Distribution and Potential Metabolic Functions of Soil Actinobacteria in Degraded Alpine Grassland on the Northern Tibetan Plateau
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Soil Sampling
2.2. DNA Extraction, PCR, and Illumina Sequencing
2.3. Sequence Processing and Diversity Analysis
2.4. Functional Prediction and Correlation Analysis
2.5. Statistical Analysis
2.6. Determination of Soil Physical and Chemical Factors
3. Results
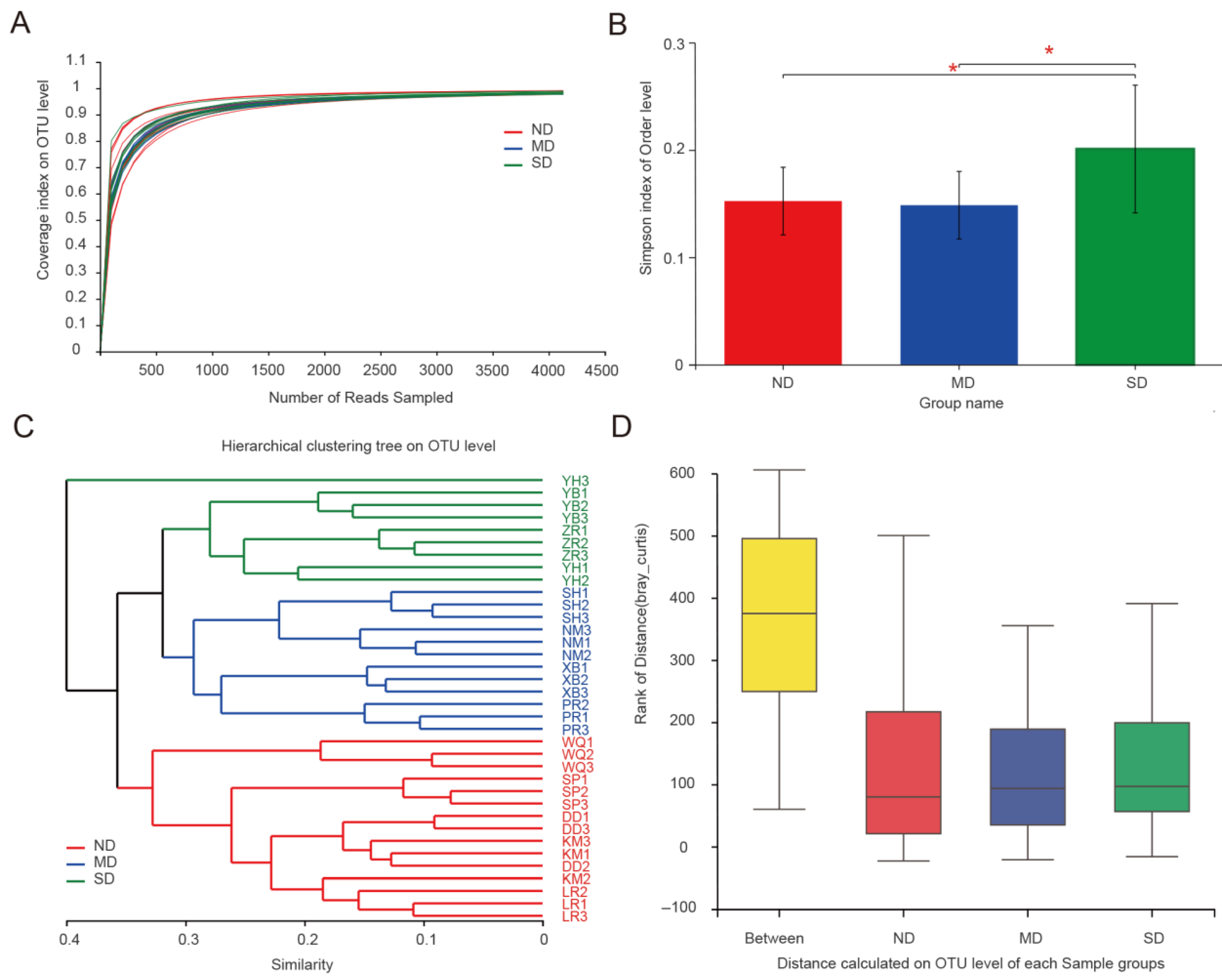
3.1. Alpha-Diversity of Actinobacteria
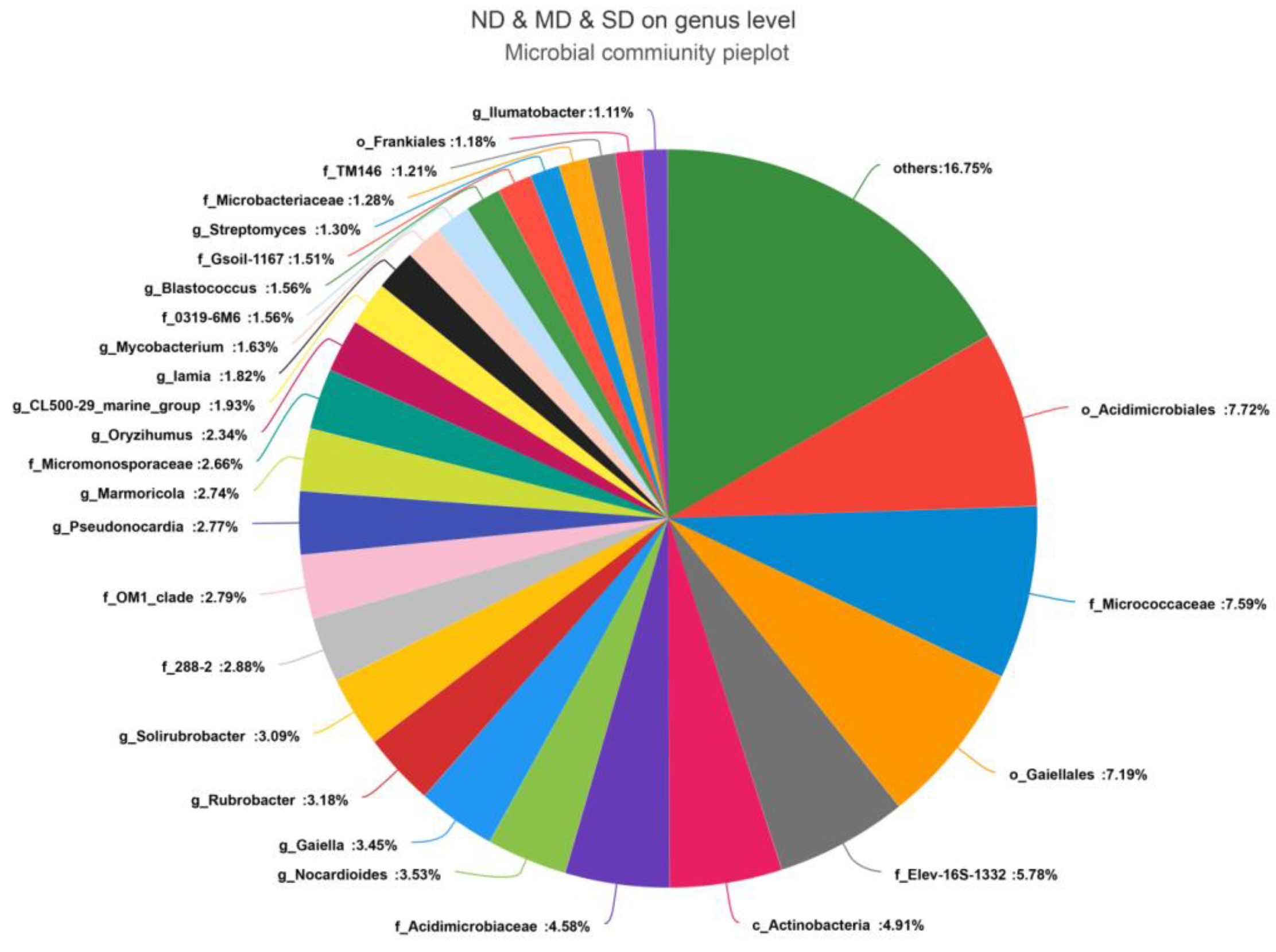
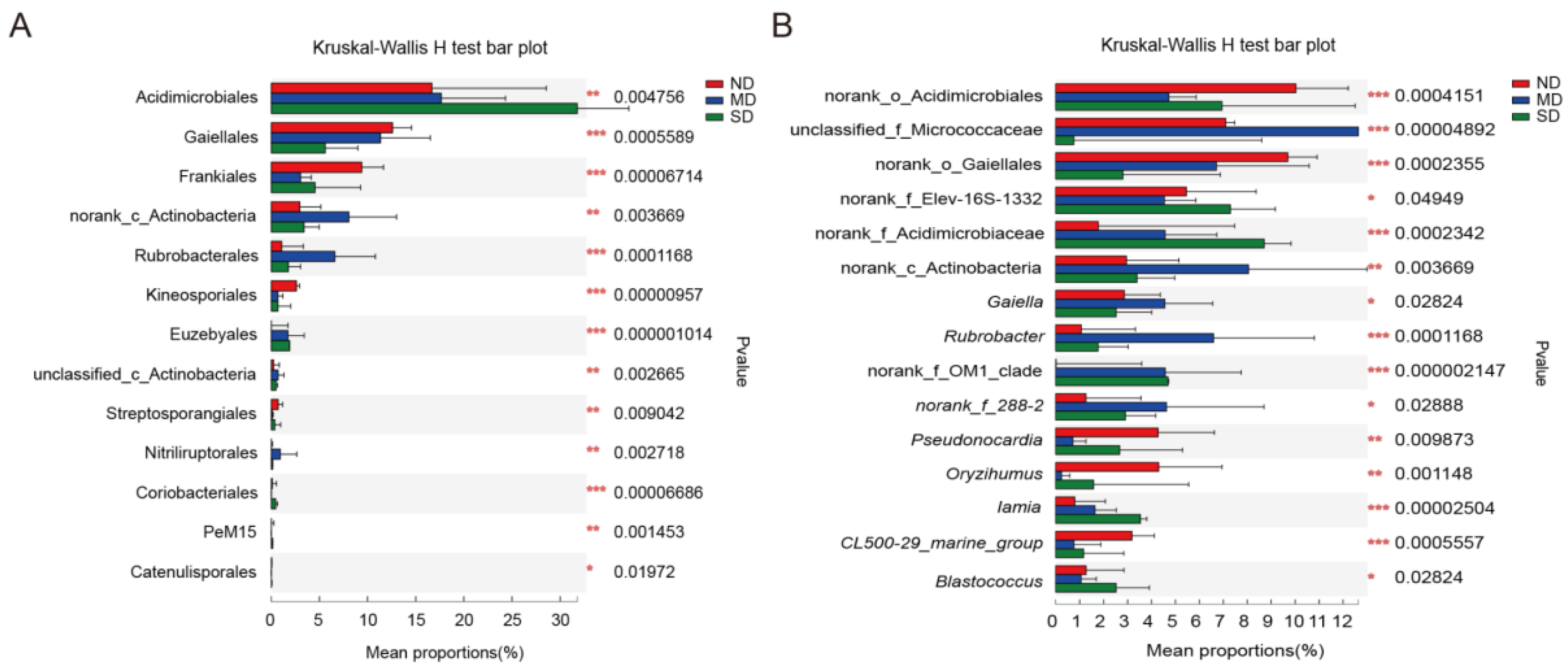
3.2. Community Composition of Actinobacteria
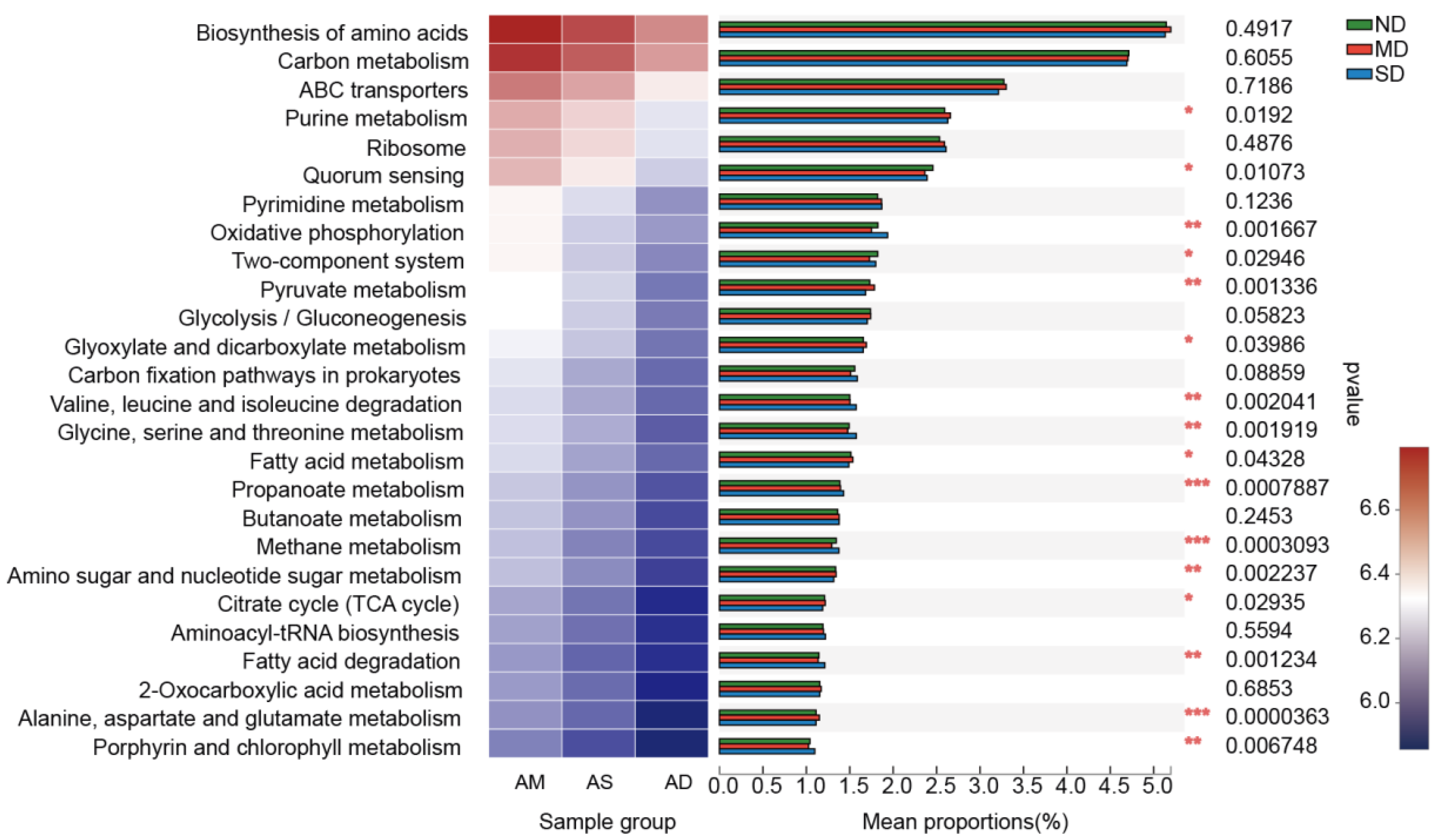
3.3. Predicting the Potential Function of Actinobacteria Communities
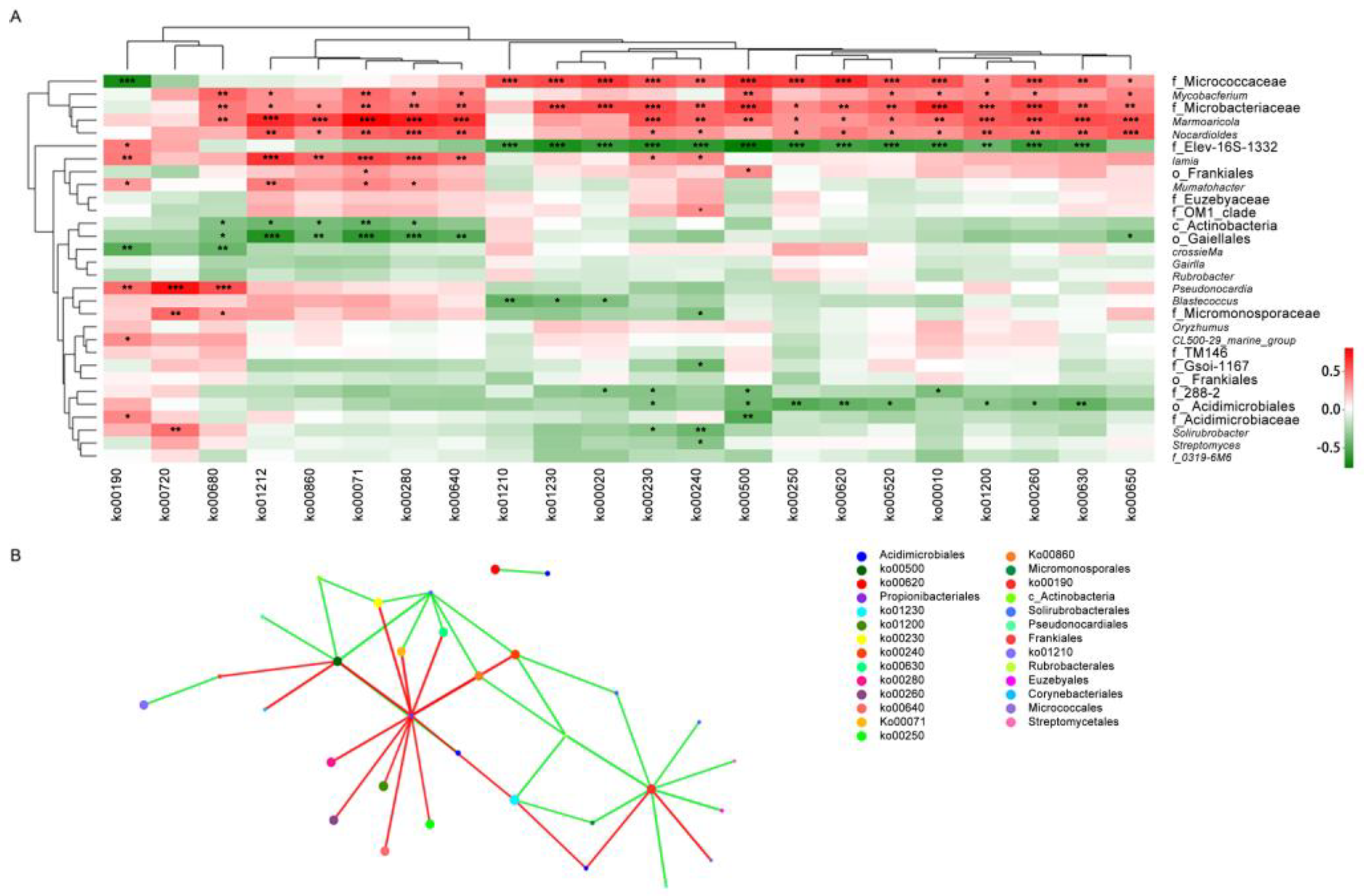
3.4. Correlation Analysis Between Metabolism Functions and Actinobacteria Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, R.J.; Wang, G.X.; Liu, W.; Wang, Q.L.; Zhang, L.; Wu, P.F. Relationship between plant communities, characters, soil physical and chemical properties, and soil microbiology in alpine meadows. Cao Ye Xue Bao 2010, 19, 25–34. (In Chinese) [Google Scholar]
- Che, R.; Wang, Y.; Li, K.; Xu, Z.; Hu, J.; Wang, F.; Rui, Y.; Li, L.; Pang, Z.; Cui, X. Degrade patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 2019, 195, 104426. [Google Scholar] [CrossRef]
- Dong, J.; Che, R.; Jia, S.; Wang, F.; Zhang, B.; Cui, X.; Wang, S.; Wang, S. Responses of ammonia-oxidizing archaea and bacteria to nitrogen and phosphorus amendments in an alpine steppe. Eur. J. Soil Sci. 2019, 71, 940–954. [Google Scholar] [CrossRef]
- Dong, J.; Wang, S.; Niu, H.; Cui, X.; Wang, K. Responses of soil microbes and their interactions with plant community after nitrogen and phosphorus addition in a Tibetan alpine steppe. J. Soils Sediments 2020, 20, 2236–2247. [Google Scholar] [CrossRef]
- Singh, J.S. Microbes: The chief ecological engineers in reinstating equilibrium in degraded ecosystems. Agric. Ecosyst. Environ. 2015, 203, 80–82. [Google Scholar] [CrossRef]
