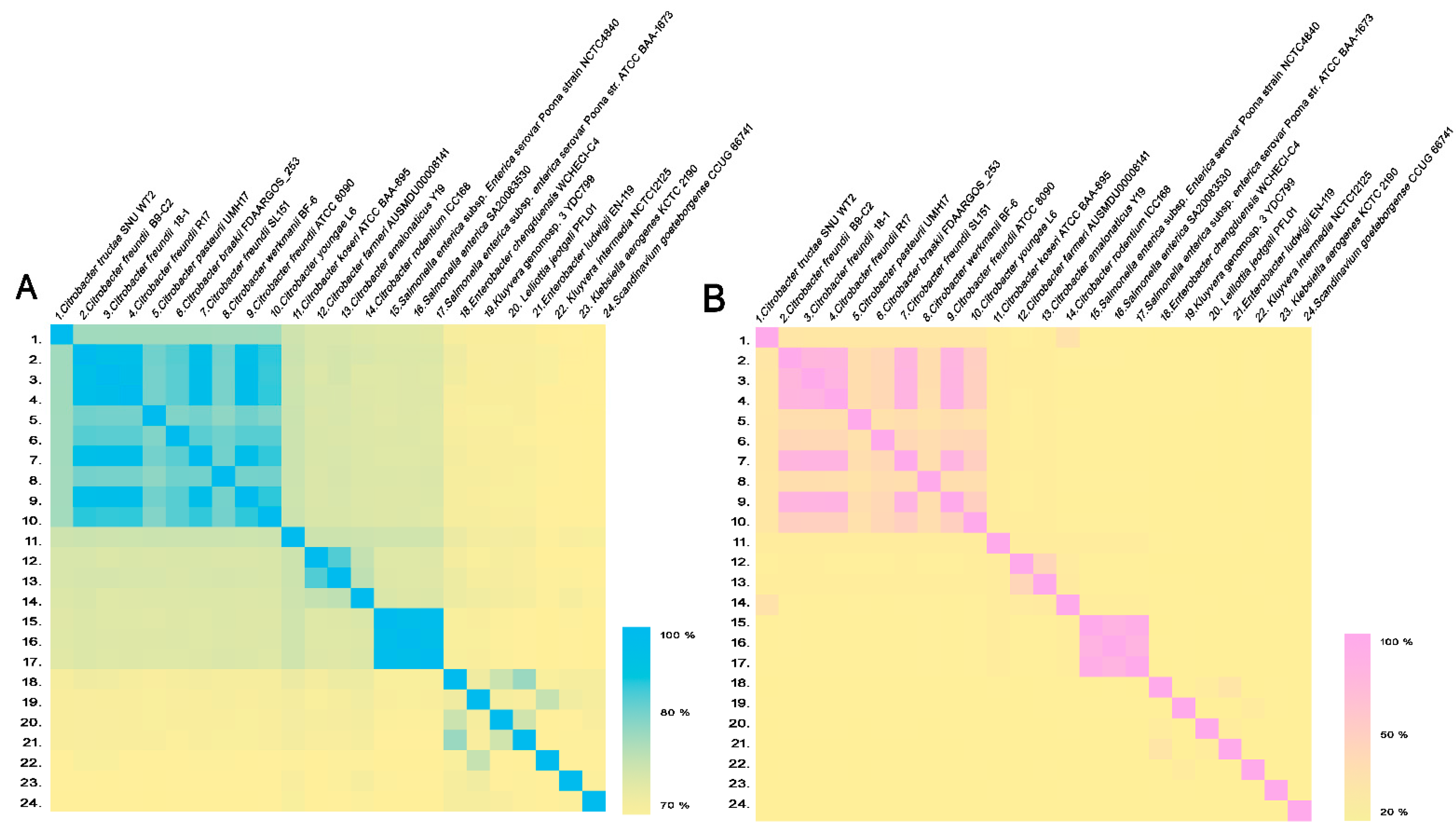



| Number | Species | Strain | C. tructae SNU WT2 (ANI Value %) | C. tructae SNU WT2 (GGDC Value %) |
|---|---|---|---|---|
| 1. | Citrobacter tructae | SNU WT2 | 100 | 100 |
| 2. | Citrobacter freundii | B9-C2 | 87.43 | 33.40 |
| 3. | Citrobacter freundii | 18-1 | 87.38 | 33.20 |
| 4. | Citrobacter freundii | R17 | 87.35 | 33.1 |
| 5. | Citrobacter pasteurii | UMH17 | 87.35 | 33.2 |
| 6. | Citrobacterbraakii | FDAARGOS_253 | 87.34 | 33.1 |
| 7. | Citrobacter freundii | SL151 | 87.33 | 33.2 |
| 8. | Citrobacter werkmanii | BF-6 | 87.25 | 32.7 |
| 9. | Citrobacter freundii | ATCC 8090 | 87.23 | 33.1 |
| 10. | Citrobacter youngae | L6 | 87.14 | 33.1 |
| 11. | Citrobacter koseri | ATCC BAA-895 | 83.49 | 27.1 |
| 12. | Citrobacter farmeri | AUSMDU00008141 | 82.46 | 25.3 |
| 13. | Citrobacter amalonaticus | Y19 | 82.44 | 25.6 |
| 14. | Citrobacter rodentium | ICC168 | 82.06 | 37.6 |
| 15. | Salmonella enterica subsp. Enterica serovar Poona | NCTC4840 | 81.79 | 24.8 |
| 16. | Salmonella enterica | SA20083530 | 81.73 | 24.8 |
| 17. | Salmonella enterica subsp. Enterica serovar Poona | ATCC BAA-1673 | 81.6 | 24.8 |
| 18. | Enterobacter chengduensis | WCHECl-C4 | 79.65 | 22.9 |
| 19. | Kluyvera genomosp. 3 | YDC799 | 79.34 | 23.1 |
| 20. | Lelliottia jeotgali | PFL01 | 79.3 | 22.9 |
| 21. | Enterobacter ludwigii | EN-119 | 79.28 | 22.6 |
| 22. | Kluyvera intermedia | NCTC12125 | 78.77 | 23 |
| 23. | Klebsiella aerogenes | KCTC 2190 | 78.6 | 22.2 |
| 24. | Scandinavium goeteborgense | CCUG 66741 | 78.38 | 22.3 |
| Query ID | Database ID | Gene Function | % Identity | Alignment Length | Mismatches | QSS A | QSE B | DSS C | DSE D | E-Value | Bit Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CP038469 | (Bla)CMY-74:JX440349:1027-2172:1146 | AmpC beta-lactamase CMY-74 | 88.88 | 1142 | 127 | 2,937,846 | 2,938,987 | 1 | 1142 | 0 | 1257 |
| CP038469 | (Bla)CMY-44:FJ437066:1-1134:1134 | class C beta-lactamase CMY-44 | 88.55 | 926 | 106 | 2,937,846 | 2,938,771 | 1 | 926 | 0 | 995 |
| CP038469 | (Bla)CFE-1:AB107899:1008-2153:1161 | AmpC beta-lactamase CFE-1 | 88.35 | 1142 | 133 | 29,378 | 2,938,987 | 1 | 1142 | 0 | 1209 |
| CP038469 | (Bla)CMY-48:HM569226:1040-2185:1146 | AmpC beta-lactamase CMY-48 | 87.96 | 1146 | 138 | 2,937,846 | 2,938,991 | 1 | 1146 | 0 | 1178 |
| CP038469 | (Bla)CMY-13:AY339625:3641-4786:1146 | class C beta-lactamase CMY-13 | 87.52 | 1146 | 143 | 2,937,846 | 2,938,991 | 1 | 1146 | 0 | 1138 |
