Abstract
Lactic acid bacteria (LAB) share several beneficial effects on human organisms, such as bioactive metabolites’ release, pathogens’ competition and immune stimulation. This study aimed at determining the probiotic potential of autochthonous lactic acid bacteria isolated from carrots. In particular, the work reported the characterization at the species level of four LAB strains deriving from carrots harvested in Fucino highland, Abruzzo (Italy). Ribosomal 16S DNA analysis allowed identification of three strains belonging to Leuconostoc mesenteroides and a Weissella soli strain. In vitro and in vivo assays were performed to investigate the probiotic potential of the different isolates. Among them, L. mesenteroides C2 and L. mesenteroides C7 showed high survival percentages under in vitro simulated gastro-intestinal conditions, antibiotic susceptibly and the ability to inhibit in vitro growth against Salmonella enterica serovar Typhimurium, Listeria monocytogenes, Pseudomonas aeruginosa and Staphylococcus aureus pathogens. In parallel, the simple model Caenorhabditis elegans was used for in vivo screenings. L. mesenteroides C2 and L. mesenteroides C7 strains significantly induced pro-longevity effects, protection from pathogens’ infection and innate immunity stimulation. Overall, these results showed that some autochthonous LAB from vegetables such as carrots have functional features to be considered as novel probiotic candidates.
1. Introduction
The Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) defined probiotics as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host [1]. According to this statement, probiotics must be safe, and not exert pathogenic effects or show antibiotic resistance genes that could be transferred. Moreover, probiotic strains should be resistant to gastrointestinal conditions, such as stomach acid pH and bile acids, produce antimicrobial compounds and compete with pathogens by stimulating immunity [2]. Furthermore, probiotics’ efficacy should be confirmed in human studies.
Among the various microbial species associated with food, some of them may share probiotic features. The main source of probiotics used in humans is represented by dairy foods, but increasing evidence has highlighted the importance to select probiotics from other sources, such as fresh fruits and vegetables [3]. Indeed, the availability of commercial milk-based products limits their consumption by people who are intolerant or allergic to lactose. Therefore, fruits and vegetables offer healthy alternatives thanks to their large distribution and nutritive value. Among them, carrot (Daucus carota L.), as well as being rich in minerals and antioxidants, is reported to be a reservoir of carotenoids, vitamins and fiber [4,5,6]. Many studies on carrots have focused on cultivation, breeding, tissue culture, nutrient content and carotenoid synthesis regulation, while few works deal with microbial composition in terms of potential probiotic bacteria [7,8]. Indeed, the most common probiotics, isolated from fruits and vegetables, include different strains belonging to the lactic acid bacteria (LAB) group. This heterogeneous group of Gram-positive and non-spore-forming bacteria are normally present in food products, involved in numerous fermentation processes and some of them are widely used in industrial processes [9]. The major representatives of this microorganism group are Lactobacillus, Streptococcus, Leuconostoc, Pediococcus, Propionibacterium, Enterococcus, Bifidobacterium and Weissella genera [10]. Although the Weissella genus is found in multiple habitats, many species were isolated from different foods, such as fermented crop products, meat and fish, along with Leuconostoc species. Moreover, many of them produce exopolysaccharides, influencing the adhesion to substrates and affecting the structure of fermented foods.
Since the direct evaluation of probiotic potentials in vivo is often expensive and timewasting, the use of simple and inexpensive model systems is needed. Caenorhabditis elegans is a powerful in vivo model to screen for probiotic bacteria. Nematodes feed only on microorganisms, which reach the intestine, influencing nematodes’ physiology [11]. Among its many advantages, the possibility to easily monitor anti-aging markers or innate immunity pathways could be used for the screening of microorganisms to identify new probiotic strains and to explore the possible molecular pathways involved. Indeed, several foodborne LAB were reported to exert positive effects in worms, and the mechanisms correlated with innate immunity and lifespan extension have been elucidated [12]. Recently, different Lactobacillus strains, isolated from vegetables or dairy products, were reported to increase nematode viability, delay the aging process and protect against foodborne S. enterica serovar typhimurium LT2 or L. monocytogenes OH pathogens [13,14]. Moreover, it has been demonstrated that Bifidobacterium isolates can also exert beneficial effects on C. elegans health and lifespan [15].
This study aimed at determining the probiotic potential of four lactic acid bacteria strains isolated from carrots. Tolerance to gastrointestinal conditions, antibiotic susceptibility and antagonism toward human pathogenic microorganisms were evaluated in vitro. The different isolates were tested in vivo using the C. elegans animal model to analyze possible beneficial effects on worm lifespan, gut colonization, the aging process and pathogen resistance.
2. Materials and Methods
2.1. Species Isolation and Identification
Carrots were provided by Aureli Mario S.S. Agricola (Ortucchio, AQ, Italy). The carrot cultivar (Daucus carota L., Nantese Dordogne, Syngenta seeds) was grown in Fucino highland (Abruzzo, Italy) and harvested at commercial maturity, as indicated by the supplier’s geneticists. Epidermis and shallow flesh of five carrots (about 20 g) were homogenized with mortar under aseptic conditions and diluted in sterile H2Odd. Dilution aliquots were plated on De Man Rogosa Sharpe (MRS) medium for 24–48 h at 30 °C, anaerobically. After that, morphologically different colonies were streaked on new MRS plates and grown at 30 °C to isolate purified strains. Each strain was then inoculated in MRS broth anaerobically and, after growth, a stock at −80 °C was carried out.
For bacterial identification, DNA was extracted and amplified according to Schifano et al. [16]. The primer pairs F8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and R1492 (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rDNA region of LAB isolates. FASTA sequences of the amplified region from each LAB isolate were submitted to GenBank, and the associated accession numbers are reported in the Results Section.
2.2. Growth Conditions of Bacterial Isolates
Bacterial strains isolated from carrots and used in this study were Leuconostoc mesenteroides C1, L. mesenteroides C2, L. mesenteroides C7 and Weissella soli T4. The LAB strains described in this work were grown in MRS medium at 30 °C under anaerobic conditions. Commercial probiotic strain L. rhamnosus GG ATCC® 53103™ (LGG), used as the LAB reference strain, was grown at 37 °C anaerobically. For C. elegans experiments, Escherichia coli OP50 strain was used as standard food. For in vitro and in vivo resistance to pathogens, Pseudomonas aeruginosa ATCC 15692, Staphylococcus aureus ATCC 25923, S. enterica serovar typhimurium LT2 and L. monocytogenes OH were used. E. coli OP50 and pathogen strains were grown in Luria-Bertani (LB) broth at 37 °C overnight, under shaking.
