Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and CDI Detection
2.2. Sample Manipulation, DNA Extraction, and Library Preparation
2.3. Bioinformatics Analyses
3. Results
3.1. Demographic and Clinical Profiles
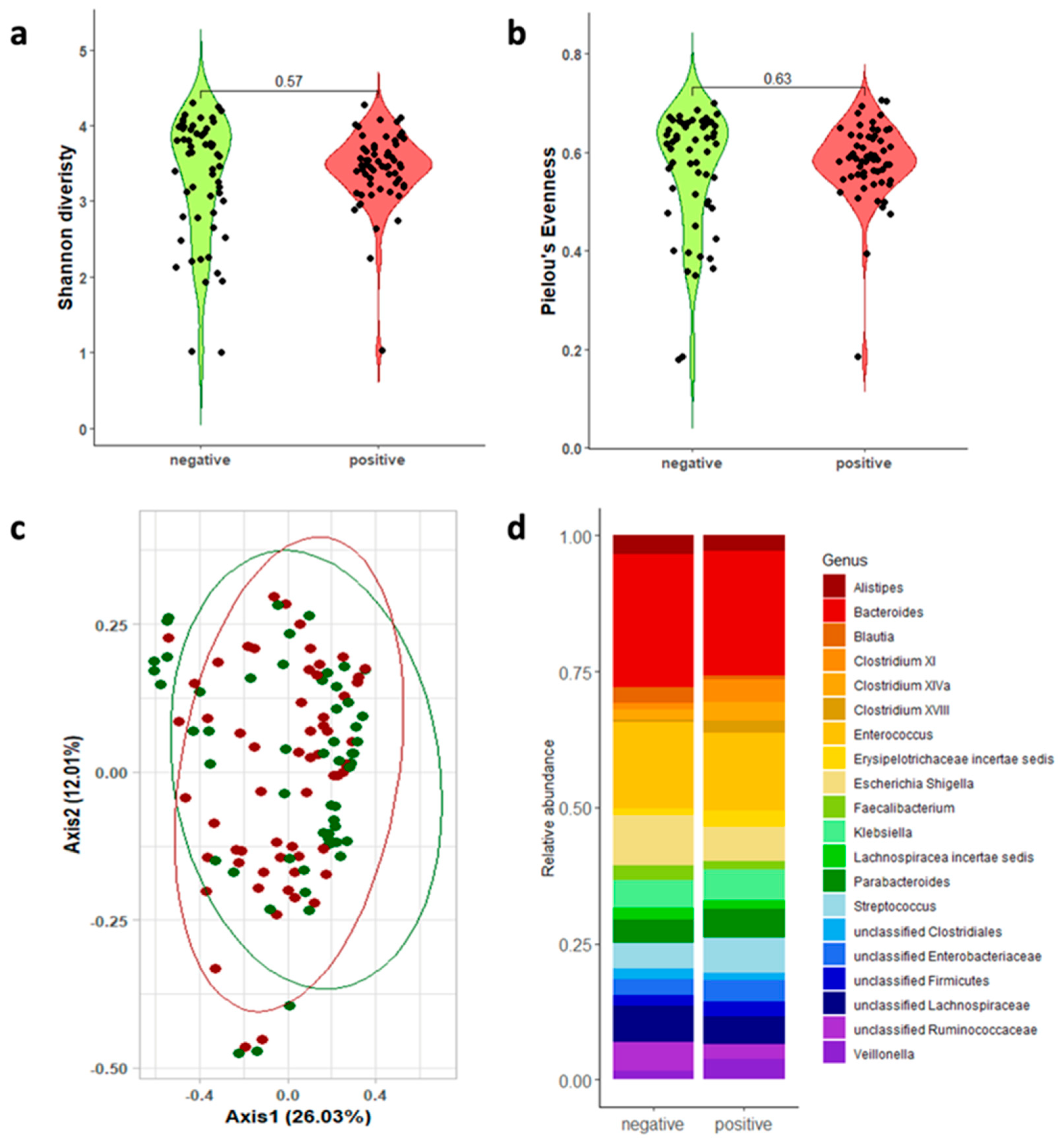
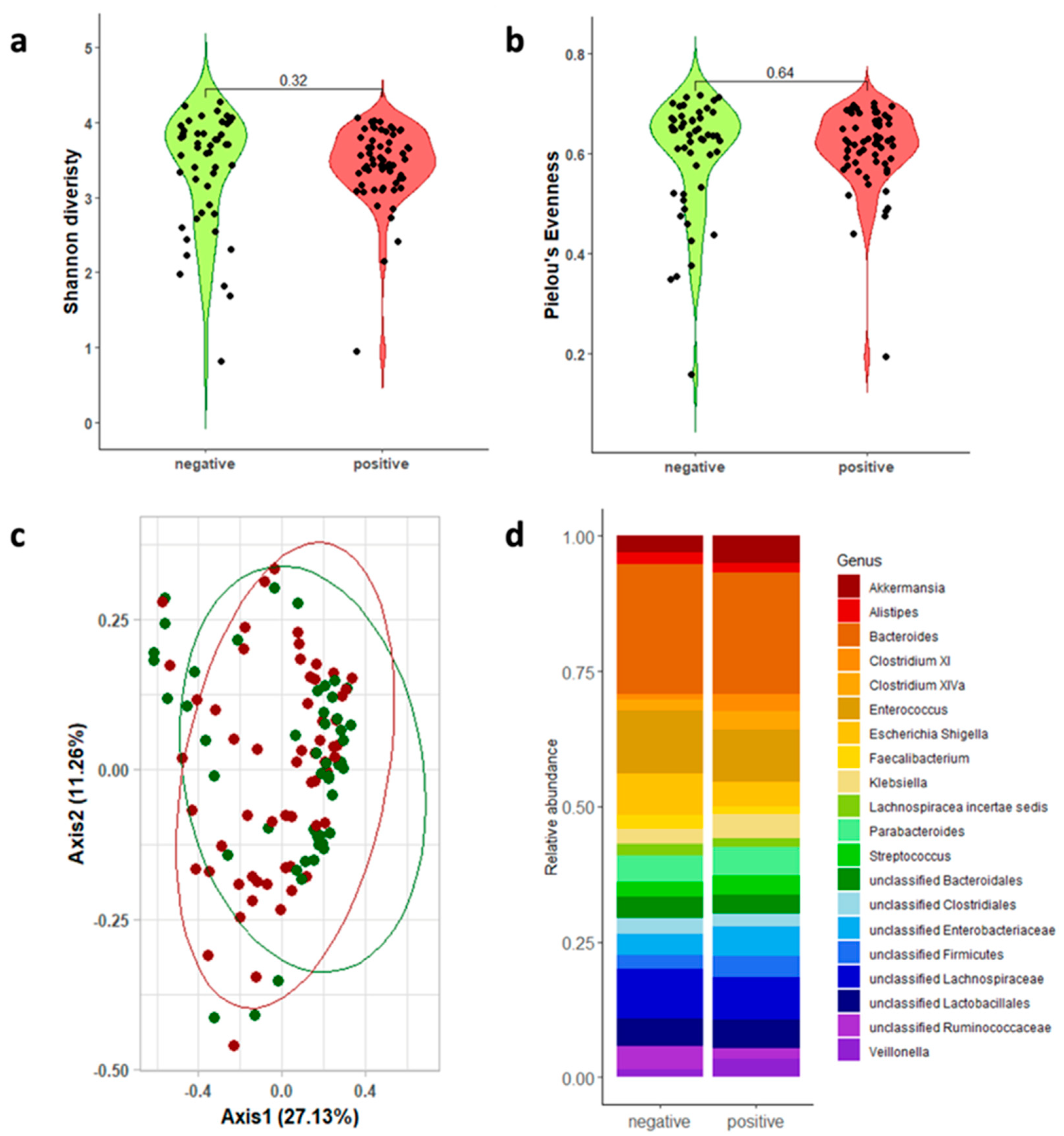
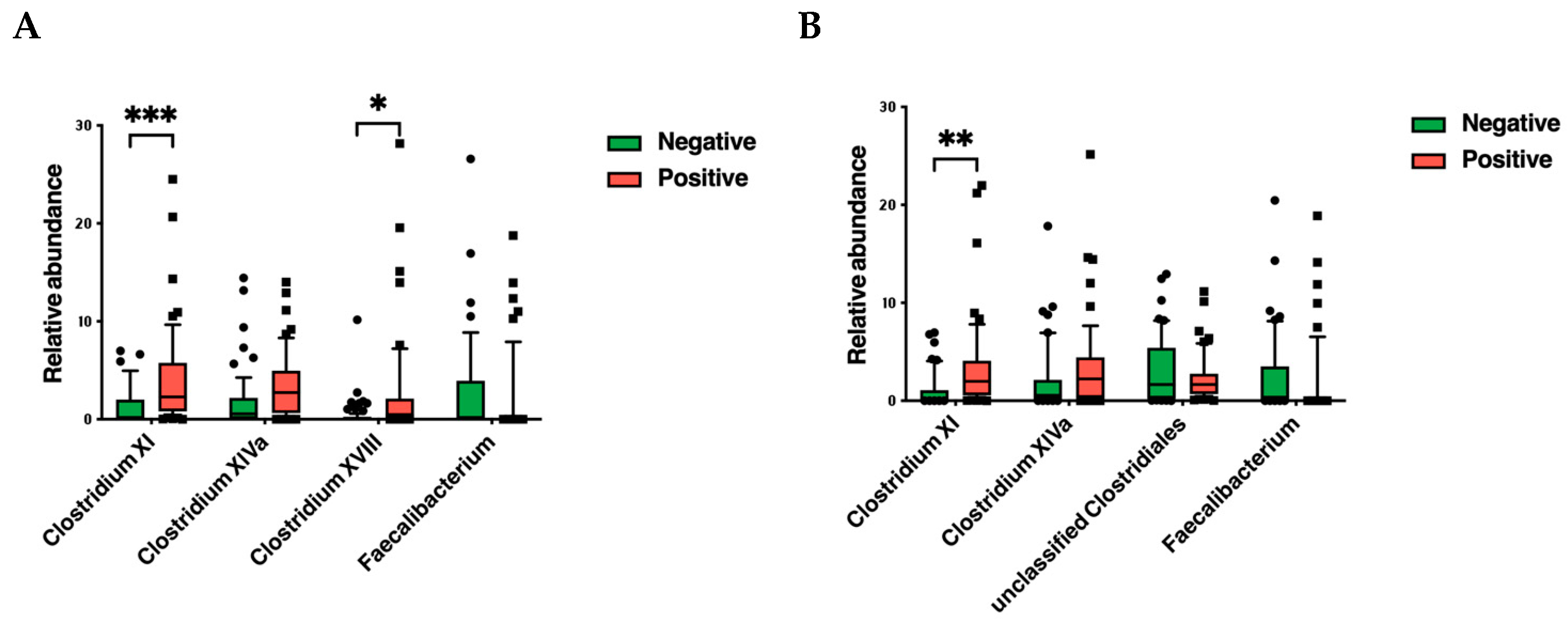
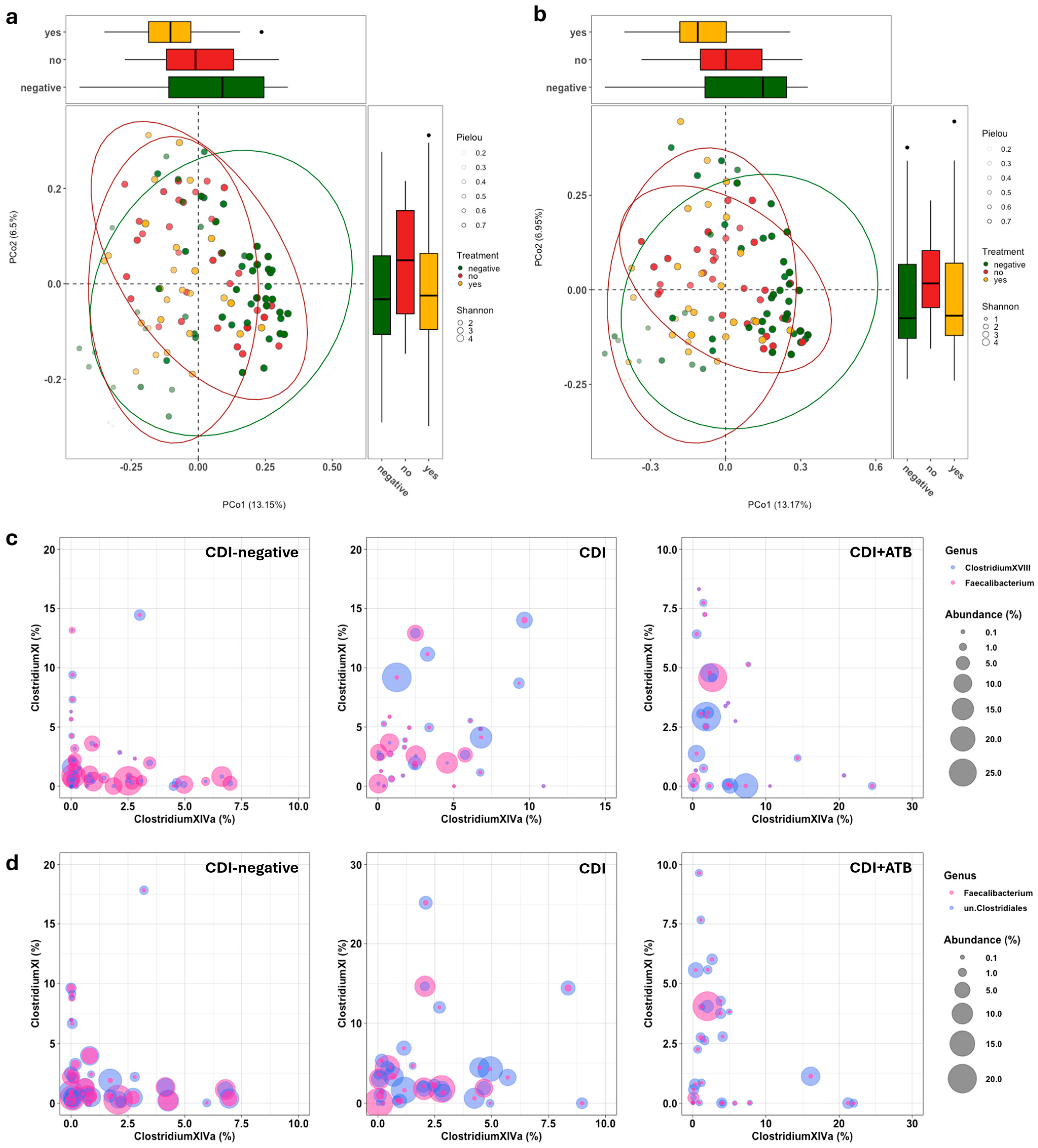
3.2. Gut Microbiota Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heinlen, L.; Ballard, J.D. Clostridium difficile infection. Am. J. Med. Sci. 2010, 340, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, A.; Rosa, F.; Manca, F.; Pignataro, G.; Zanza, C.; Savioli, G.; Covino, M.; Ojetti, V.; Gasbarrini, A.; Franceschi, F.; et al. Gut Microbiota and Clostridium difficile: What We Know and the New Frontiers. Int. J. Mol. Sci. 2022, 23, 13323. [Google Scholar] [CrossRef] [PubMed]
- Rajack, F.; Medford, S.; Naab, T. Clostridioides difficile infection leading to fulminant colitis with toxic megacolon. Autops. Case Rep. 2023, 13, e2023457. [Google Scholar] [CrossRef]
