Abstract
The association of the lower respiratory tract microbiome in pigs with that of other tissues and environment is still unclear. This study aimed to describe the microbiome of tracheal and oral fluids, air, and feces in the late stage of Mycoplasma hyopneumoniae infection in pigs, and assess the association between the tracheal microbiome and those from air, feces, and oral fluids. Tracheal fluids (n = 73), feces (n = 71), oropharyngeal fluids (n = 8), and air (n = 12) were collected in seeder pigs (inoculated with M. hyopneumoniae) and contact pigs (113 days post exposure to seeder pigs). After DNA extraction, the V4 region from 16S rRNA gene was sequenced and reads were processed using Divisive Amplicon Denoising Algorithm (DADA2). Clostridium and Streptococcus were among the top five genera identified in all sample types. Mycoplasma hyopneumoniae in tracheal fluids was associated with a reduction of diversity and increment of M. hyorhinis, Glaesserella parasuis, and Pasteurella multocida in tracheal fluids, as well as a reduction of Ruminiclostridium, Barnesiella, and Lactobacillus in feces. Air contributed in a greater proportion to bacteria in the trachea compared with feces and oral fluids. In conclusion, evidence suggests the existence of complex interactions between bacterial communities from distant and distinct niches.
1. Introduction
The collection of microorganisms and their genetic elements, persistently or transiently present in a certain ecological niche, is known as its microbiome. The microbiome can be decomposed in different dimensions depending on the type of microorganisms of interest, e.g., the viral microbiome (virome), the fungal microbiome (mycobiome), or the bacterial microbiome. The latter is the most commonly studied and is associated with a plethora of effects in animal hosts, such as breakdown of otherwise indigestible feedstuff (influencing feed efficiency [1]), competitive exclusion of pathogens [2], priming and regulation of the mucosal immune system [3,4], and disease [5].
The most studied microbiome by far is the gut microbiome. Each segment of the gut has a specific associated microbiome, which can be shaped by diet, host genetics, and environmental factors [6,7,8]. Zhao et al. [9] documented that aerobic and facultative anaerobic bacteria dominate in the small intestine of adult pigs, while anaerobic bacteria dominate in the large intestine. In pigs, the fecal microbiome is more related to the large intestine microbiome [9] and some of the genera commonly present in high abundance include Bacteroides, Lactobacillus, Prevotella, Clostridium, Escherichia, and Megasphaera, with evidence of bacterial succession during the pig’s life [10,11,12,13,14,15]. Changes in the fecal microbiome are associated with infections from major swine pathogens, such as Salmonella, Lawsonia, Brachyspira, and porcine epidemic diarrhea virus (PEDV) [16,17,18,19].
The microbiome of the oropharyngeal section of the digestive system in pigs is composed predominantly of Streptococcus, Lactobacillus, and Actinobacillus [20]. Shared between the digestive and respiratory systems, the oropharyngeal section is attractive for sampling due to their accessibility. In fact, oral fluids, saliva, and oral mucosal transudate [21] collected in ropes gnawed on by pigs in pens, are widely used for the detection of respiratory viruses in swine herds [22,23]. Although Murase, et al. [24] described the microbiome in pig saliva, the microbiome associated with oral fluids was not described.
The microbiome from certain sections of the respiratory tract in pigs was documented [15,25,26]. However, the extent of evidence is far less than that for the gut microbiome. More specifically, the lung microbiome in pigs affected by Mycoplasma hyopneumoniae was described to be dominated by bacteria members of the families Mycoplasmataceae, Flavobacteriaceae, and Pasteurellaceae if there are signs of pneumonia, or by members of Mycoplasmataceae, Bradyrhizobiaceae, and Flavobacteriaceae otherwise [25]. Mycoplasma hyopneumoniae is a major respiratory pathogenic bacterium in pigs, causing a chronic disease characterized by transient dry cough and increased susceptibility to other respiratory pathogens [27,28,29]. By contrast, the microbiome associated with the trachea in pigs has not yet been described, nor has its association with the extent of M. hyopneumoniae colonization. This is of interest, since deep tracheal catheter samples are nowadays commonly used for the detection of M. hyopneumoniae in live pigs, as the trachea represents an important site of colonization [30].
Recently, researchers strived to characterize the microbiome of the environment of swine facilities [31,32,33,34]. The air microbiome is of special interest as it may serve as an epidemiological vehicle for the transmission of microorganisms between different environments. A core of 19 bacterial families were shown to increase in the air microbiome of swine farms and in the noses of pigs and workers in comparison with the microbiome of the noses of people not exposed to live pigs [32].
Currently, there is little evidence of the association of the microbiome residing in the lower respiratory tract in pigs with that of other environments. The lower respiratory tract has long been considered a pristine and privileged section of the host, regarding bacterial colonization. However, recent data uncovered microbial communities that colonize the lower respiratory tract and appear to influence host responses to disease. Therefore, the objectives of this study were to describe the microbiome associated with tracheal and oral fluids, air, and feces during late stages of M. hyopneumoniae infection in pigs, to assess their potential association with infection status, and to infer to what extent the tracheal microbiome is related to microbial communities from air, feces, and oral fluids.
2. Materials and Methods
All pigs in this study were cared for following the guidelines of the Institutional Animal Care and Use Committee of Pipestone Applied Research (IACUC #3–18). Crossbred pigs were sourced from a commercial sow farm with high health status, known to be positive for influenza A virus, and negative for Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and M. hyopneumoniae infection for the last five years. Pigs were weaned at approximately three weeks of age, then transported and placed in a Midwestern United States commercial wean-to-finish facility designed for research [30]. Pigs were separated into groups of seeders (experimentally infected) and contacts (naturally infected via contact with seeders) via systematic assignment after blocking by sow parity range and ordering them by sex and weight. Seeders and contacts were randomly allocated to pens based on a 1:6 seeder-to-contact ratio, for a total of 28 pigs per pen. At eight weeks of age, seeders were intratracheally inoculated with M. hyopneumoniae and contacts were exposed to seeders following inoculation [30]. Samples included in this study were collected randomly from a subset of seeders (n = 47) and contacts (n = 26), or the environment at 113 days postexposure to M. hyopneumoniae. The majority of contact pigs were confirmed infected at least one month prior to sampling. Therefore, they were considered to be at a late infection stage [24]. Pigs were not exposed to antimicrobials known to be effective against Mycoplasma species during the study. Five out of the 73 selected pigs (one seeder and four contacts) were treated for clinical signs suggestive of Streptococcus suis infection using penicillin or ceftiofur, as indicated, at least two months prior to sample collection.