| CP038469 | (Bla)CMY-5:Y17716:2374-3519:1146 | beta-lactamase CMY-5 | 87.43 | 1146 | 144 | 2,937,846 | 2,938,991 | 1 | 1146 | 0 | 1130 |
| CP038469 | (Bla)LAT-1:X78117:122-1287:1146 | beta-lactamase precursor blaLAT-1 | 87 | 1146 | 149 | 2,937,846 | 2,938,991 | 1 | 1146 | 0 | 1090 |
| CP038469 | (Bla)BIL-1:X74512:127-1272:1146 | beta-lactamase bla BIL-1 | 86.65 | 1146 | 153 | 29,378 | 29,389 | 1 | 1146 | 0 | 1059 |
| CP038469 | (Bla)Penicillin_Binding_Protein_Ecoli: CP002291:6 64439-666340:1902 | Penicillin-binding protein 2 mrdA | 82.35 | 1898 | 335 | 1,975,761 | 1,977,658 | 1 | 1898 | 0 | 1106 |
| CP038469 | (Bla)AmpH:CP003785:4208384-4209544:1161 | Penicillin-binding protein AmpH | 81.62 | 729 | 134 | 2,267,261 | 2,267,989 | 385 | 1113 | 3 × 10−103 | 383 |
| CP038469 | (Bla)AMPH_Ecoli:AP012030:395554-396711:1158 | Beta-lactamase class C and penicillin binding proteins | 80.49 | 687 | 134 | 2,266,877 | 2,267,563 | 1 | 687 | 4 × 10−78 | 299 |
| CP038468 | (Tet)TetD:AB089602:1521-2705:1185 | tetracycline resistant tetD | 100 | 1185 | 0 | 47,708 | 48,892 | 1 | 1185 | 0 | 2349 |
| CP038468 | (AGly)StrB:FJ474091:264-1100:837 | streptomycin resistance protein B | 100 | 837 | 0 | 38,821 | 39,657 | 1 | 837 | 0 | 1659 |
| CP038468 | (Sul)SulII:EU360945:1617-2432:816 | SulII gene resistant to beta lactam | 100 | 816 | 0 | 37,142 | 37,957 | 1 | 816 | 0 | 1618 |
| CP038468 | (Phe)CatB4:EU935739:59054-59602:549 | chloramphenicol acetyltransferase cat B4 | 100 | 108 | 0 | 34,213 | 34,320 | 549 | 442 | 4 × 10−54 | 214 |
| CP038468 | (AGly)Aph3-Ia:HQ840942:23569-24384:816 | aphA1a confers resistance to kanamycin and neomycin | 99.88 | 816 | 1 | 32,597 | 33,412 | 816 | 1 | 0 | 1610 |
| CP038468 | (AGly)StrA:AB366441:22458-23261:804 | streptomycin resistance protein A | 99.88 | 804 | 1 | 38,018 | 38,821 | 1 | 804 | 0 | 1586 |
| CP038468 | (Phe)FloR:AKLJ01000508:383-1597:1215 | floR | 99.84 | 1215 | 2 | 40,613 | 41,827 | 1215 | 1 | 0 | 2393 |
| Query ID | Database ID | Gene Function | % Identity | Alignment Length | QSS A | QSE B | DSS C | DSE D |
|---|---|---|---|---|---|---|---|---|
| CP038469 | VFG049144 | (acrB) acriflavine resistance protein B (AcrAB) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 85 | 1572 | 2,164,436 | 2,166,007 | 1528 | 3099 |