- Binyamin, D.; Nitzan, O.; Azrad, M.; Hamo, Z.; Koren, O.; Peretz, A. The microbial diversity following antibiotic treatment of Clostridioides difficile infection. BMC Gastroenterol. 2021, 21, 166. [Google Scholar] [CrossRef]
- Martinez-Melendez, A.; Camacho-Ortiz, A.; Morfin-Otero, R.; Maldonado-Garza, H.J.; Villarreal-Trevino, L.; Garza-Gonzalez, E. Current knowledge on the laboratory diagnosis of Clostridium difficile infection. World J. Gastroenterol. 2017, 23, 1552–1567. [Google Scholar] [CrossRef]
- Cheng, J.W.; Xiao, M.; Kudinha, T.; Xu, Z.P.; Sun, L.Y.; Hou, X.; Zhang, L.; Fan, X.; Kong, F.; Xu, Y.C. The Role of Glutamate Dehydrogenase (GDH) Testing Assay in the Diagnosis of Clostridium difficile Infections: A High Sensitive Screening Test and an Essential Step in the Proposed Laboratory Diagnosis Workflow for Developing Countries like China. PLoS ONE 2015, 10, e0144604. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.C.; Mizusawa, M. Laboratory Tests for the Diagnosis of Clostridium difficile. Clin. Colon. Rectal Surg. 2020, 33, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, K.; Khanna, S. Gut microbiome and Clostridioides difficile infection: A closer look at the microscopic interface. Therap Adv. Gastroenterol. 2021, 14, 1756284821994736. [Google Scholar] [CrossRef]
- Rafey, A.; Jahan, S.; Farooq, U.; Akhtar, F.; Irshad, M.; Nizamuddin, S.; Parveen, A. Antibiotics Associated With Clostridium difficile Infection. Cureus 2023, 15, e39029. [Google Scholar] [CrossRef] [PubMed]
- Dysangco, A.T.; Pierce, T.M.; Bravata, D.M. Risk of antibiotics associated with Clostridioides difficile infection: Antibiotic stewardship in action. Antimicrob. Steward. Healthc. Epidemiol. 2022, 2, e146. [Google Scholar] [CrossRef]
- Kachrimanidou, M.; Tsintarakis, E. Insights into the Role of Human Gut Microbiota in Clostridioides difficile Infection. Microorganisms 2020, 8, 200. [Google Scholar] [CrossRef]