Deep tracheal catheters were used to collect tracheal fluids in pigs [30]. Fecal samples were obtained by digital insertion immediately following tracheal fluid sample collection. Oral fluid samples (n = 8) were collected as described by Prickett et al. [23], a procedure standardized to be performed at the pen level. Briefly, cotton ropes were hung using new gloves on the front gate of pens at pig shoulder height and pigs were allowed to chew on the ropes for 30 min. New gloves were donned for each rope collection and a new Ziploc bag was placed around the rope. The rope was squeezed from outside of the Ziploc bag while pulling away from the wall hook. The oral fluid sample was poured off from the Ziploc bag into a sterile snap cap Falcon tube, without touching the sample. Pens for sample collection were selected using a random number generator.
Six cyclonic filter collection devices (ACD-200 BOBCAT Air Sampler, Drexel, MO, USA) distributed throughout the entire air space where pens were located were used to collect air samples (n = 12). Air samples were collected for one hour during each of two samplings. Once the filtration run was complete, the filter was removed and rinsed using the manufacturer provided filter reagent into a sterile cup. All samples were refrigerated immediately and submitted to the Health Management Center (HMC; Boehringer Ingelheim Animal Health, Ames, IA, USA) for analysis.
DNA extraction for oral fluid and air samples (MagMAXTM Pathogen DNA/RNA Kit, Life Technologies, Grand Island, NY, USA) [35,36,37] or tracheal fluid samples (MagMax-96 Viral RNA Isolation Kit, Life Technologies, Grand Island, NY, USA) [38] was performed using an extraction robot (KingFisherTM, Life Technologies, Grand Island, NY, USA). A species-specific real-time PCR was run for M. hyopneumoniae detection (VetMAXTM qPCR Master Mix and VetMAXTM M. hyopneumoniae Reagents Kit, Life Technologies, Grand Island, NY, USA) using a Roche LightCycler® (480 Roche Life Science, Indianapolis, IN, USA). Both extraction and PCR methods were performed at the HMC. Real-time PCR samples with a Ct value < 38 were considered positive for M. hyopneumoniae. The M. hyopneumoniae infection status of each pig was based on the real-time PCR result in tracheal fluid samples, as those were individual samples that could be matched to a pig, contrary to environment and pen-based samples, such as air and oral fluids.
Fecal samples were submitted to the University of Minnesota Genomic Center (UMGC) for DNA extraction (DNAeasy PowerSoil kit, Qiagen, Germantown, MD, USA) and sequencing. The V4 region from 16S rRNA gene was amplified for 25 cycles in fecal samples and for 30 cycles in tracheal fluid, air, and oral fluid samples to account for the expected lower biomass in the latter. Samples were also dual indexed during library preparation. Sequencing of the amplicons was performed using MiSeq 2 × 300 bp platform.
All bioinformatics analyses were performed using R version 3.6.2 [39]. The quality profiles of the reads for each sample were visually inspected separately for forward and reverse reads. Primers and adapters were trimmed from all reads (Trim Galore version 0.6.4_dev for adapter removal, URL: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The last 15 bases of forward reads and the last 50 bases of reverse reads were also trimmed. Denoising of reads, merging of paired reads, and removal of chimera were performed using the Divisive Amplicon Denoising Algorithm (DADA2) package (version 1.14.0) [40]. DADA2-formatted training fasta file derived from the Silva Project’s version 132 release [41] was used to assign taxonomy to the amplicon sequence variants (ASV) obtained from the previous steps. Amplicon sequence variants from Eukarya, chloroplasts, and mitochondria were removed. Similarly, ASVs present in less than 1% of the samples were removed. This last step was performed separately by sample type. Samples with fewer reads than negative DNA extraction controls were removed as well.
The inverse Simpson alpha diversity index was computed and used in linear regression models to assess its association with sample type and M. hyopneumoniae Ct values, either as a continuous variable or as a binary variable (M. hyopneumoniae status). Detection of M. hyopneumoniae was not evaluated in the fecal samples due to the intrinsic restricted respiratory nature of this bacterium. Thus, M. hyopneumoniae Ct values from the tracheal fluid samples were used for analysis.
The following beta diversity dissimilarity indices were computed: Bray–Curtis, Jaccard, Unifrac, weighted Unifrac, and Aitchison. The beta diversity dissimilarity indices were used in principal coordinate analysis (PCoA) plots to explore the clustering of samples. The association of beta diversity dissimilarity indices with M. hyopneumoniae status, seeder/contact status, sample type, and the interaction of sample type and M. hyopneumoniae status was evaluated in tracheal fluids and fecal samples, using PERMANOVA stratified by pig [42] when appropriate.
With the aim of assessing the association between the ASVs corresponding to M. hyopneumoniae with the rest of the ASVs in tracheal fluids, the counts for all ASVs determined as M. hyopneumoniae through the search of the NCBI 16S ribosomal RNA sequences database (Bacteria and Archaea) using the nucleotide BLAST tool were aggregated. The centered log relative abundance (CLR) for all ASVs was calculated. In order to select the ASVs more strongly associated with M. hyopneumoniae, an elastic net model was built using glmnetUtils and glmnet packages in R [43,44], selecting the alpha and lambda parameters via leave-one-out cross-validation (LOOC).
A similar penalized approach was used to assess the association of the ASVs corresponding to M. hyopneumoniae in tracheal fluids with the ASVs in paired fecal samples, as well as the association of M. hyopneumoniae Ct values in tracheal fluids with the ASVs in the same samples and with the ASVs in paired fecal samples. Furthermore, ASVs differentially abundant according to M. hyopneumoniae status were detected in tracheal and fecal samples using ANCOM package in R [45].
A Bayesian finite mixture model (SourceTracker package) [46] was used to infer the proportions of the tracheal fluid microbiome potentially originating from the other microbial communities, i.e., fecal, oral fluids, and air microbiome, as putative sources. The Kolmogorov–Smirnov test was used to assess whether the empirical cumulative distribution functions (ecdf) of the proportions inferred for each microbial community differed according to M. hyopneumoniae status.