| CP038469 | VFG048830 | (gnd) 6-phosphogluconate dehydrogenase [capsule) [Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 83 | 1403 | 440,817 | 442,219 | 1 | 1403 |
| CP038469 | VFG001443 | (ompA) outer membrane protein A (OmpA) (Escherichia coli O18:K1:H7 str. RS218) | 89 | 762 | 1,696,932 | 1,697,693 | 280 | 1041 |
| CP038469 | VFG048639 | (vipB/tssC) type VI secretion system contractile sheath large subunit VipB (T6SS) (Klebsiella pneumoniae subsp. pneumoniae HS11286) | 83 | 1175 | 1,677,973 | 1,679,147 | 1529 | 355 |
| CP038469 | VFG049018 | (rcsB) transcriptional regulator RcsB (RcsAB) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 87 | 623 | 213,022 | 213,644 | 623 | 1 |
| CP038469 | VFG048693 | (clpV/tssH) type VI secretion system ATPase TssH (T6SS) (Klebsiella pneumoniae subsp. pneumoniae HS11286) | 83 | 887 | 1,672,108 | 1,672,994 | 1454 | 568 |
| CP038469 | VFG000917 | (chuA) outer membrane heme/hemoglobin receptor ChuA (Chu) (Escherichia coli CFT073) | 82 | 963 | 684,051 | 685,013 | 1947 | 985 |
| CP038469 | VFG000923 | (fepA) ferrienterobactin outer membrane transporter (enterobactin) (Escherichia coli CFT073) | 84 | 777 | 2,033,028 | 2,033,804 | 94 | 870 |
| CP038469 | VFG048518 | (fepA) outer membrane receptor FepA (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 82 | 915 | 2,033,030 | 2,033,944 | 111 | 1025 |
| CP038469 | VFG002329 | (fliG) flagellar motor switch protein G (flagella) (Yersinia enterocolitica subsp. enterocolitica 8081) | 83 | 748 | 1,516,371 | 1,517,118 | 244 | 991 |
| CP038469 | VFG013064 | (shuA) outer membrane heme/hemoglobin receptor ShuA (Shu) (Shigella dysenteriae Sd197) | 81 | 963 | 684,051 | 685,013 | 1983 | 1021 |
| CP038469 | VFG000462 | (csgG) curli production assembly/transport protein CsgG (Agf) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 82 | 749 | 1,596,224 | 1,596,972 | 1 | 749 |
| CP038469 | VFG044165 | (entS) enterobactin exporter, iron-regulated (enterobactin) (Escherichia coli CFT073) | 80 | 834 | 2,022,167 | 2,023,000 | 842 | 9 |
| CP038469 | VFG048409 | (entA) 2,3-dihydroxybenzoate-2,3-dehydrogenase (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 84 | 548 | 2,016,160 | 2,016,707 | 786 | 239 |
| CP038469 | VFG048429 | (entE) enterobactin synthase subunit E (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 80 | 902 | 2,017,805 | 2,018,706 | 1589 | 688 |
| CP038469 | VFG000925 | (fepC) ferrienterobactin ABC transporter ATPase (enterobactin) (Escherichia coli CFT073) | 82 | 689 | 2,025,211 | 2,025,899 | 100 | 788 |