- Tatsuhiro, E.; Katsubaru, S. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol. 2018, 220, 1116–1121. [Google Scholar] [CrossRef]
- Piao, Z.; Yang, L.; Zhao, L.; Yin, S. Actinobacterial community structure in soils receiving long-term organic and inorganic amendments. Appl. Environ. Microbiol. 2008, 74, 526–530. [Google Scholar] [CrossRef][Green Version]
- Bérdy, J. Bioactive microbial metabolites: A personal review. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Miao, V.; Davies, J. Actinobacteria: The good, the bad, and the ugly. Antonie Van Leeuwenhoek 2010, 98, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, Y.; Du, X.; Li, Y.; Wang, Z.; Jiang, S.; Li, Q. Effect of grassland degradation on soil quality and soil biotic community in a semi-arid temperate steppe. Ecol. Process. 2020, 9, 63. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Jiang, L.; Zhang, L.; Cui, S.; Meng, F.; Wang, Q.; Li, X.; Zhou, Y. Changes of soil microbial community under different degraded gradients of alpine meadow. Agric. Ecosyst. Environ. 2016, 222, 213–222. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Tai, X.; Sun, L.; Wu, M.; Zhan, W.; Chen, X.; Zhang, G.; Chen, T.; Liu, G.; et al. Variation in Actinobacterial community composition and potential function in different soil ecosystems belonging to the arid Heihe River Basin of Northwest China. Front. Microbiol. 2019, 10, 2209. [Google Scholar] [CrossRef]
- Salomé, C.; Nunan, N.; Pouteau, V.; Lerch, T.Z.; Chenu, C. Carbon dynamics in topsoil and in subsoil may be controlled by different regulatory mechanisms. Glob. Change Biol. 2010, 16, 416–426. [Google Scholar] [CrossRef]
- Che, R.; Wang, F.; Wang, W.; Zhang, J.; Zhao, X.; Rui, Y.; Xu, Z.; Wang, Y.; Hao, Y.; Cui, X. Increase in ammonia-oxidizing microbe abundance during degradation of alpine meadows may lead to greater soil nitrogen loss. Biogeochemistry 2017, 136, 341–352. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, X.; Li, B.; Yao, Y. Biodiversity and conservation in the Tibetan Plateau. J. Geogr. Sci. 2002, 12, 135–143. [Google Scholar] [CrossRef]