3. Results
The distribution of M. hyopneumoniae-positive samples categorized by sample type is shown in Table 1. Detection of M. hyopneumoniae was not evaluated in feces due to the respiratory nature of M. hyopneumoniae pathogenesis. The range and median of the sequencing depth (in reads) per sample type were 6079–23,346 and 15,486 for tracheal fluids, 4525–26,202 and 16,323 for air, 10,486–20,337 and 19,188 for oral fluids, and 6925–32,307 and 22,649 for feces, respectively. Rarefaction curves showed that the asymptote of the number of ASVs was reached in all sample types (Figure S1). The total numbers of detected ASVs were 3601 in tracheal fluids, 2758 in feces, 1843 in oral fluids, and 1348 in air. The range and median of ASVs detected per sample type were 115–368 and 229 ASVs for tracheal fluids, 65–371 and 263 ASVs for air, 104–362 and 276 ASVs for feces, and 217–397 and 371 ASVs for oral fluids, respectively. Overall, 26 percent of the ASVs detected in tracheal fluids were shared with feces, 25% with oral fluids, and 22% with air. The common presence of ASVs among sample types is represented in Figure 1. Furthermore, the set of ASVs exclusively shared between tracheal and oral fluids was the greatest nonoverlapping set (412 shared ASVs), followed by the set of ASVs exclusively shared between tracheal fluids and feces (363 shared ASVs). A set of shared ASVs, represented by those shared among all sample types, was composed of 245 ASVs. Clostridium and Streptococcus were consistently among the top five most abundant bacterial genera in all sample types (Table 2, Figures S2–S6).

Table 1.
Distribution of Mycoplasma hyopneumoniae detection in study fluids and samples.
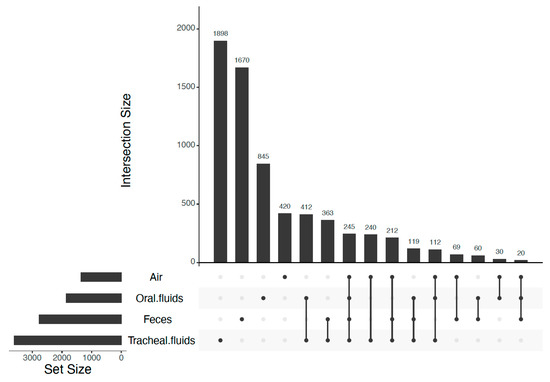
Figure 1.
Amplicon sequence variants (ASVs) shared among sample types. Intersection size: number of ASVs comprising a specific nonoverlapping set. Dark circles indicate the sample types that are part of the set. Dark circles connected by a line indicate that two or more sample types are part of the set. The plot was made using UpSetR package in R [47] with the layout described by Lex and Gehlenborg [48].

Table 2.
Five most abundant bacterial genera identified in microbial communities from various fluids and samples.
There was evidence of an overall effect of sample type on the alpha diversity (p = 0.0014), measured with the inverse Simpson index, when M. hyopneumoniae status was used as a covariate (Figure S7). No statistically significant overall effect for M. hyopneumoniae status was observed (p = 0.1672). Both overall effects were statistically significant when M. hyopneumoniae Ct values were used as covariate values instead (p < 0.0001 for each overall effect). Since M. hyopneumoniae DNA was detected in one of eight oral fluid samples and undetected in air samples, the linear model assessing the association among alpha diversity, sample types, and M. hyopneumoniae Ct values was reduced to include tracheal fluids and fecal samples. No association between alpha diversity of fecal samples and M. hyopneumoniae Ct values in tracheal fluids was observed (p = 0.6538). However, a quadratic association between alpha diversity of tracheal fluids and M. hyopneumoniae Ct values was detected (p < 0.0001), with a faster reduction in alpha diversity as the Ct values decreased (Figure 2).
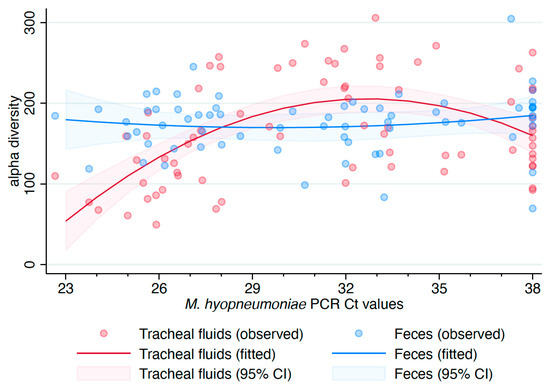
Figure 2.
Association of alpha diversity (inverse Simpson index) with sample type and Mycoplasma hyopneumoniae real-time PCR Ct values in tracheal fluids. 95% CI: 95% confidence interval.
All sample types were analyzed using several dissimilarity measures in PCoA plots for the assessment of clustering, namely Aitchison, Bray–Curtis, Jaccard, Unifrac, and weighted Unifrac. Three clusters were consistently distinguished in the PCoA plots, one cluster composed of fecal samples, another composed of air samples, and a third cluster composed of tracheal and oral fluid samples (Figure 3). A significant association between sample type and beta diversity was observed irrespective of the dissimilarity index used (Table 3). Mycoplasma hyopneumoniae status was significantly associated with beta diversity in tracheal fluids, except when the Unifrac index was used. However, M. hyopneumoniae was not significantly associated with beta diversity in fecal samples regardless of the index used. Similarly, M. hyopneumoniae modified the association between sample type and beta diversity when either Bray–Curtis or the weighted Unifrac index were used (Table 3). Seeder/contact status was not significantly associated with beta diversity in tracheal fluids or fecal samples, except when the weighted Unifrac index was used in tracheal fluids.
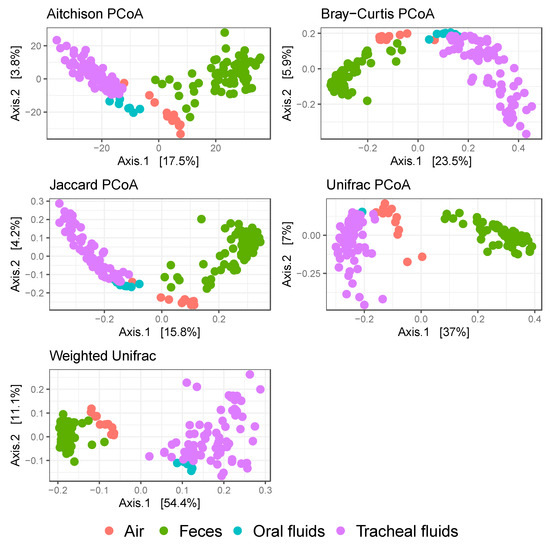
Figure 3.