| CP038469 | VFG002356 | (flhA) flagellar biosynthesis protein FlhA (flagella) (Yersinia enterocolitica subsp. enterocolitica 8081) | 80 | 927 | 1,461,459 | 1,462,385 | 975 | 49 |
| CP038469 | VFG048683 | (hcp/tssD) type VI secretion system protein, Hcp family (T6SS) (Klebsiella pneumoniae subsp. pneumoniae HS11286) | 84 | 465 | 1,673,742 | 1,674,206 | 465 | 1 |
| CP038469 | VFG000446 | (fimD) usher protein FimD (type 1 fimbriae) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 82 | 643 | 2,096,402 | 2,097,044 | 1124 | 482 |
| CP038469 | VFG000932 | (entE) 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex (enterobactin) (Escherichia coli CFT073) | 84 | 458 | 2,017,799 | 2,018,256 | 1598 | 1141 |
| CP038469 | VFG048797 | (ugd) UDP-glucose 6-dehydrogenase (capsule) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 80 | 733 | 442,849 | 443,581 | 428 | 1160 |
| CP038469 | VFG002331 | (fliI) flagellum-specific ATP synthase FliI (flagella) (Yersinia enterocolitica subsp. enterocolitica 8081) | 80 | 695 | 1,518,261 | 1,518,955 | 406 | 1100 |
| CP038469 | VFG048478 | (fepG) iron-enterobactin transporter permease (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 79 | 862 | 2,024,255 | 2,025,115 | 133 | 993 |
| CP038469 | VFG000928 | (fepG) iron-enterobactin ABC transporter permease (enterobactin) (Escherichia coli CFT073) | 82 | 512 | 2,024,204 | 2,024,715 | 82 | 593 |
| CP038469 | VFG048459 | (ybdA) enterobactin exporter EntS (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 81 | 564 | 2,022,446 | 2,023,008 | 563 | 1 |
| CP038469 | VFG000930 | (entF) enterobactin synthase multienzyme complex component, ATP-dependent (enterobactin) (Escherichia coli CFT073) | 82 | 482 | 2,028,929 | 2,029,410 | 2279 | 1798 |
| CP038469 | VFG004125 | (csgD) DNA-binding transcriptional regulator CsgD (curli fibers/thin aggregative fimbriae (AGF)) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 86 | 329 | 1,594,712 | 1,595,040 | 1 | 329 |
| CP038469 | VFG000460 | (csgE) curli production assembly/transport protein CsgE (Agf) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 86 | 321 | 1,595,439 | 1,595,759 | 76 | 396 |
| CP038469 | VFG000920 | (chuX) putative heme-binding protein ChuX (Chu) (Escherichia coli CFT073) | 82 | 458 | 2,127,828 | 2,128,285 | 1 | 458 |