2.3. Resistance to Lysozyme, Acid pH and Bile Salts
Isolates were grown in MRS broth overnight at 30 °C. For the lysozyme tolerance assay, 10 mL of overnight culture was centrifuged at 5000 rpm at 4 °C and suspended in the same volume of SES buffer (0.22 g/L CaCl2, 6.2 g/L NaCl, 2.2 g/L KCl, 1.2 g/L NaHCO3) containing 0.1 mg/mL of lysozyme (Sigma-Aldrich, St. Louis, MO, USA) [17]. After 30 min and 2 h of incubation at 37 °C, 100 μL of each suspension was plated on MRS agar plates and further incubated at 37 °C for 24 h, under anaerobic conditions. SES without lysozyme was used as a control.
The acid tolerance assay was performed according to [18], with some modifications. A 1 mL aliquot of overnight culture (109 cfu/mL) was inoculated into 10 mL of sterile phosphate-buffered saline (9 g/L NaCl, 9 g/L Na2HPO4 2H2O, 1.5 g/L KH2PO4) adjusted to pH 2.5 and pH 3.0 with 8M HCl. pH 5 buffer was used as a control. The tubes were incubated at 37 °C and the viable organisms were recovered after 3 h of incubation on MRS agar incubated for 48 h at 30 °C. For resistance to bile salts, the same protocol was performed, using phosphate-buffered saline with 0.3% bile salts (Sigma-Aldrich).
The viability was measured as percent viability = [(CFUtreated/mL)/(CFUuntreated/mL)] × 100. The untreated value corresponds to plate counts of inoculated bacteria in control phosphate-buffered saline, and the treated value corresponds to the bacterial counts obtained after incubation in simulated GI conditions.
2.4. Antibiotic Resistance
For the susceptibility test, antibiotic discs (Biolab Zrt., Budapest, Hungary) were used. The experiment was performed as described in [13]. Briefly, 100 μL of overnight cultures of different isolates or LGG were plated onto MRS agar plates; then, the antibiotic discs were gently placed on the plates and incubated under anaerobic conditions for 24 h at 37 °C. The zones of inhibition were measured from the center of the disc, recorded and compared with those of the reference strain.
2.5. Antimicrobial Activity
The agar diffusion test was performed using, as indicator strains, P. aeruginosa ATCC 15692, S. aureus ATCC 25923, S. enterica serovar typhimurium LT2 and L. monocytogenes OH. To evaluate the antagonistic activity of LAB isolates against the different pathogens, 100 μL each of LAB overnight cultures was spotted onto MRS agar and coated with 5 mL of TSA soft agar (0.7%), previously inoculated with 500 μL of each pathogen indicator strain. Plates were incubated at 37 °C for 24 h. The antagonist activity was recorded as the diameter (mm) of growth inhibition halo around each spot.
2.6. C. elegans Strain and Lifespan Assay
The wild-type C. elegans strain, Bristol N2, was grown at 16 °C on Nematode Growth Medium (NGM) plates plated with E. coli OP50. Fertile N2 adults were placed to lay embryos for 8 h on peptone-free NGM plates, plated with LAB strains, LGG or E. coli OP50, and then sacrificed. For the preparation of the bacterial lawns, overnight cultures were centrifuged for 15 min at 6000 rpm. The pellet was weighed and suspended in M9 buffer in order to obtain a final concentration of 400 mg/mL. Then, 25 µL of each type of bacterial lawn was plated on mNGM, as described in [19]. When the progeny became fertile (t0), 60 worms per condition were transferred to new plates plated with fresh bacterial cultures and monitored daily. A worm was considered dead when it did not respond to touch.
2.7. Fertility Assay
As described in [20], synchronized worms obtained as above were incubated at 16 °C on mNGM plates plated with different strains, allowing embryo laying. Animals were transferred onto new plates every day and the number of progeny was documented until the mother worms became infertile.
2.8. Colonization Assay of C. elegans Gut
For each condition, 10 L4 larvae or 8-day-old adults were washed in M9 buffer and lysed, as described in [21]. Whole worm lysates and serial dilutions were plated onto MRS-agar plates. The number of colony-forming units (CFU) was counted after 24 h of incubation at 37 °C, anaerobically. Instead, OP50-fed worm lysates were plated onto LB-agar and incubated at 37 °C.
2.9. Aging Markers’ Analysis
For the pharyngeal pumping rate, the number of grinder contractions was counted under a Zeiss Axiovert 25 microscope in 10-day-old adult animals fed different bacteria from embryo hatching. Ten worms were analyzed for each treatment, during a period of 30 s. The locomotion ability of nematodes was analyzed by body bending counting after 30 s. In particular, as described in [14], 10 worms for each treatment were washed in M9 buffer to remove bacteria, and then placed in 10 μL of M9 buffer to facilitate the locomotion measure. For lipofuscin accumulation analysis, 10-day-old adult worms, after washes in M9 buffer, were placed onto a 3% agar pad containing 20 mM of sodium azide. Afterwards, nematodes were observed with the Axio Observer Z1 inverted microscope, equipped with an ApoTome.2 System (Carl Zeiss Inc., Oberkochen, Germany). Digital images were acquired with the AxioCam MRm high-resolution digital camera (Zeiss) and processed with the AxioVision 4.8.2 software (Zeiss). ApoTome optical sectioning images of animals were recorded under a 40 Å~/0.75 objective (Zeiss). Median fluorescence intensity was analyzed using the ImageJ software, measuring the ratio of pixels per area of the worm.
2.10. Resistance to Pathogens in C. elegans
For the killing assay, 35 mm NGM plates were seeded with 60 μL of L. mesenteroides C2 or L. mesenteroides C7 mixed with different pathogens, in a 1:1 ratio. Pseudomonas aeruginosa ATCC 15692 and Staphylococcus aureus ATCC 25923 were chosen as representative pathogens of Gram-negative and Gram-positive bacteria, respectively. The assay was performed as described in [22]. Synchronous L4 larvae were placed onto the different co-cultures and incubated at 25 °C. Lifespan was monitored daily and worms fed with pathogen alone were taken as the control. A worm was considered dead when it failed to respond to touch.