Microbial communities clustering analysis. Each dot represents an individual sample. Five different dissimilarity indices were performed and are shown in the following order (left to right, top to bottom): Aitchison, Bray–Curtis, Jaccard, Unifrac, and weighted Unifrac. Samples are color-coded by type. Orange: air; green: feces; teal: oral fluids; purple: tracheal fluids.

Table 3.
Association of beta diversity dissimilarity indices with sample type (tracheal fluids and feces) and the interaction of sample type and Mycoplasma hyopneumoniae status.
A total of 205 ASVs in tracheal fluids were associated with M. hyopneumoniae ASVs in tracheal fluid samples (Table S1). One ASV belonging to the genus Ruminiclostridium in feces displayed a negative association with M. hyopneumoniae ASVs in tracheal fluids (Figure S8). Two ASVs in tracheal fluids, from genera Clostridium and Parabacteroides, were positively associated with M. hyopneumoniae PCR Ct values, whereas two ASVs from the genus Pasteurella and the ASVs corresponding to M. hyopneumoniae in tracheal fluids were negatively associated with Ct values (Figures S9–S12). No ASVs in feces were associated with M. hyopneumoniae PCR Ct values in tracheal fluids. Furthermore, no ASVs in tracheal fluids were significantly associated with M. hyopneumoniae-positive status, except for those belonging to M. hyopneumoniae. In contrast, two ASVs from the genera Barnesiella and Lactobacillus in feces were shown to have a significant negative association with M. hyopneumoniae-positive status (FDR = 0.05; Figure 4). Mycoplasma hyopneumoniae ASVs were not detected in feces.
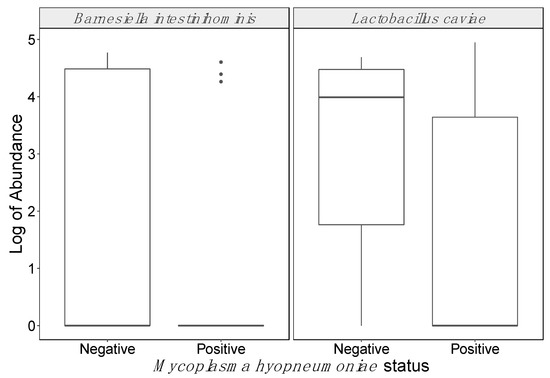
Figure 4.
Amplicon sequence variants differentially abundant in fecal samples according to Mycoplasma hyopneumoniae status of paired tracheal fluid samples. Real-time PCR in tracheal samples was used to establish M. hyopneumoniae status.
The heterogeneity in the proportions of the tracheal fluid microbial community potentially originating from fecal, air, or oral fluids sources is depicted in Figure 5. The characteristics of the distribution of the estimated proportions of ASVs by source are shown in Table 4. There was no statistical evidence that the distribution of the proportions of ASVs potentially originating from each microbial community differed according to M. hyopneumoniae status (p = 0.1815, 0.9903, and 0.4273 for air, fecal, and oral fluid sources, respectively; Figures S13–S15).
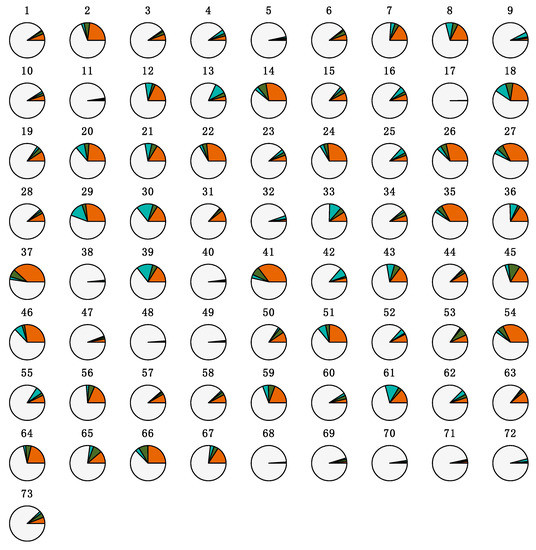
Figure 5.
Proportions of the tracheal fluid microbial community potentially originating from fecal, air, or oral fluid sources. Each pie represents the tracheal fluid of one individual pig (n = 73). Sources are color-coded. Orange: air; green: feces; teal: oral fluids; light gray: unknown.

Table 4.
Characteristics of the distribution of the proportions of tracheal fluid microbial communities potentially originating from fecal, air, or oral fluid sources.
4. Discussion
This study described the microbiome identified in tracheal and oral fluids, air, and feces of pigs reared under commercial conditions during late-stage infection with M. hyopneumoniae. Results of this study showed that both alpha and beta diversity significantly differed among sample types. The presence of M. hyopneumoniae in tracheal samples was associated with several bacterial species in those samples, as well as in fecal samples. Moreover, this research assessed whether the tracheal microbiome could originate from microbial communities from air, feces, and oral fluids in pigs. Our results suggest that a relatively small but higher proportion of the tracheal microbiome community may have originated from the air, compared to the other two studied potential sources, namely, feces and oral fluids.
Infections caused by M. hyopneumoniae are highly prevalent worldwide and constitute the most important bacterial disease affecting the respiratory tract in swine [29]. Pigs affected with M. hyopneumoniae generally do not succumb to the disease. However, they remain infectious for extremely long periods, reaching up to seven months [28], which identifies this infection as chronic in nature, leading to endemic situations. Infected pigs usually exhibit growth retardation and increased susceptibility to other respiratory pathogens of bacterial and viral origin [29]. Thus, M. hyopneumoniae is a key player in the development of the porcine respiratory disease complex. Therefore, the M. hyopneumoniae infection model recreated in this study is considered ideal to assess the effect of bacterial infections in the respiratory microbial communities.