- Xue, K.; Zhang, B.; Zhou, S.T.; Ran, Q.W.; Tang, L.; Che, R.X.; Pang, Z.; Wang, F.; Wang, Y.; Zhang, J.; et al. Soil microbial communities in alpine grasslands on the Tibet Plateau and their influencing factors (in Chinese). Chin. Sci. Bull. 2019, 64, 2915–2927. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Wang, S.; Xu, D.; Yu, H.; Wu, L.; Lin, Q.; Hu, Y.; Li, X.; He, Z.; et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014, 8, 430–440. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Yang, Y.; Zhou, W.; Gang, C.; Zhang, Y.; Li, J.; An, R.; Wang, K.; Odeh, I.; et al. Quantitative assess the driving forces on the grassland degradation in the Qinghai–Tibet Plateau, in China. Ecol. Inform. 2016, 33, 32–44. [Google Scholar] [CrossRef]
- Wu, G.L.; Ren, G.H.; Dong, Q.M.; Shi, J.J.; Wang, Y.L. Above- and belowground response along degradation gradient in an Alpine Grassland of the Qinghai-Tibetan Plateau. Clean-Soil Air Water 2014, 42, 319–323. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, H.; Jing, Q.; Wu, Y.; Ma, S. Differences in species diversity, biomass, and soil properties of five types of alpine grasslands in the Northern Tibetan Plateau. PLoS ONE 2020, 15, e0228277. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.K.; Tang, L.; Zhang, X.F.; Liu, S.L.; Liu, Q.R.; Su, X.K.; Zhang, Y.; Wu, X.Y.; Zhao, Z.; Li, Y.; et al. Relationship between plant species diversity and functional diversity in alpine grasslands. Acta Ecol. Sin. 2017, 37, 1472–1483. [Google Scholar] [CrossRef]
- Li, Y.; Dong, S.; Wen, L.; Wang, X.; Wu, Y. Assessing the soil quality of alpine grasslands in the Qinghai-Tibetan Plateau using a modified soil quality index. Environ. Monit. Assess. 2013, 185, 8011–8022. [Google Scholar] [CrossRef]
- Zhang, B.L.; Wu, X.K.; Zhang, G.S.; Li, S.; Zhang, W.; Chen, X.; Sun, L.; Zhang, B.; Liu, G.; Chen, T. The diversity and biogeography of the communities of Actinobacteria in the forelands of glaciers at a continental scale. Environ. Res. Lett. 2016, 11, 054012. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 15 November 2024).