2.11. Real-Time qPCR
At the stage of 1-day-old adults, 200 worms for each condition were lysed and total RNA was extracted as described in [23]. pmk-1, skn-1, daf-16, sod-3 and hsf-1 mRNA levels were analyzed. The differences between the mean CT value of each sample and the CT value of the housekeeping gene (act-1) were calculated. Primers used in this study are reported in Table 1. The experiment was carried out in triplicate.

Table 1.
Primers for real-time qPCR analysis.
2.12. Statistical Analysis
All experiments were performed at least in triplicate. Data are presented as mean ± SD. The statistical significance was determined by Student’s t test or one-way ANOVA coupled with a Bonferroni post-test (GraphPad Prism 5.0 software, GraphPad Software Inc., La Jolla, CA, USA). Differences with p-values < 0.05 were considered significant and were indicated as follows: * p < 0.05, ** p < 0.01 and *** p < 0.001.
3. Results
3.1. Bacteria Isolation
In this study, potential probiotic properties of four bacterial isolates were evaluated in vitro and in vivo. Different bacterial colonies were isolated from carrots and identified at the molecular level by the amplification of 16S rDNA. The sequences obtained from sequencing were compared with those in the BLAST database, so that three strains belonging to Leuconostoc mesenteroides and a Weissella soli strain were identified. The Gram-positive strains isolated were identified as L. mesenteroides C1 (accession number OK513088), L. mesenteroides C2 (accession number OK513089), L. mesenteroides C7 (accession number OK513090) and Weissella soli T4 (accession number OK513091).
3.2. In Vitro Tests
3.2.1. Resistance to Lysozyme, Low pH and Bile Salts
To perform a selection of possible probiotic candidates, their resistance to the extreme conditions of the gastrointestinal tract, such as low pH in stomach and bile in the upper intestine, was evaluated. The high concentration of lysozyme present in the mouth represents the first barrier. Figure 1A reports bacteria survival data after 30 and 120 min of treatment with 1 mg/mL of lysozyme. All LAB strains showed high resistance to lysozyme after 30 min of incubation, with percentages of survival of 100%. Notably, L. mesenteroides C2 was able to resist even after 120 min of incubation with lysozyme, which can be considered a severe treatment. On the other hand, W. soli T4 and L. mesenteroides C7 showed a percent survival of 60% and 80% respectively, after 2 h of incubation. L. mesenteroides C1, instead, did not resist lysozyme treatment. Tolerance to low pH conditions was performed to simulate microbial flux along the mammalian gastrointestinal tract. As shown in Figure 1B, 3 h of incubation in pH 2.5 exerted a strong reduction (about 80%) of L. mesenteroides C1, L. mesenteroides C7 and W. soli T4 counts, as compared to the control. This result suggested that these strains were not able to endure acidic environments. Notably, L. mesenteroides C2 cell recovery showed a higher ability to survive in low pH conditions, similarly to the probiotic LGG strain. Similar results were obtained after testing the ability of different strains to resist at pH 3.0.
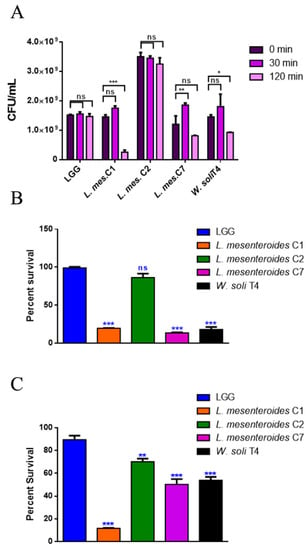
Figure 1.
In vitro tolerance to lysozyme, pH 2.5 and 0.3% bile salts. (A) Cell counts of viable bacteria recovered at the initial time point (t0), following 30 or 120 min of incubation in 1 mg/mL of lysozyme SES buffer. (B) Recovery of viable bacteria after 3 h of incubation in phosphate buffer adjusted to pH 2.5 or (C) 0.3% bovine bile salts. LGG was taken as the LAB reference strain. Columns represent the mean ± SD of three independent experiments. Statistical analysis was performed by one-way ANOVA, followed by the Bonferroni post-test. Asterisks indicate significant differences (* p < 0.05; ** p < 0.01; *** p < 0.001), ns: not significant.
The human bile concentration ranges from 0.3% to 0.5%, and a high bile tolerance improves probiotics’ colonization in the host GI tract [24]. To investigate the ability of the different isolates to survive in the presence of bile, the percent survival in the presence of 0.3% bile was tested. As shown in Figure 1C, L. mesenteroides C2, L. mesenteroides C7 and W. soli T4 strains showed a growth percentage above 50% in the presence of bile. As expected, the reference probiotic strain LGG exhibited a resistance of about 90%. On the other hand, L. mesenteroides C1 was not able to resist the treatment, showing a decrease in viability of about 90%.
3.2.2. Antibiotic Susceptibility and Antagonistic Activity to Pathogens
Antibiotic susceptibility was determined by the disc diffusion assay, analyzing a panel of 20 antibiotics. Among them, there are inhibitors of synthesis of cell wall, DNA and RNA, proteins and inhibitors of membrane function. As reported in Table 1, LAB isolates showed an antibiotic susceptibility pattern very similar to that of the LGG control strain. L. mesenteroides C2 displayed resistance to only five antibiotics, and in some cases, the inhibition halo was larger as compared to the control (Table 2).

Table 2.
Resistance to antibiotics of different isolates. The zones of inhibition were measured from the center of the disc and recorded in mm ± SD. Absence of inhibition halo was indicated as (+), ns: not significant.
The antagonistic activity was evaluated through the agar double-layer diffusion test, against four pathogen test strains: the Gram-positive Staphylococcus aureus and Listeria monocytogenes, and the Gram-negative Pseudomonas aeruginosa and S. enterica serovar typhimurium LT2. The antagonistic activity was less variable among the different isolates, as shown by inhibition halo diameters on all four pathogen test strains (Table 3). Indeed, the inhibition halo diameters produced by the different isolates were comparable to that of the probiotic strain LGG.