Clostridium and Streptococcus were consistently among the top five most abundant bacterial genera in all sample types. Streptococcus is commonly documented in several pig niches. Streptococcus is part of the core tonsillar microbiome in newborn piglets [26], and is also part of the core oropharyngeal microbiome in piglets, together with Lactobacillus and Actinobacillus [20]. Streptococcus is one of the most abundant genera in pig saliva, along with Actinobacillus, Moraxella, and Rothia [24], and was reported as the major bacterial genus in aerosols from pig confinement buildings [33]. Additionally, Clostridium and Escherichia were documented to be among the most abundant genera in feces during the preweaning phase, while Megasphaera and Lactobacillus seem to be more abundant during the postweaning phase in pigs [12,13,14,15]. Clostridium was also shown to have a high relative abundance in the noses of pigs before weaning, being replaced by Lactobacillus thereafter [15].
The alpha diversity of a bacteriological niche is comprised of the number of bacterial species and the evenness of the distribution of those species [49]. The alpha diversity of the tracheal microbiome decreased with greater concentrations of M. hyopneumoniae, which is in agreement with other studies, where pathogens, such as PEDV in pigs and Enterococcus faecalis in humans, induce the reduction of the evenness and richness in their ecological niche [50,51]. The dominance of M. hyopneumoniae in the tracheas of affected pigs could have led to an imbalance of the proportions of the bacterial members of the tracheal microbiome, resulting in a lower evenness and alpha diversity in comparison with the trachea of pigs not infected with this bacterium. Mycoplasma hyopneumoniae-induced shift in the tracheal microbiome alpha diversity may not only reflect its fitness at colonizing the tracheal epithelium, but also its ability to modulate the microbiome to its advantage.
Differences in the microbiome composition among samples collected in this study were reflected in the association of beta diversity and sample type, which was robust to the dissimilarity index used. Similarly, beta diversity was shown to reflect age effects in the nasal and fecal microbiomes of pre- and postweaning pigs [15], and litter effects in the tonsillar microbiomes of newborn piglets [26].
Beta diversity is intrinsically related to differentially abundant bacteria. Using different approaches to parameterize the presence of M. hyopneumoniae in tracheal samples, M. hyorhinis, Niastella hibisci [52], Glaesserella parasuis, Terrimonas rubra [53], and Pasteurella multocida were observed to increase in tracheal samples alongside M. hyopneumoniae. It is remarkable that M. hyopneumoniae was associated with three major swine pathogens. Mycoplasma hyorhinis may cause arthritis and/or polyserositis [54], Glaesserella (Haemophilus) parasuis is the etiologic agent of Glässer’s disease characterized by polyserositis [55], and P. multocida is associated with atrophic rhinitis and the porcine respiratory disease complex [56]. However, all three bacteria can be present in the respiratory tracts of healthy pigs, which highlights the complexity of the pathogen–host-microbiome interaction. Furthermore, these findings support the current perception of M. hyopneumoniae as a pathogen capable of inducing dysbiosis in the respiratory tract, potentiating opportunistic pathogens in situ.
Negative associations were detected between M. hyopneumoniae in tracheal samples and bacteria from the genera Ruminiclostridium, Barnesiella, and Lactobacillus in fecal samples from the same pigs. Ruminiclostridium is member of the family Ruminococcaceae. Interestingly, the relative abundance of Ruminococcus, another genus of the family Ruminococcaceae, in feces was previously documented to be negatively associated with severity of lung lesions caused by M. hyopneumoniae [57]. Since Barnesiella and Lactobacillus are associated with a healthy gut microbiome [58,59], the findings of this study suggest that M. hyopneumoniae-induced dysbiosis in the respiratory tract microbiome may be associated through yet unknown mechanisms to changes in gut bacterial colonization patterns.
The analysis using a Bayesian finite mixture model allowed the estimation of the proportions of the tracheal microbial community originating from fecal, air, or oral fluid sources [46], providing deeper insights than analyses based on the presence/absence of ASVs and PCoA of the Jaccard dissimilarity index. Despite great heterogeneity, the air microbial community was more closely related to that of the trachea, hence, the estimated proportion for air as a source was greater than that inferred for feces and oral fluids. However, the distribution of the proportions was different from that observed in previous research [31], in which the inferred contribution of air as a source of the nasal microbiome in pigs and workers was higher than 40% throughout the year. Those differences could stem from the fact that Kraemer, Aebi, Oppliger, and Hilty [31] sampled the upper respiratory tract of pigs instead of the lower respiratory tract, as in this study.
This was a cross-sectional study, which was not suited to address the temporality of the associations under scrutiny. Since the type of samples collected was part of the design, the authors consider the effect of sample type on the microbiome not to be prone to confounding, i.e., bias due to common causes of the type of samples collected and the microbiome composition. Conversely, M. hyopneumoniae status at day 113 post contact was not randomized, but followed experimental infection in seeders and the inherent transmission of the pathogen in contacts, which could potentially affect the associations detected in this study between M. hyopneumoniae and microbiome composition. However, it is important to highlight that common confounders, such as age, breed, biosecurity practices, and diet, were similar for all pigs in the study. Therefore, the residual confounding was considered to be small. It is important to note that the differential number of 16S PCR cycles used to account for low biomass in air, oral, and tracheal fluid samples could have affected the number of ASVs detected in those specimens. This is especially relevant for comparisons between low (tracheal fluids) and high (feces) biomass specimens. Caution is warranted in the interpretation of the results from the Bayesian finite mixture model approach, since the directionality of the associations, referring to which samples are potential sources and which are the sink, are assumed to be known. In the case of this study, the authors assessed one possible scenario, in which the air, oral fluids, and feces were the potential sources and the tracheal microbiome was the sink. However, the true directionality is unknown. Moreover, the potential contribution of the microbiome from other sites of the respiratory tract, such as nasal microbiome, was not explored in this study. Future research assessing the contribution of nasopharyngeal microbiome to the bacterial communities of the lower respiratory tract is warranted.
An important aspect to mention is the fact that M. hyopneumoniae was mainly detected in the tracheal fluid samples, while remaining undetected in the majority of the oral fluid samples. This situation is commonly encountered in infection with this bacterium, even under experimental conditions [60], and may be related to the target tissue of the pathogen, which attaches to the cilia of the respiratory tract and may be detected in the oral cavity only after being coughed up by the host. For this reason, establishing the M. hyopneumoniae status of pigs was performed based on detection of the bacterium in tracheal fluid samples.