- Jin, Z.W.; Zhong, W.H.; Wu, S.S.; Han, C. Effect of vegetation degradation on microbial communities in alpine grassland soils in northwest Yunnan. Acta Microbiol. Sin. 2018, 58, 2174–2185. (In Chinese) [Google Scholar]
- Whitman, W.B.; Suzuki, K. Solirubrobacterales. Bergey’s Man. Syst. Archaea Bact. 2015, 1–3. [Google Scholar] [CrossRef]
- Albuquerque, L.; da Costa, M.S. The Family Gaiellaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 357–360. [Google Scholar] [CrossRef]
- Li, H.; Gao, Y. Plant Community Structure and Species Diversity Characteristics of Meadow Grasslands with Different Degrees of Degradation in Xiwuzhumuqin. Sci. Soil Water Conserv. 2025, in press. [Google Scholar]
- Li, P.; Zhang, Y.; Meng, Q.; Liu, Y.; Tuyiringire, D.; Chen, Z.; Liang, S. Effects of trichloroethylene stress on the microbiological characteristics of Mollisol. Ecotoxicol. Environ. Saf. 2019, 184, 109595. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Li, X.Z.; Yin, Y.B.; Luo, S.C.; Wang, D.X.; Zheng, H.; Liu, Y.X. High-throughput sequencing-based analysis of the composition and diversity of the endophyte community in roots of Stellera chamaejasme. Sci. Rep. 2024, 14, 8607. [Google Scholar] [CrossRef]
- You, D.; Yin, B.C.; Li, Z.H.; Zhou, Y.; Yu, W.B.; Zuo, P.; Ye, B.C. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. USA 2016, 113, 6653–6658. [Google Scholar] [CrossRef]
- Lee, S.M.; Kong, H.G.; Song, G.C.; Ryu, C.M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2020, 15, 330–347. [Google Scholar] [CrossRef]
- Yin, Y.; Geng, Y.; Han, X.; Fan, Q.; Chen, L.; Wang, H. Effects of vegetation types on soil microbial community structure in alpine loess region. Chin. J. Appl. Environ. Biol. 2025, 1–13. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Hall, E.K. A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 2012, 109, 35–47. [Google Scholar] [CrossRef]