Table 3.
Antagonistic activity in vitro. The diameter of inhibition halos was recorded in mm and the data were expressed as average ± SD.
3.3. In Vivo Tests
3.3.1. Effects on C. elegans Lifespan and Colonization Capability
In vivo screening of the four strains was performed in the C. elegans model system, to test possible beneficial effects exerted by LAB. For this purpose, worms were separately fed each of the isolated strains starting from embryo hatching, using animals fed LGG or standard E. coli OP50 as control populations. Among the tested strains, L. mesenteroides C2 and L. mesenteroides C7 induced a relevant increase in C. elegans viability (Figure 2A), showing similar survival as compared to those fed the probiotic strain LGG. Indeed, 50% of viability was recorded at days 17, 18 and 21 in L. mesenteroides C1, L. mesenteroides C2 and L. mesenteroides C7 fed nematodes respectively, in comparison with day 22 in LGG-fed nematodes. On the other hand, only W. soli T4 showed a similar worm lifespan when compared to the control OP50 diet, with 50% of viability recorded at days 14 and 12, respectively. To test possible effects on fertility, progeny production was evaluated. Like probiotic LGG, the brood size of worms fed different isolates showed a reduction of about 60% of the progeny number compared to the OP50 control (Figure 2B). Afterwards, the gut colonization capability was explored by plating worm lysates at different time points and by CFU counting. Results highlighted the increase of all bacterial strains along the lifespan (Figure 2C). At the L4 stage, the CFU number relative to L. mesenteroides C1 resulted to be about 2-fold higher than that relative to controls. At the stage of 8 days of adulthood, instead, the gut colonization capability of L. mesenteroides C1 was about 80% lower than that of probiotic LGG. On the other hand, in adult worms, L. mesenteroides C2 resulted to be able to colonize C. elegans gut, similarly to the LGG control. Along worms’ lifespan, L. mesenteroides C7 and W. soli T4 showed a colonization capability similar to the OP50 strain.
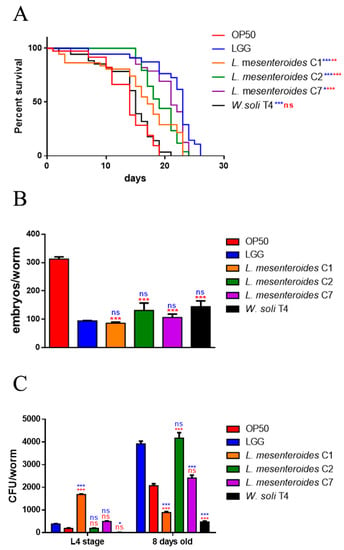
Figure 2.
Effects of different isolates on C. elegans lifespan. (A) Kaplan–Meier survival plot of N2 worms fed L. mesenteroides and W. soli strains. Lifespans of OP50- and LGG-fed animals are reported as controls; n = 60 for each data point of single experiments. (B) Average embryos’ production per worm of nematodes fed different bacterial isolates. (C) Bacterial colony-forming units (CFU) recovered from L4 larvae and 8-day-old adults fed the four isolates or LGG (LAB reference strain) and OP50 controls. Bars represent the mean of three independent experiments. Asterisks indicate significant differences (* p < 0.05, ** p < 0.01, *** p < 0.001) as compared to LGG (blue asterisks) or OP50 (red asterisks) controls, ns: not significant.
3.3.2. Effects on C. elegans Aging Processes
To investigate whether the pro-longevity effects exerted by LAB strains positively correlated to a delay in aging, age-related biomarkers, such as pumping, locomotion and lipofuscin accumulation, were analyzed. The pharyngeal pumping rate measures grinder contractions associated with food intake ability, normally declining with age. Figure 3A showed that L. mesenteroides C7-fed worms share a significantly high pumping rate, at 10 days of adulthood, similarly to LGG-fed worms. Moreover, nematodes fed with L. mesenteroides C2 and W. soli T4 showed an increase in grinder contraction of 12% and 8%, as compared to OP50-fed worms, respectively (Figure 3A). On the other hand, in L. mesenteroides C1-fed worms, a reduction in pumping rate of about 40% with respect to LGG was observed. Then, the locomotion rate of C. elegans was evaluated at day 10 of adulthood. In this case, nematodes fed different L. mesenteroides strains displayed a higher motility than OP50-fed worms, while locomotion of W. soli T4-fed worms was similar to the control (Figure 3B). Furthermore, accumulation of auto-fluorescent lipofuscin is also an aging marker of cellular impairment. Indeed, 10-day-old adult nematodes, when fed L. mesenteroides isolates, showed a reduced fluorescence compared to OP50-fed adults, while W. soli T4-fed animals showed a higher accumulation of fluorescent granules along the intestine, usual in old animals (Figure 4A,B).
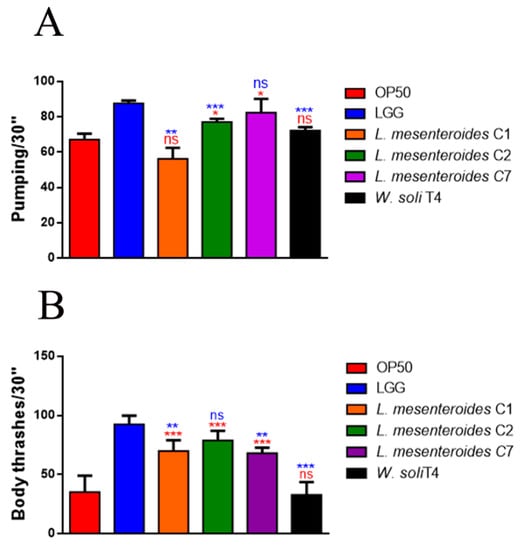
Figure 3.
Impact of the different isolates on C. elegans aging markers. (A) Pumping rate of 10-day-old worms measured for 30 s. 10 worms were used for each condition. Worms fed OP50 or LGG were taken as controls. (B) Body bending of C. elegans-fed isolates as compared to LAB reference strain LGG or OP50, measured for 30 s. Bars represent the mean of three independent experiments. Statistical analysis was evaluated by one-way ANOVA with the Bonferroni post-test. Asterisks indicate significant differences (* p < 0.05, ** p < 0.01, *** p < 0.001) as compared to LGG (blue asterisks) or OP50 (red asterisks) controls, ns: not significant.