In recent years, a greater appreciation of the complex pathogen–host-microbiome interactions resulted from studies assessing the effects of the commensal bacterial microbiome not only in situ, but also outside its immediate ecological niche. For instance, protective effects of diverse fecal microbiome were shown against systemic viral diseases in pigs, specifically against PRRSV and PCV2 [61,62], and also against bacterial diseases, such as M. hyopneumoniae [63]. In this context, the findings of this study further our knowledge about the interplay between respiratory pathogens, M. hyopneumoniae in this case, and the local microbiome. More importantly, this study provides additional evidence of long-reaching effects of M. hyopneumoniae infection and the fecal microbiome. Future research should dissect these associations to better understand data directionality and temporality. In an era of increasing antibiotic stewardship, the prospect of manipulating the microbiome to prevent and/or mitigate infectious diseases fuels the momentum of the current microbiome research.
5. Conclusions
All studied bacterial niches, tracheal and oral fluids, air, and feces, showed intrinsic differences in their bacterial communities. However, Clostridium and Streptococcus were consistently among the top five most abundant bacterial genera across all bacterial niches. Mycoplasma hyopneumoniae infection status was associated with a shift in the tracheal microbiome, reducing its diversity and increasing the relative abundance of the major swine pathogens M. hyorhinis, G. parasuis, and P. multocida. Furthermore, M. hyopneumoniae infection was associated with a decrease in the relative abundance of Ruminiclostridium, Barnesiella, and Lactobacillus in feces. Finally, the air microbial community may have a greater contribution to the tracheal microbiome in comparison with feces and oral fluids.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/2/252/s1: Figures S1–S15, Table S1, File 1.txt: Top five results of the BLAST search for the ASVs associated with M. hyopneumoniae ASVs in tracheal fluid samples; File 2.txt: Top five results of the BLAST search for the ASVs in tracheal fluids associated with M. hyopneumoniae PCR Ct values; File 3.txt: Top five results of the BLAST search for the ASVs in feces associated with M. hyopneumoniae-positive status.
Author Contributions
Conceptualization, A.S., E.F., F.L.L., and M.P.; funding acquisition, A.S., E.F., and F.L.L.; methodology, R.V.-C., A.S., E.F., R.I., F.L.L., and M.P.; investigation, R.V.-C., A.S., E.F., J.N., F.L.L., and M.P.; formal analysis, R.V.-C., R.S.S., F.L.L., and M.P.; visualization, R.V.-C., R.S.S., F.L.L., and M.P.; supervision, F.L.L. and M.P.; writing—original draft preparation, R.V.-C., A.S., and M.P.; writing—review and editing, R.V.-C., A.S., E.F., R.I., R.S.S., J.N., F.L.L., and M.P. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support for this investigation was provided by Boehringer Ingelheim Animal Health USA Inc.
Institutional Review Board Statement
The study was approved by the Institutional Animal Care and Use Committee of Pipestone Applied Research (IACUC #3–18 on 22 May 2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors. The data are not publicly available due to being proprietary to Boehringer Ingelheim Animal Health (BIAH).
Acknowledgments
Special thanks to Boehringer Ingelheim Animal Health and Pipestone Applied Research field support personnel who helped with sample collection for the study.
Conflicts of Interest
A.S., E.F., and F.L.L. are employed by Boehringer Ingelheim Animal Health USA Inc. However, the evaluation of commercial products was not part of this investigation.
References
- McCormack, U.M.; Curiao, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D.; et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017, 83, e00380-17. [Google Scholar] [CrossRef] [PubMed]
- Argüello, H.; Estellé, J.; Leonard, F.; Crispie, F.; Cotter, P.D.; O’Sullivan, O.; Lynch, H.; Walia, K.; Duffy, G.; Lawlor, P.G.; et al. Influence of the intestinal microbiota on colonization resistance to Salmonella and the shedding pattern of naturally exposed pigs. mSystems 2019, 4, e00021-19. [Google Scholar] [CrossRef] [PubMed]
- Zwirzitz, B.; Pinior, B.; Metzler-Zebeli, B.; Handler, M.; Gense, K.; Knecht, C.; Ladinig, A.; Dzieciol, M.; Wetzels, S.U.; Wagner, M.; et al. Microbiota of the Gut-Lymph Node Axis: Depletion of Mucosa-Associated Segmented Filamentous Bacteria and Enrichment of Methanobrevibacter by Colistin Sulfate and Linco-Spectin in Pigs. Front. Microbiol. 2019, 10, 599. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Niederwerder, M.C. Role of the microbiome in swine respiratory disease. Vet. Microbiol. 2017, 209, 97–106. [Google Scholar] [CrossRef]
- Pollock, J.; Hutchings, M.R.; Hutchings, K.E.K.; Gally, D.L.; Houdijk, J.G.M. Changes in the Ileal, but Not Fecal, Microbiome in Response to Increased Dietary Protein Level and Enterotoxigenic Escherichia coli Exposure in Pigs. Appl. Environ. Microbiol. 2019, 85, e01252-19. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. The pig gut microbial diversity: Understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015, 177, 242–251. [Google Scholar] [CrossRef]
- Chen, C.; Huang, X.; Fang, S.; Yang, H.; He, M.; Zhao, Y.; Huang, L. Contribution of Host Genetics to the Variation of Microbial Composition of Cecum Lumen and Feces in Pigs. Front. Microbiol. 2018, 9, 2626. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef]
- Han, G.G.; Lee, J.Y.; Jin, G.D.; Park, J.; Choi, Y.H.; Kang, S.K.; Chae, B.J.; Kim, E.B.; Choi, Y.J. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 2018, 8, 6012. [Google Scholar] [CrossRef]
- Kim, H.B.; Borewicz, K.; White, B.A.; Singer, R.S.; Sreevatsan, S.; Tu, Z.J.; Isaacson, R.E. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 2011, 153, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Alain, B.P.E.; Chae, J.P.; Balolong, M.P.; Bum Kim, H.; Kang, D.K. Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J. Gen. Appl. Microbiol. 2014, 60, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Kubasova, T.; Davidova-Gerzova, L.; Babak, V.; Cejkova, D.; Montagne, L.; Le-Floc’h, N.; Rychlik, I. Effects of host genetics and environmental conditions on fecal microbiota composition of pigs. PLoS ONE 2018, 13, e0201901. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Slifierz, M.J.; Friendship, R.M.; Weese, J.S. Longitudinal study of the early-life fecal and nasal microbiotas of the domestic pig. BMC Microbiol. 2015, 15, 184. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, L.; Zhai, Z.; Zhao, W.; Ding, J.; Dai, R.; Sun, T.; Meng, H. Porcine Epidemic Diarrhea Virus Infection Induced the Unbalance of Gut Microbiota in Piglets. Curr. Microbiol. 2015, 71, 643–649. [Google Scholar] [CrossRef]