- Krause, S.; Le Roux, X.; Niklaus, P.A.; Van Bodegom, P.M.; Lennon, J.T.; Bertilsson, S.; Grossart, H.P.; Philippot, L.; Bodelier, P.L.E. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 2014, 5, 251. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.L.; Tang, C.; Franks, A.E. Competitive traits are more important than stress-tolerance traits in a cadmium-contaminated rhizosphere: A role for trait theory in microbial ecology. Front. Microbiol. 2018, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Martiny, J.; Brodie, E.L.; Martiny, A.C.; Treseder, K.K.; Allison, S.D. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 2020, 14, 1–9. [Google Scholar] [CrossRef]
- Laughlin, D.C. The intrinsic dimensionality of plant traits and its relevance to community assembly. J. Ecol. 2014, 102, 186–193. [Google Scholar] [CrossRef]
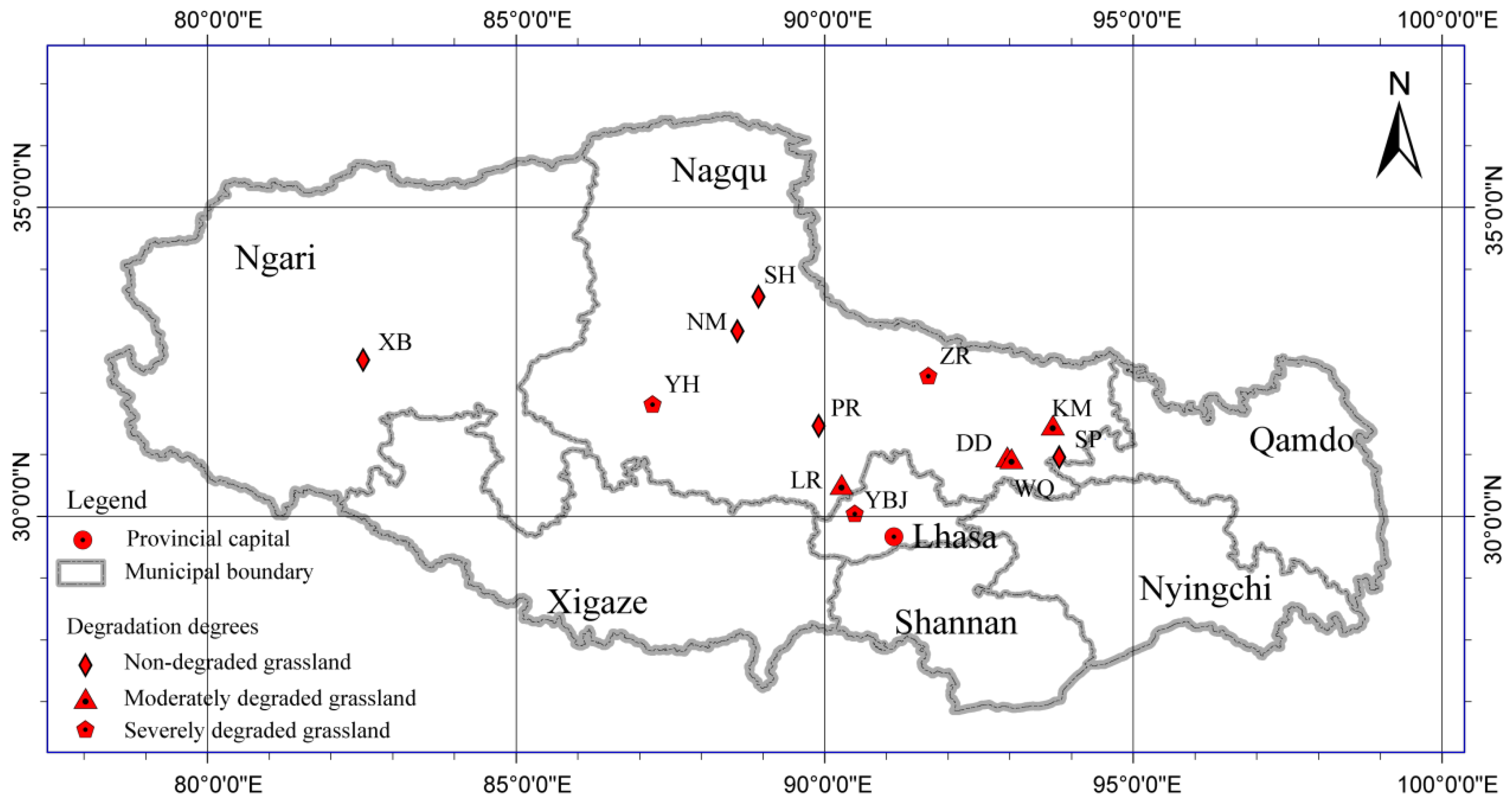

| Degradation Degrees | Symbol | Location | Latitude | Longitude | Altitude (m) |
|---|---|---|---|---|---|
| Non-degraded grassland (ND) | KM | Kongma Township, Nagqu | 31°28′50.85″ | 93°41′31.28″ | 4472 |
| LR | Longren Township, Nagqu | 30°31′23.44″ | 90°16′19.71″ | 4292 | |
| DD | Dongde Lake, Jiali County, Nagqu | 30°57′59.58″ | 92°57′04.80″ | 4893 | |
| WQ | Wuqiong Lake, Jiali County, Nagqu | 30°57′59.54″ | 92°57′04.84″ | 4691 | |
| Moderately degraded grassland (MD) | PR | Puruogangri Glacier, Shuanghu County, Nagqu | 31°28′18.33″ | 89°53′55.7″ | 5262 |
| SP | Sapu Glacier, Biru County, Nagqu | 30°57′26.67″ | 93°48′01.64″ | 4722 | |
| SH | Shuanghu County, Nagqu | 33°33′33.61″ | 88°55′34.61″ | 4815 | |
| XB | Xiongba Township, Bange County, Ngari | 32°32′07.74″ | 82°31′13.42″ | 4475 | |
| NM | Nima County, Nagqu | 32°59′58.62″ | 88°35′15.29″ | 4784 | |
| Severely degraded grassland (SD) | YH | Yanhu Township, Geji County, Nagqu | 31°49′15.54″ | 87°12′51.07″ | 4201 |
| YB | Yangbajing Town, Dangxion County, Lhasa | 30°03′14.51″ | 90°29′19.90″ | 4173 | |
| ZR | Zaren Township, Anduo County, Nagqu | 32°16′52.22″ | 91°40′54.96″ | 4566 |