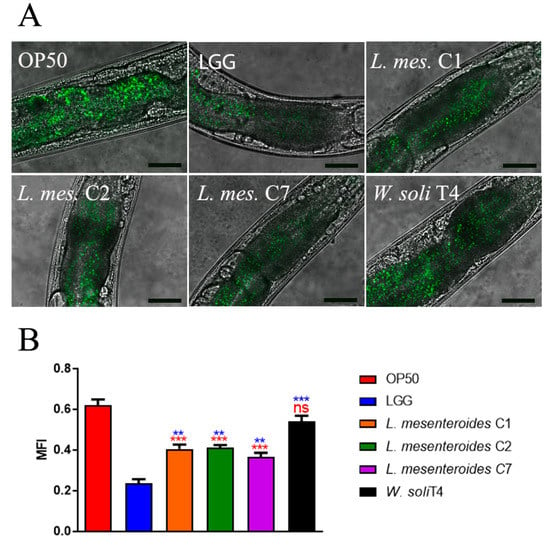
Figure 4.
Evaluation of lipofuscin accumulation in C. elegans. (A) Autofluorescence of lipofuscin granules in C. elegans fed different LAB on day 10. Ten worms were used for each measurement. LGG- (LAB reference strain) and OP50-fed worms were used as controls. Scale bar = 50 μm. (B) Median fluorescence intensity of nematodes’ lipofuscin. Statistical analysis was evaluated by one-way ANOVA with the Bonferroni post-test. Asterisks indicate significant differences (** p < 0.01, *** p < 0.001) as compared to LGG (blue asterisks) or OP50 (red asterisks) controls, ns: not significant. Bars represent the mean of three independent experiments.
3.3.3. Pathogen Resistance and Innate Immunity Stimulation
Among the four LAB isolates, L. mesenteroides C2 and L. mesenteroides C7 strains resulted as the most promising candidates in terms of beneficial features. Since a good probiotic is reported to compete with pathogens, protecting the host from infections, the C. elegans killing assay was performed to test possible protection from infection by the two isolates. S. aureus and P. aeruginosa were chosen as representatives of the Gram-positive and Gram-negative group, respectively. As shown in Table 4, nematodes displayed reduced survival when fed pathogens alone, as compared to nematodes fed co-cultures of the same pathogen with L. mesenteroides C2 and L. mesenteroides C7. Interestingly, 50% of viability was recorded at day 6 in nematodes fed co-cultures of L. mesenteroides strain P. aeruginosa, in comparison with day 3 of nematodes fed the pathogen alone. Similarly, L. mesenteroides C2 and L. mesenteroides C7 were able to protect worms from S. aureus infection, with 50% of viability recorded at day 7, with respect to day 5 in worms fed S. aureus alone.

Table 4.
In vivo resistance to pathogens. Survival assay of N2 worms fed co-cultures of LAB and P. aeruginosa or S. aureus. The lifespan of worms fed pathogens alone was reported as the control. Three experiments were performed in triplicate for each condition.
To study whether L. mesenteroides C2 and C7 could stimulate nematodes’ innate immunity, transcript levels were analyzed for pmk-1, skn-1, sod-3, daf-16 and hsf-1 genes, whose activation after probiotic feeding have been described [15,25]. Interestingly, in L. mesenteroides C2- and C7-fed nematodes, significantly increased expressions of pmk-1 and hsf-1 transcripts were observed, similarly to LGG (Figure 5). On the other hand, while in LGG-fed worms DAF-16 also seemed to be activated, the two isolates were not able to stimulate this pathway. The expression of genes involved in detoxification processes (skn-1 and sod-3) were instead very low, as compared to the OP50 control, but similar to LGG.
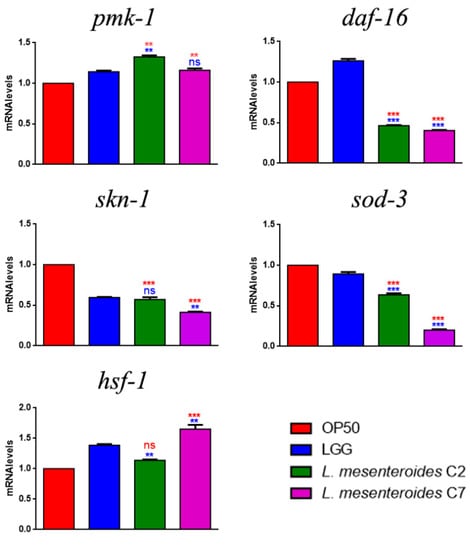
Figure 5.
Transcript levels of genes involved in C. elegans immunity. Expression of pmk-1, skn-1, sod-3, daf-16 and hsf-1 genes in 1-day-old adults fed with different bacterial strains. Experiments were performed in triplicate. LGG (LAB reference strain) and OP50 were used as controls. Data are presented as mean ± SD. Asterisks indicate significant differences (** p < 0.01, *** p < 0.001) as compared to LGG (blue asterisks) or OP50 (red asterisks) controls; ns: not significant.