- Costa, M.O.; Chaban, B.; Harding, J.C.; Hill, J.E. Characterization of the fecal microbiota of pigs before and after inoculation with “Brachyspira hampsonii”. PLoS ONE 2014, 9, e106399. [Google Scholar] [CrossRef]
- Borewicz, K.A.; Kim, H.B.; Singer, R.S.; Gebhart, C.J.; Sreevatsan, S.; Johnson, T.; Isaacson, R.E. Changes in the Porcine Intestinal Microbiome in Response to Infection with Salmonella enterica and Lawsonia intracellularis. PLoS ONE 2015, 10, e0139106. [Google Scholar] [CrossRef]
- Leite, F.L.L.; Singer, R.S.; Ward, T.; Gebhart, C.J.; Isaacson, R.E. Vaccination Against Lawsonia intracellularis Decreases Shedding of Salmonella enterica serovar Typhimurium in Co-Infected Pigs and Alters the Gut Microbiome. Sci. Rep. 2018, 8, 2857. [Google Scholar] [CrossRef]
- Wang, Q.; Cai, R.; Huang, A.; Wang, X.; Qu, W.; Shi, L.; Li, C.; Yan, H. Comparison of Oropharyngeal Microbiota in Healthy Piglets and Piglets with Respiratory Disease. Front. Microbiol. 2018, 9, 3218. [Google Scholar] [CrossRef]
- Prickett, J.R.; Zimmerman, J.J. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim. Health Res. Rev. 2010, 11, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Prickett, J.; Simer, R.; Christopher-Hennings, J.; Yoon, K.-J.; Evans, R.B.; Zimmerman, J.J. Detection of porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: A longitudinal study under experimental conditions. J. Vet. Diagn. Investig. 2008, 20, 156–163. [Google Scholar] [CrossRef]
- Prickett, J.R.; Kim, W.; Simer, R.; Yoon, K.-J.; Zimmerman, J.J. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J. Swine Health Prod. 2008, 16, 86–91. [Google Scholar]
- Murase, K.; Watanabe, T.; Arai, S.; Kim, H.; Tohya, M.; Ishida-Kuroki, K.; Vo, T.H.; Nguyen, T.P.B.; Nakagawa, I.; Osawa, R.; et al. Characterization of pig saliva as the major natural habitat of Streptococcus suis by analyzing oral, fecal, vaginal, and environmental microbiota. PLoS ONE 2019, 14, e0215983. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, F.M.; Perez-Wohlfeil, E.; Carvalho, F.M.; Trelles, O.; Schrank, I.S.; Vasconcelos, A.T.R.; Zaha, A. Microbiome overview in swine lungs. PLoS ONE 2017, 12, e0181503. [Google Scholar] [CrossRef] [PubMed]
- Pena Cortes, L.C.; LeVeque, R.M.; Funk, J.; Marsh, T.L.; Mulks, M.H. Development of the tonsillar microbiome in pigs from newborn through weaning. BMC Microbiol. 2018, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Thacker, E.; Halbur, P.; Ross, R.; Thanawongnuwech, R.; Thacker, B. Mycoplasma hyopneumoniae potentiation of Porcine Reproductive and Respiratory Syndrome Virus-induced pneumonia. J. Clin. Microbiol. 1999, 37, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Pieters, M.; Pijoan, C.; Fano, E.; Dee, S. An assessment of the duration of Mycoplasma hyopneumoniae infection in an experimentally infected population of pigs. Vet. Microbiol. 2009, 134, 261–266. [Google Scholar] [CrossRef]
- Pieters, M.; Maes, D. Mycoplasmosis. In Diseases of Swine; Zimmerman, J., Karriker, L., Ramirez, A., Schwartz, K., Stevenson, G., Zhang, J., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 863–883. [Google Scholar]
- Sponheim, A.; Alvarez, J.; Fano, E.; Schmaling, E.; Dee, S.; Hanson, D.; Wetzell, T.; Pieters, M. Comparison of the sensitivity of laryngeal swabs and deep tracheal catheters for detection of Mycoplasma hyopneumoniae in experimentally and naturally infected pigs early and late after infection. Vet. Microbiol. 2020, 241, 108500. [Google Scholar] [CrossRef]
- Kraemer, J.G.; Aebi, S.; Oppliger, A.; Hilty, M. The Indoor-Air Microbiota of Pig Farms Drives the Composition of the Pig Farmers’ Nasal Microbiota in a Season-Dependent and Farm-Specific Manner. Appl. Environ. Microbiol. 2019, 85, e03038-18. [Google Scholar] [CrossRef]
- Kraemer, J.G.; Ramette, A.; Aebi, S.; Oppliger, A.; Hilty, M. Influence of Pig Farming on the Human Nasal Microbiota: Key Role of Airborne Microbial Communities. Appl. Environ. Microbiol. 2018, 84, e02470-17. [Google Scholar] [CrossRef]
- Kristiansen, A.; Saunders, A.M.; Hansen, A.A.; Nielsen, P.H.; Nielsen, J.L. Community structure of bacteria and fungi in aerosols of a pig confinement building. FEMS Microbiol. Ecol. 2012, 80, 390–401. [Google Scholar] [CrossRef]
- Mbareche, H.; Veillette, M.; Pilote, J.; Letourneau, V.; Duchaine, C. Bioaerosols Play a Major Role in the Nasopharyngeal Microbiota Content in Agricultural Environment. Int. J. Environ. Res. Public Health 2019, 16, 1375. [Google Scholar] [CrossRef]
- Lehtoranta, L.; Hibberd, A.A.; Reimari, J.; Junnila, J.; Yeung, N.; Maukonen, J.; Crawford, G.; Ouwehand, A.C. Recovery of Vaginal Microbiota After Standard Treatment for Bacterial Vaginosis Infection: An Observational Study. Microorganisms 2020, 8, 875. [Google Scholar] [CrossRef]
- Vangroenweghe, F.; Karriker, L.; Main, R.; Christianson, E.; Marsteller, T.; Hammen, K.; Bates, J.; Thomas, P.; Ellingson, J.; Harmon, K.; et al. Assessment of litter prevalence of Mycoplasma hyopneumoniae in preweaned piglets utilizing an antemortem tracheobronchial mucus collection technique and a real-time polymerase chain reaction assay. J. Vet. Diagn. Investig. 2015, 27, 606–610. [Google Scholar] [CrossRef]
- Simner, P.J.; Miller, H.B.; Breitwieser, F.P.; Pinilla Monsalve, G.; Pardo, C.A.; Salzberg, S.L.; Sears, C.L.; Thomas, D.L.; Eberhart, C.G.; Carroll, K.C. Development and Optimization of Metagenomic Next-Generation Sequencing Methods for Cerebrospinal Fluid Diagnostics. J. Clin. Microbiol. 2018, 56, e00472-18. [Google Scholar] [CrossRef]
- Chaban, B.; Albert, A.; Links, M.G.; Gardy, J.; Tang, P.; Hill, J.E. Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLoS ONE 2013, 8, e69559. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B. Silva Taxonomic Training Data Formatted for DADA2 (Silva Version 132); Zenodo: Genève, Switzerland, 2018. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5-6; Vienna, Austria, 2019. Available online: https://cran.r-project.org; https://github.com/vegandevs/vegan (accessed on 1 August 2020).