| CP038469 | VFG049133 | (acrA) acriflavine resistance protein A (AcrAB) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 82 | 449 | 2,162,149 | 2,162,597 | 460 | 908 |
| CP038469 | VFG048419 | (entB) 2,3-dihydro-2,3-dihydroxybenzoate synthetase, isochroismatase (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 82 | 473 | 2,017,165 | 2,017,637 | 608 | 136 |
| CP038469 | VFG043209 | (cheD) methyl-accepting chemotaxis protein CheD (peritrichous flagella) (Yersinia enterocolitica subsp. enterocolitica 8081) | 80 | 608 | 2,726,131 | 2,726,738 | 1517 | 910 |
| CP038469 | VFG048808 | (manB) phosphomannomutase (capsule) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 85 | 364 | 439,986 | 440,347 | 715 | 1076 |
| CP038469 | VFG002304 | (misL) putative autotransporter (MisL) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 83 | 413 | 3,490,020 | 3,490,431 | 1880 | 2291 |
| CP038469 | VFG048498 | (entF) enterobactin synthase subunit F (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 81 | 471 | 2,028,580 | 2,029,050 | 2625 | 2155 |
| CP038469 | VFG000926 | (fepD) ferrienterobactin ABC transporter permease (enterobactin) (Escherichia coli CFT073) | 79 | 599 | 2,023,128 | 2,023,726 | 22 | 620 |
| CP038469 | VFG013067 | (shuX) shu locus protein ShuX (Shu) (Shigella dysenteriae Sd197) | 81 | 458 | 2,127,828 | 2,128,285 | 1 | 458 |
| CP038469 | VFG000933 | (entB) isochorismatase (enterobactin) (Escherichia coli CFT073) | 86 | 248 | 2,017,165 | 2,017,412 | 608 | 361 |
| CP038469 | VFG000918 | (chuT) periplasmic heme-binding protein ChuT (Chu) (Escherichia coli CFT073) | 79 | 633 | 2,125,737 | 2,126,369 | 271 | 903 |
| CP038469 | VFG000924 | (fepB) ferrienterobactin ABC transporter periplasmic binding protein (enterobactin) (Escherichia coli CFT073) | 80 | 468 | 2,020,841 | 2,021,308 | 73 | 540 |
| CP038469 | VFG000919 | (chuW) putative oxygen independent coproporphyrinogen III oxidase (Chu) (Escherichia coli CFT073) | 78 | 671 | 2,126,763 | 2,127,433 | 286 | 956 |
| CP038469 | VFG048468 | (fepD) iron-enterobactin transporter membrane protein (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 80 | 449 | 2,023,260 | 2,023,708 | 142 | 590 |
| CP038469 | VFG000461 | (csgF) curli production assembly/transport protein CsgF (Agf) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 81 | 413 | 1,595,785 | 1,596,194 | 1 | 413 |
| CP038469 | VFG000457 | (csgB) minor curlin subunit precursor, curli nucleator protein CsgB (Agf) (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2) | 91 | 155 | 1,593,794 | 1,593,948 | 155 | 1 |