4. Discussion
The human microbiota represents the first defense barrier against gut colonization by pathogens. This defense is due to microbiota competition with pathogenic bacteria, preventing their adhesion and subsequent internalization. Indeed, some commensal strains share adhesion receptors with higher affinity than pathogens, also competing for the availability of nutrients and trophic substances. Another strategy adopted by microbiota species is the production of antimicrobial substances called bacteriocins. Therefore, probiotics commonly share the ability to compete for receptor sites, nutrients and trophic substances, and they are able to synthesize bacteriocins. Recently, growing attention in pro-longevity effects exerted by different LAB strains allowed the development of several probiotic products [26]. In particular, fermented foods are the main source of LAB [14,19,27,28], but fruits have also been cited several times for the isolation of interesting LAB [29,30,31]. Due to the probiotics’ relevance to human health, it is pivotal to characterize bacterial isolates to be used in alternative food products. In this context, the main objective of this work was the isolation of LAB from carrots and the selection of strains with potential to be used as probiotic microorganisms. The root carrot (Daucus carota L.) is one of the most important vegetables cultivated and consumed worldwide, rich in bioactive compounds, such as provitamin A [5,7]. It is also rich in dietary fiber, antioxidants and other nutrients, but especially in carotenoids. In this work, a combination of in vitro and in vivo methods was used to screen for new potential probiotic Leuconostoc and Weissella strains deriving from carrots grown in Fucino highland (Abruzzo, Italy). Characterization of LAB isolates at the species level identified three strains belonging to Leuconostoc mesenteroides and a Weissella soli strain. Indeed, a large percentage of probiotic microorganisms belongs to the LAB group. Leuconostoc and Weissella, together with Lactobacillus and Pediococcus, are important genera of LAB associated with foods and fermented products, such as meat, vegetables, dairy and bakery products, and also act as flavoring and texturizing agents [32]. This work showed that, among all tested strains, L. mesenteroides C2 and L. mesenteroides C7 could survive against the stress conditions assayed in this study. Survival to the adverse environment of the stomach is a key pre-requisite for effective colonization by a probiotic strain [33]. At first, tolerance to gastrointestinal conditions was carried out by evaluating the viability of each isolate in comparison with the commercial reference strain L. rhamnosus GG. Notably, L. mesenteroides C2 and L. mesenteroides C7 displayed survival rates equal to or higher than that of the commercial probiotic control LGG after the different treatments. Only in the case of long exposure to lysozyme and low pH resistance did L. mesenteroides C7 show a reduced recovery of viable cells. If this strain will result as positive in subsequent tests, this problem could be solved by encapsulating the bacteria cells, making them viable in the human gut. Furthermore, antibiotic resistance profiling of all isolates resulted similar to probiotic LGG, and this is an important trait to be verified for safety purposes [34]. In this study, different groups of antibiotics were used: cell wall inhibitors, inhibitors of protein synthesis and inhibitors of DNA and RNA synthesis. A recurrent ingestion of these types of antibiotics may cause imbalance in the intestinal sensitive microbiota. Moreover, the antibiotic resistance in probiotics usually does not constitute a safety issue, since resistance genes could be transferred to potential pathogens. It has been demonstrated that probiotics are able to prevent infections by foodborne pathogens, through different mechanisms, such as competitive exclusion or antimicrobial molecules’ production [35,36]. Indeed, LAB show various antimicrobial features, such as the production of organic acids, cyclic dipeptides, phenylacetic acid, hydrogen peroxide, low molecular weight compounds, protein compounds, bacteriocins and fatty acids [37,38,39,40]. We therefore tested the antimicrobial activity exerted by L. mesenteroides C2 and L. mesenteroides C7 strains against common pathogens, such as Pseudomonas aeruginosa and Staphylococcus aureus. The isolates resulted to counteract the pathogens in vitro and in vivo. In parallel, the isolated strains were also analyzed evaluating possible beneficial properties in the in vivo model of C. elegans. Nematodes commonly feed on bacteria, but a significant number of bacterial cells escape the grinder contractions and can proceed to colonize the worm gut [41]. The L. mesenteroides C2 and C7 strains were able to significantly increase C. elegans lifespan as compared to the OP50 control strain, similar to the effect exerted by the reference strain LGG. The impact of L. mesenteroides C2 and C7 on C. elegans physiology could be due to the high gut colonization capacity of the bacterial strains. Moreover, the pro-longevity effects observed in lifespan experiments were associated with the anti-aging effects, highlighted by analyzing different aging markers, such as pharyngeal pumping rate, brood size and lipofuscin. These data further demonstrate the ability of specific LAB strains to prolong nematodes’ lifespan, as described in previous studies [41,42,43,44]. As discussed above, the different isolates also displayed health-promoting activities in host defense against Gram-positive and Gram-negative pathogens in vivo, increasing the survival of infected worms. In C. elegans, host–pathogen interactions have been studied for a number of pathogens of human and animal origin [45], including P. aeruginosa and S. aureus, which colonize the worm gut and infect the nematode [46]. Moreover, innate immunity responses and lifespan were strongly correlated in nematodes [15]. Indeed, worms do not have adaptative immunity, but only innate immune defenses that have many aspects similar to human pathways [47]. Among them, the p38 MAPK and IIS pathways, the transforming growth factor-beta (TGF-beta) and the beta-catenin signaling pathways, are more conserved in humans and nematodes and they can be induced by probiotics [48]. In agreement with these works, real-time analysis highlighted the activation of pmk-1 and hsf-1 pathways, suggesting a stimulation of C. elegans immunity, which correlates with a reduction of oxidative stress, leading to the pro-longevity and anti-aging effects.
5. Conclusions
Among the different strains isolated from carrots, L. mesenteroides C2 and L. mesenteroides C7 showed interesting probiotic characteristics, such as greater lysozyme, pH and bile tolerance, in vitro suppression of pathogen growth and in vivo beneficial effects, exerted on the C. elegans animal model. However, in vivo analysis on animal or human systems should be performed to further test their potential beneficial properties for human health.
Author Contributions
Conceptualization, A.M., A.T., W.A. and D.U.; methodology, E.S., J.M. and A.P.; software, E.S., J.M. and A.P.; investigation, E.S. and D.U.; data curation, E.S.; writing—original draft preparation, E.S.; writing—review and editing, D.U. and A.M.; visualization, E.S.; supervision, D.U., A.M. and P.M.; project administration, A.M. and D.U.; funding acquisition, W.A. All authors have read and agreed to the published version of the manuscript.
Funding
The present work has been carried out under the project “INNOPAQ”, funded by POR-FESR Abruzzo 2014–2020, CUP number: C73D18000320007.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO/WHO Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation 2006. Available online: http://www.fao.org/food/food-safety-quality/a-z-index/probiotics/en/ (accessed on 16 September 2021).