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Microsoft; Ooi, H. glmnetUtils: Utilities for “glmnet”. 1.1.5. 2020. Available online: https://cran.r-project.org/web/packages/glmnetUtils/index.html (accessed on 1 August 2020).
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbo, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [PubMed]
- Gehlenborg, N. UpSetR: A More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. 1.4.0. 2019. Available online: http://github.com/hms-dbmi/UpSetR (accessed on 1 August 2020).
- Lex, A.; Gehlenborg, N. Points of view: Sets and intersections. Nat. Methods 2014, 11, 779. [Google Scholar] [CrossRef]
- Finotello, F.; Mastrorilli, E.; Di Camillo, B. Measuring the diversity of the human microbiota with targeted next-generation sequencing. Brief. Bioinform. 2018, 19, 679–692. [Google Scholar] [CrossRef]
- Ravi, A.; Halstead, F.D.; Bamford, A.; Casey, A.; Thomson, N.M.; van Schaik, W.; Snelson, C.; Goulden, R.; Foster-Nyarko, E.; Savva, G.M.; et al. Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb. Genom. 2019, 5, e000293. [Google Scholar] [CrossRef]
- Tan, Z.; Dong, W.; Ding, Y.; Ding, X.; Zhang, Q.; Jiang, L. Changes in cecal microbiota community of suckling piglets infected with porcine epidemic diarrhea virus. PLoS ONE 2019, 14, e0219868. [Google Scholar] [CrossRef]
- Yan, Z.F.; Lin, P.; Wang, Y.S.; Gao, W.; Li, C.T.; Kook, M.C.; Yi, T.H. Niastella hibisci sp. nov., isolated from rhizosphere soil of mugunghwa, the Korean national flower. Int. J. Syst. Evol. Microbiol. 2016, 66, 5218–5222. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, T.; Zhou, Y.; He, J.; Zheng, L.Q.; Li, W.J.; Huang, X.; Li, S.P. Terrimonas rubra sp. nov., isolated from a polluted farmland soil and emended description of the genus Terrimonas. Int. J. Syst. Evol. Microbiol. 2012, 62, 2593–2597. [Google Scholar] [CrossRef]
- Roos, L.R.; Surendran Nair, M.; Rendahl, A.K.; Pieters, M. Mycoplasma hyorhinis and Mycoplasma hyosynoviae dual detection patterns in dams and piglets. PLoS ONE 2019, 14, e0209975. [Google Scholar] [CrossRef]
- Macedo, N.; Rovira, A.; Torremorell, M. Haemophilus parasuis: Infection, immunity and enrofloxacin. Vet. Res. 2015, 46, 128. [Google Scholar] [CrossRef]
- Pors, S.E.; Hansen, M.S.; Christensen, H.; Jensen, H.E.; Petersen, A.; Bisgaard, M. Genetic diversity and associated pathology of Pasteurella multocida isolated from porcine pneumonia. Vet. Microbiol. 2011, 150, 354–361. [Google Scholar] [CrossRef][Green Version]
- Surendran Nair, M.; Eucker, T.; Martinson, B.; Neubauer, A.; Victoria, J.; Nicholson, B.; Pieters, M. Influence of pig gut microbiota on Mycoplasma hyopneumoniae susceptibility. Vet. Res. 2019, 50, 86. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Bucci, V.; Caballero, S.; Djukovic, A.; Toussaint, N.C.; Equinda, M.; Lipuma, L.; Ling, L.; Gobourne, A.; No, D.; et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013, 81, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Suo, C.; Yin, Y.; Wang, X.; Lou, X.; Song, D.; Wang, X.; Gu, Q. Effects of lactobacillus plantarum ZJ316 on pig growth and pork quality. BMC Vet. Res. 2012, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Pieters, M.; Daniels, J.; Rovira, A. Comparison of sample types and diagnostic methods for in vivo detection of Mycoplasma hyopneumoniae during early stages of infection. Vet. Microbiol. 2017, 203, 103–109. [Google Scholar] [CrossRef]
- Ober, R.A.; Thissen, J.B.; Jaing, C.J.; Cino-Ozuna, A.G.; Rowland, R.R.R.; Niederwerder, M.C. Increased microbiome diversity at the time of infection is associated with improved growth rates of pigs after co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2). Vet. Microbiol. 2017, 208, 203–211. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Jaing, C.J.; Thissen, J.B.; Cino-Ozuna, A.G.; McLoughlin, K.S.; Rowland, R.R. Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2). Vet. Microbiol. 2016, 188, 1–11. [Google Scholar] [CrossRef]
- Schachtschneider, K.M.; Yeoman, C.J.; Isaacson, R.E.; White, B.A.; Schook, L.B.; Pieters, M. Modulation of systemic immune responses through commensal gastrointestinal microbiota. PLoS ONE 2013, 8, e53969. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).