| CP038469 | VFG048449 | (fepB) iron-enterobactin transporter periplasmic binding protein (Ent) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 81 | 373 | 2,020,902 | 2,021,274 | 134 | 506 |
| CP038469 | VFG000931 | (entC) isochorismate synthase 1 (enterobactin) (Escherichia coli CFT073) | 79 | 598 | 2,019,406 | 2,019,944 | 1188 | 650 |
| CP038469 | VFG048990 | (galF) UTP-glucose-1-phosphate uridylyltransferase subunit GalF (capsule) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 81 | 380 | 424,787 | 425,166 | 13 | 392 |
| CP038469 | VFG002365 | (gmd) GDP-mannose 4,6-dehydratase (O-antigen) (Yersinia enterocolitica subsp. enterocolitica 8081) | 78 | 527 | 434,829 | 435,355 | 268 | 794 |
| CP038469 | VFG048885 | (gmd) GDP-D-mannose dehydratase (capsule) (Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044) | 78 | 590 | 434,577 | 435,166 | 25 | 614 |
| CP038469 | VFG000936 | (iutA) ferric aerobactin receptor precursor IutA (aerobactin) (Escherichia coli CFT073) | 82 | 290 | 546,850 | 547,139 | 545 | 256 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, W.J.; Kim, H.J.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kang, J.W.; Kwon, J.; Lee, S.B.; Oh, W.T.; Jun, J.W.; et al. Citrobacter tructae sp. nov. Isolated from Kidney of Diseased Rainbow Trout (Oncorhynchus mykiss). Microorganisms 2021, 9, 275. https://doi.org/10.3390/microorganisms9020275
Jung WJ, Kim HJ, Giri SS, Kim SG, Kim SW, Kang JW, Kwon J, Lee SB, Oh WT, Jun JW, et al. Citrobacter tructae sp. nov. Isolated from Kidney of Diseased Rainbow Trout (Oncorhynchus mykiss). Microorganisms. 2021; 9(2):275. https://doi.org/10.3390/microorganisms9020275
Chicago/Turabian StyleJung, Won Joon, Hyoun Joong Kim, Sib Sankar Giri, Sang Guen Kim, Sang Wha Kim, Jeong Woo Kang, Jun Kwon, Sung Bin Lee, Woo Taek Oh, Jin Woo Jun, and et al. 2021. "Citrobacter tructae sp. nov. Isolated from Kidney of Diseased Rainbow Trout (Oncorhynchus mykiss)" Microorganisms 9, no. 2: 275. https://doi.org/10.3390/microorganisms9020275
APA StyleJung, W. J., Kim, H. J., Giri, S. S., Kim, S. G., Kim, S. W., Kang, J. W., Kwon, J., Lee, S. B., Oh, W. T., Jun, J. W., & Park, S. C. (2021). Citrobacter tructae sp. nov. Isolated from Kidney of Diseased Rainbow Trout (Oncorhynchus mykiss). Microorganisms, 9(2), 275. https://doi.org/10.3390/microorganisms9020275