- Markowiak-Kopeć, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Roselli, M.; Natella, F.; Zinno, P.; Guantario, B.; Canali, R.; Schifano, E.; De Angelis, M.; Nikoloudaki, O.; Gobbetti, M.; Perozzi, G.; et al. Colonization Ability and Impact on Human Gut Microbiota of Foodborne Microbes from Traditional or Probiotic-Added Fermented Foods: A Systematic Review. Front. Nutr. 2021, 8, 689084. [Google Scholar] [CrossRef] [PubMed]
- Sciubba, F.; Tomassini, A.; Giorgi, G.; Brasili, E.; Pasqua, G.; Capuani, G.; Aureli, W.; Miccheli, A. NMR-Based Metabolomic Study of Purple Carrot Optimal Harvest Time for Utilization as a Source of Bioactive Compounds. Appl. Sci. 2020, 10, 8493. [Google Scholar] [CrossRef]
- Tomassini, A.; Sciubba, F.; Di Cocco, M.E.; Capuani, G.; Delfini, M.; Aureli, W.; Miccheli, A. 1H NMR-Based Metabolomics Reveals a Pedoclimatic Metabolic Imprinting in Ready-to-Drink Carrot Juices. J. Agric. Food Chem. 2016, 64, 5284–5291. [Google Scholar] [CrossRef] [PubMed]
- Arscott, S.A.; Tanumihardjo, S.A. Carrots of Many Colors Provide Basic Nutrition and Bioavailable Phytochemicals Acting as a Functional Food. Compr. Rev. Food Sci. Food Saf. 2010, 9, 223–239. [Google Scholar] [CrossRef]
- Que, F.; Hou, X.-L.; Wang, G.-L.; Xu, Z.-S.; Tan, G.-F.; Li, T.; Wang, Y.-H.; Khadr, A.; Xiong, A.-S. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic. Res. 2019, 6, 69. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, E.A.; Vieira, M.A.; Vieira, E.A.; Amboni, R.D.D.M.C.; Amante, E.R.; Teixeira, E. Chemical, Physical and Sensory Parameters of Different Carrot Varieties (Daucus carota L.). J. Food Process. Eng. 2007, 30, 746–756. [Google Scholar] [CrossRef]
- Forssten, S.D.; Sindelar, C.W.; Ouwehand, A. Probiotics from an industrial perspective. Anaerobe 2011, 17, 410–413. [Google Scholar] [CrossRef]
- Sornplang, P.; Piyadeatsoontorn, S. Probiotic isolates from unconventional sources: A review. J. Anim. Sci. Technol. 2016, 58, 26. [Google Scholar] [CrossRef] [Green Version]
- Poupet, C.; Chassard, C.; Nivoliez, A.; Bornes, S. Caenorhabditis elegans, a Host to Investigate the Probiotic Properties of Beneficial Microorganisms. Front. Nutr. 2020, 7, 135. [Google Scholar] [CrossRef]
- Kwon, G.; Lee, J.; Lim, Y.-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016, 6, 31713. [Google Scholar] [CrossRef] [PubMed]
- Schifano, E.; Zinno, P.; Guantario, B.; Roselli, M.; Marcoccia, S.; Devirgiliis, C.; Uccelletti, D. The Foodborne Strain Lactobacillus fermentum MBC2 Triggers pept-1-Dependent Pro-Longevity Effects in Caenorhabditis elegans. Microorganisms 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guantario, B.; Zinno, P.; Schifano, E.; Roselli, M.; Perozzi, G.; Palleschi, C.; Uccelletti, D.; Devirgiliis, C. In Vitro and In Vivo Selection of Potentially Probiotic Lactobacilli from Nocellara del Belice Table Olives. Front. Microbiol. 2018, 9, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roselli, M.; Schifano, E.; Guantario, B.; Zinno, P.; Uccelletti, D.; Devirgiliis, C. Caenorhabditis Elegans and Probiotics Interactions from a Prolongevity Perspective. Int. J. Mol. Sci. 2019, 20, 5020. [Google Scholar] [CrossRef] [Green Version]
- Schifano, E.; Cavallini, D.; De Bellis, G.; Bracciale, M.P.; Felici, A.C.; Santarelli, M.L.; Sarto, M.S.; Uccelletti, D. Antibacterial Effect of Zinc Oxide-Based Nanomaterials on Environmental Biodeteriogens Affecting Historical Buildings. Nanomaterials 2020, 10, 335. [Google Scholar] [CrossRef] [Green Version]
- Samedi, L.; Charles, A.L. Isolation and characterization of potential probiotic Lactobacilli from leaves of food plants for possible additives in pellet feeding. Ann. Agric. Sci. 2019, 64, 55–62. [Google Scholar] [CrossRef]
- Erkkilä, S.; Petäjä, E. Screening of commercial meat starter cultures at low pH and in the presence of bile salts for potential probiotic use. Meat Sci. 2000, 55, 297–300. [Google Scholar] [CrossRef]
- Zanni, E.; Laudenzi, C.; Schifano, E.; Palleschi, C.; Perozzi, G.; Uccelletti, D.; Devirgiliis, C. Impact of a Complex Food Microbiota on Energy Metabolism in the Model Organism Caenorhabditis elegans. BioMed Res. Int. 2015, 2015, 621709. [Google Scholar] [CrossRef] [Green Version]
- Zanni, E.; Schifano, E.; Motta, S.; Sciubba, F.; Palleschi, C.; Mauri, P.; Perozzi, G.; Uccelletti, D.; Devirgiliis, C.; Miccheli, A. Combination of Metabolomic and Proteomic Analysis Revealed Different Features among Lactobacillus delbrueckii Subspecies bulgaricus and lactis Strains While In Vivo Testing in the Model Organism Caenorhabditis elegans Highlighted Probiotic Properties. Front. Microbiol. 2017, 8, 1206. [Google Scholar] [CrossRef] [Green Version]
- Schifano, E.; Marazzato, M.; Ammendolia, M.G.; Zanni, E.; Ricci, M.; Comanducci, A.; Goldoni, P.; Conte, M.P.; Uccelletti, D.; Longhi, C. Virulence behavior of uropathogenic Escherichia colistrains in the host model Caenorhabditis elegans. Microbiologyopen 2019, 8, e00756. [Google Scholar] [CrossRef]
- Schifano, E.; Ficociello, G.; Vespa, S.; Ghosh, S.; Cipollo, J.F.; Talora, C.; Lotti, L.V.; Mancini, P.; Uccelletti, D. Pmr-1 gene affects susceptibility of Caenorhabditis elegans to Staphylococcus aureus infection through glycosylation and stress response pathways’ alterations. Virulence 2019, 10, 1013–1025. [Google Scholar] [CrossRef] [Green Version]