| Degree of Degeneration | Plants Cover (%) | Proportion of Primary Plant (%) | Height of Plant (cm) |
|---|---|---|---|
| ND | 80–95 | 70 | 25 |
| MD | 50–70 | 30–50 | 5 |
| SD | <30 | <15 | - |
| Ecotype | Sampling Site | pH | Soil Moisture (%) | Electrical Conductivity (ms/cm) | Organic Matter (mg/kg) | Available K (mg/kg) | Available P (mg/kg) | Available N (mg/kg) | Illumination Intensity (lx) | Relative Humidity (%) | CO2 Concentration (ppm) | Dew-Point Temperature (°C) | Atmospheric Temperature (°C) | Soil Temperature (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-degraded grassland (ND) | KM | 7.74 ± 0.15 | 0.47 ± 0.03 | 0.75 ± 0.13 | 214.03 ± 18.94 | 352.7 ± 11.65 | 34.82 ± 10.2 | 740.5 ± 41.36 | 56,656 | 50.6 | 928 | 8.3 | 17.8 | 13.6 |
| LR | 5.64 ± 0.1 | 0.14 ± 0.01 | 0.01 ± 0.01 | 238.17 ± 19.89 | 73.25 ± 6.69 | 5.03 ± 4.11 | 915.03 ± 33.96 | 141,344 | 28.9 | 817 | 12.3 | 31.7 | 23.7 | |
| DD | 5.06 ± 0.02 | 0.29 ± 0 | 0.01 ± 0.01 | 170.23 ± 21.11 | 160.43 ± 29.51 | 22.65 ± 4.6 | 853.4 ± 32.19 | 133,650 | 25.4 | 827 | 5.8 | 27.4 | 12.9 | |
| WQ | 4.24 ± 0.05 | 0.31 ± 0.02 | 0.02 ± 0.01 | 160.67 ± 21.97 | 273.03 ± 36.98 | 12.04 ± 0.26 | 894.8 ± 54.77 | 155,000 | 43.8 | 783 | 6.1 | 17.9 | 11.4 | |
| PR | 8.18 ± 0.13 | 0.12 ± 0.01 | 0.08 ± 0.02 | 150 ± 17.06 | 8.22 ± 3.63 | 2.44 ± 1.39 | 590.07 ± 44.51 | 42,976 | 64.9 | 846 | 5.3 | 12 | 13.9 | |
| Moderately degraded grassland (MD) | SP | 5.81 ± 0.04 | 0.22 ± 0.03 | 0.01 ± 0 | 200.97 ± 35.08 | 135.77 ± 31.12 | 7.07 ± 2.62 | 644.43 ± 59.05 | 43,600 | 41.6 | 799 | 4.4 | 16.9 | 16 |
| SH | 8.12 ± 0.04 | 0.12 ± 0 | 0.05 ± 0.01 | 211.3 ± 14.25 | 148.3 ± 38.9 | 6.2 ± 1.85 | 768.63 ± 76.08 | 59,296 | 66.4 | 932 | 6.6 | 12.8 | 10.1 | |
| XB | 9.4 ± 0.03 | 0.1 ± 0.01 | 0.27 ± 0.02 | 68.28 ± 9.13 | 207.5 ± 22.51 | 7.1 ± 4.06 | 477.3 ± 29.88 | 31,644 | 30.8 | 712 | 5.2 | 22.6 | 18.8 | |
| NM | 8.56 ± 0.02 | 0.14 ± 0.01 | 0.22 ± 0 | 165.33 ± 16.97 | 196.8 ± 40.74 | 6.14 ± 0.88 | 672.77 ± 64.07 | 40,208 | 37.7 | 880 | 13.4 | 28.5 | 18.4 | |