- Schifano, E.; Cicalini, I.; Pieragostino, D.; Heipieper, H.J.; Del Boccio, P.; Uccelletti, D. In vitro and in vivo lipidomics as a tool for probiotics evaluation. Appl. Microbiol. Biotechnol. 2020, 104, 8937–8948. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386s–392s. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, L.; Laghi, L.; Correani, V.; Schifano, E.; Landi, C.; Uccelletti, D.; Mattei, B. A Combined Proteomics, Metabolomics and In Vivo Analysis Approach for the Characterization of Probiotics in Large-Scale Production. Biomolecules 2020, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Terpou, A.; Papadaki, A.; Lappa, I.K.; Kachrimanidou, V.; Bosnea, L.A.; Kopsahelis, N. Probiotics in Food Systems: Significance and Emerging Strategies Towards Improved Viability and Delivery of Enhanced Beneficial Value. Nutrients 2019, 11, 1591. [Google Scholar] [CrossRef] [Green Version]
- Domingos-Lopes, M.; Stanton, C.; Ross, P.; Dapkevicius, M.; Silva, C. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017, 63, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P.; Mutanda, T.; Olaniran, A.O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 2016, 60, 29630. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, L.G.R.; Gasga, V.M.Z.; Pescuma, M.; Van Nieuwenhove, C.; Mozzi, F.; Burgos, J.A.S. Fruits and fruit by-products as sources of bioactive compounds. Benefits and trends of lactic acid fermentation in the development of novel fruit-based functional beverages. Food Res. Int. 2021, 140, 109854. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Filannino, P.; Gobbetti, M. Vegetable and Fruit Fermentation by Lactic Acid Bacteria. Biotechnol. Lact. Acid Bact. 2015, 216–230. [Google Scholar] [CrossRef]
- Ngea, G.L.N.; Yang, Q.; Tchabo, W.; Castoria, R.; Zhang, X.; Zhang, H. Leuconostoc mesenteroides subsp. mesenteroides LB7 isolated from apple surface inhibits P. expansum in vitro and reduces patulin in fruit juices. Int. J. Food Microbiol. 2021, 339, 109025. [Google Scholar] [CrossRef]
- O’Sullivan, L.; Ross, R.; Hill, C. Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie 2002, 84, 593–604. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Botes, M. Probiotic lactic acid bacteria in the gastro-intestinal tract: Health benefits, safety and mode of action. Benef. Microbes 2010, 1, 11–29. [Google Scholar] [CrossRef]
- Imperial, I.C.V.; Ibana, J.A. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [PubMed]
- Rafieian-Kopaei, M.; Karami, S.; Roayaei, M.; Hamzavi, H.; Bahmani, M.; Hassanzad-Azar, H.; Leila, M. Isolation and identification of probiotic Lactobacillus from local dairy and evaluating their antagonistic effect on pathogens. Int. J. Pharm. Investig. 2017, 7, 137–141. [Google Scholar] [CrossRef]
- Mathipa, M.G.; Thantsha, M.S. Probiotic engineering: Towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 2017, 9, 28. [Google Scholar] [CrossRef]
- Pawlowska, A.M.; Zannini, E.; Coffey, A.; Arendt, E.K. “Green Preservatives”: Combating Fungi in the Food and Feed Industry by Applying Antifungal Lactic Acid Bacteria. Nutr. Benefits Kiwifruit 2012, 66, 217–238. [Google Scholar] [CrossRef]
- Peyer, L.C.; Zannini, E.; Arendt, E.K. Lactic acid bacteria as sensory biomodulators for fermented cereal-based beverages. Trends Food Sci. Technol. 2016, 54, 17–25. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Lactic Acid Bacteria as Antifungal and Anti-Mycotoxigenic Agents: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef] [Green Version]
- Silva, H.; Balthazar, C.F.; Silva, R.; Vieira, A.; Costa, R.; Esmerino, E.; Freitas, M.; Cruz, A. Sodium reduction and flavor enhancer addition in probiotic prato cheese: Contributions of quantitative descriptive analysis and temporal dominance of sensations for sensory profiling. J. Dairy Sci. 2018, 101, 8837–8846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, H.; Shiozaki, T.; Kobatake, E.; Hosoya, T.; Moriya, T.; Sakai, F.; Taru, H.; Miyazaki, T. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell 2015, 15, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, T.; Yasui, C.; Hoshino, K.; Arikawa, K.; Nishikawa, Y. Influence of Lactic Acid Bacteria on Longevity of Caenorhabditis elegans and Host Defense against Salmonella enterica Serovar Enteritidis. Appl. Environ. Microbiol. 2007, 73, 6404–6409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komura, T.; Ikeda, T.; Yasui, C.; Saeki, S.; Nishikawa, Y. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 2013, 14, 73–87. [Google Scholar] [CrossRef] [Green Version]
- Oh, A.; Daliri, E.B.-M.; Oh, D.H. Screening for potential probiotic bacteria from Korean fermented soybean paste: In vitro and Caenorhabditis elegans model testing. LWT 2018, 88, 132–138. [Google Scholar] [CrossRef]
- Clark, L.C.; Hodgkin, J. Commensals, probiotics and pathogens in the Caenorhabditis elegansmodel. Cell. Microbiol. 2013, 16, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Irazoqui, J.E.; Troemel, E.R.; Feinbaum, R.L.; Luhachack, L.G.; Cezairliyan, B.O.; Ausubel, F.M. Distinct Pathogenesis and Host Responses during Infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010, 6, e1000982. [Google Scholar] [CrossRef] [Green Version]
- Marsh, E.; May, R.C. Caenorhabditis elegans, a Model Organism for Investigating Immunity. Appl. Environ. Microbiol. 2012, 78, 2075–2081. [Google Scholar] [CrossRef] [Green Version]
- Kwon, G.; Lee, J.; Koh, J.-H.; Lim, Y.-H. Lifespan Extension of Caenorhabditis elegans by Butyricicoccus pullicaecorum and Megasphaera elsdenii with Probiotic Potential. Curr. Microbiol. 2017, 75, 557–564. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).