| Severely degraded grassland (SD) | YH | 7.21 ± 0.06 | 0.24 ± 0.03 | 0.11 ± 0.01 | 147.37 ± 10.6 | 211.37 ± 9.86 | 17.48 ± 2.29 | 804.4 ± 67.59 | 153,280 | 36.3 | 860 | 11.7 | 27.3 | 14.6 |
| YB | 6.27 ± 0.14 | 0.14 ± 0 | 0.03 ± 0.01 | 125.23 ± 15.51 | 138.8 ± 28.14 | 12.22 ± 0.1 | 859.87 ± 45.61 | 60,896 | 31.3 | 806 | 9.1 | 26.3 | 22.8 | |
| ZR | 7.29 ± 0.02 | 0.47 ± 0.04 | 0.18 ± 0.03 | 277.03 ± 57.85 | 539.6 ± 7.31 | 35.28 ± 6.81 | 923.5 ± 79.1 | 28,037 | 62.9 | 850 | 13.4 | 13.5 | 11.1 |
| Sample\Estimators | Shannon | Simpson | ACE | Chao1 |
|---|---|---|---|---|
| DD1 | 4.76 ± 0.01 | 0.02 | 397.38 ± 0.03 | 396.29 ± 0.04 |
| DD2 | 4.84 ± 0.01 | 0.02 | 410.15 ± 0.02 | 397.66 ± 0.02 |
| DD3 | 4.63 ± 0.02 | 0.03 | 396.08 ± 0.05 | 388.88 ± 0.04 |
| KM1 | 4.90 ± 0.01 | 0.02 | 409.75 ± 0.01 | 421.34 ± 0.03 |
| KM2 | 4.94 ± 0.01 | 0.01 | 405.51 ± 0.05 | 401.75 ± 0.01 |
| KM3 | 4.53 ± 0.03 | 0.02 | 373.09 ± 0.02 | 379.08 ± 0.01 |
| LR1 | 5.20 ± 0.01 | 0.01 | 430.85 ± 0.03 | 453.08 ± 0.04 |
| LR2 | 4.75 ± 0.01 | 0.02 | 426.90 ± 0.03 | 436.92 ± 0.03 |
| LR3 | 5.12 ± 0.02 | 0.01 | 475.39 ± 0.04 | 484.47 ± 0.01 |
| SP1 | 4.48 ± 0.01 | 0.03 | 342.91 ± 0.02 | 333.78 ± 0.05 |
| SP2 | 3.90 ± 0.01 | 0.08 | 337.85 ± 0.02 | 348.16 ± 0.03 |
| SP3 | 4.20 ± 0.02 | 0.06 | 334.37 ± 0.01 | 328.00 ± 0.01 |
| WQ1 | 3.94 ± 0.01 | 0.03 | 202.57 ± 0.05 | 203.00 ± 0.04 |
| WQ2 | 4.04 ± 0.03 | 0.03 | 225.79 ± 0.04 | 229.00 ± 0.03 |
| WQ3 | 3.97 ± 0.01 | 0.03 | 208.88 ± 0.03 | 209.22 ± 0.02 |
| NM1 | 4.73 ± 0.01 | 0.02 | 389.70 ± 0.02 | 392.19 ± 0.02 |
| NM2 | 4.84 ± 0.01 | 0.01 | 413.57 ± 0.01 | 436.41 ± 0.04 |
| NM3 | 4.58 ± 0.03 | 0.02 | 369.20 ± 0.04 | 386.29 ± 0.04 |
| PR1 | 4.46 ± 0.01 | 0.05 | 396.70 ± 0.01 | 389.02 ± 0.05 |
| PR2 | 4.83 ± 0.01 | 0.02 | 421.27 ± 0.01 | 425.02 ± 0.01 |
| PR3 | 4.71 ± 0.03 | 0.03 | 407.24 ± 0.03 | 406.42 ± 0.03 |
| SH1 | 4.87 ± 0.01 | 0.02 | 404.72 ± 0.01 | 404.23 ± 0.02 |
| SH2 | 4.66 ± 0.04 | 0.02 | 387.82 ± 0.02 | 388.00 ± 0.01 |
| SH3 | 4.78 ± 0.01 | 0.02 | 423.81 ± 0.04 | 429.29 ± 0.03 |
| XB1 | 4.66 ± 0.03 | 0.03 | 400.43 ± 0.02 | 408.00 ± 0.01 |
| XB2 | 3.92 ± 0.02 | 0.11 | 384.06 ± 0.05 | 381.00 ± 0.02 |
| XB3 | 4.39 ± 0.01 | 0.06 | 403.95 ± 0.01 | 398.16 ± 0.03 |
| YB1 | 4.61 ± 0.04 | 0.04 | 438.42 ± 0.04 | 437.75 ± 0.05 |
| YB2 | 4.90 ± 0.01 | 0.01 | 422.04 ± 0.02 | 445.45 ± 0.03 |
| YB3 | 4.57 ± 0.02 | 0.02 | 374.50 ± 0.04 | 383.36 ± 0.03 |
| YH1 | 4.62 ± 0.01 | 0.02 | 331.78 ± 0.05 | 337.78 ± 0.02 |
| YH2 | 4.67 ± 0.01 | 0.02 | 295.72 ± 0.02 | 298.56 ± 0.04 |
| YH3 | 3.29 ± 0.01 | 0.10 | 230.08 ± 0.01 | 226.20 ± 0.01 |
| ZR1 | 4.34 ± 0.03 | 0.04 | 343.02 ± 0.03 | 365.72 ± 0.02 |
| ZR2 | 4.76 ± 0.01 | 0.02 | 375.14 ± 0.02 | 379.13 ± 0.01 |
| ZR3 | 4.68 ± 0.01 | 0.02 | 371.94 ± 0.05 | 369.56 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Luo, S.; Wang, Y.; Yin, Y.; Li, Y.; Zhao, W.; Zheng, S.; Xu, G.; Ma, H.; Cao, P.; et al. Distribution and Potential Metabolic Functions of Soil Actinobacteria in Degraded Alpine Grassland on the Northern Tibetan Plateau. Microorganisms 2025, 13, 2230. https://doi.org/10.3390/microorganisms13102230
Zhang J, Luo S, Wang Y, Yin Y, Li Y, Zhao W, Zheng S, Xu G, Ma H, Cao P, et al. Distribution and Potential Metabolic Functions of Soil Actinobacteria in Degraded Alpine Grassland on the Northern Tibetan Plateau. Microorganisms. 2025; 13(10):2230. https://doi.org/10.3390/microorganisms13102230
Chicago/Turabian StyleZhang, Junze, Sicen Luo, Yanying Wang, Yebing Yin, Yu Li, Wenxiang Zhao, Shirui Zheng, Guoqi Xu, Hongmei Ma, Pengxi Cao, and et al. 2025. "Distribution and Potential Metabolic Functions of Soil Actinobacteria in Degraded Alpine Grassland on the Northern Tibetan Plateau" Microorganisms 13, no. 10: 2230. https://doi.org/10.3390/microorganisms13102230
APA StyleZhang, J., Luo, S., Wang, Y., Yin, Y., Li, Y., Zhao, W., Zheng, S., Xu, G., Ma, H., Cao, P., & Liu, Y. (2025). Distribution and Potential Metabolic Functions of Soil Actinobacteria in Degraded Alpine Grassland on the Northern Tibetan Plateau. Microorganisms, 13(10), 2230. https://doi.org/10.3390/microorganisms13102230