- Khan, I.; Bai, Y.; Zha, L.; Ullah, N.; Ullah, H.; Shah, S.R.H.; Sun, H.; Zhang, C. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front. Cell Infect. Microbiol. 2021, 11, 716299. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, I.M.; Chifiriuc, M.C.; Pircalabioru, G.G.; Filip, R.; Bolocan, A.; Lazar, V.; Ditu, L.M.; Bleotu, C. Gut Dysbiosis and Clostridioides difficile Infection in Neonates and Adults. Front. Microbiol. 2021, 12, 651081. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Han, S.; Kwon, J.; Ju, S.; Choi, T.G.; Kang, I.; Kim, S.S. Roles of Short-Chain Fatty Acids in Inflammatory Bowel Disease. Nutrients 2023, 15, 4466. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Jiang, C.; Li, Z.; Wang, X.; Peng, Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 2015, 34, 1–7. [Google Scholar] [CrossRef]
- Han, S.H.; Yi, J.; Kim, J.H.; Lee, S.; Moon, H.W. Composition of gut microbiota in patients with toxigenic Clostridioides (Clostridium) difficile: Comparison between subgroups according to clinical criteria and toxin gene load. PLoS ONE 2019, 14, e0212626. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.R. Gut check: In vitro diagnostics for gut microbiome analysis. Clin. Microbiol. Newsl. 2019, 41, 57–62. [Google Scholar] [CrossRef]
- Jeon, Y.D.; Ann, H.W.; Lee, W.J.; Kim, J.H.; Seong, H.; Kim, J.H.; Ahn, J.Y.; Jeong, S.J.; Ku, N.S.; Yeom, J.S.; et al. Characteristics of Faecal Microbiota in Korean Patients with Clostridioides difficile-associated Diarrhea. Infect. Chemother. 2019, 51, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Sangster, W.; Hegarty, J.P.; Schieffer, K.M.; Wright, J.R.; Hackman, J.; Toole, D.R.; Lamendella, R.; Stewart, D.B., Sr. Bacterial and Fungal Microbiota Changes Distinguish C. difficile Infection from Other Forms of Diarrhea: Results of a Prospective Inpatient Study. Front. Microbiol. 2016, 7, 789. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Maciel-Fiuza, M.F.; Muller, G.C.; Campos, D.M.S.; do Socorro Silva Costa, P.; Peruzzo, J.; Bonamigo, R.R.; Veit, T.; Vianna, F.S.L. Role of gut microbiota in infectious and inflammatory diseases. Front. Microbiol. 2023, 14, 1098386. [Google Scholar] [CrossRef]
- Stewart, D.B., Sr.; Wright, J.R.; Fowler, M.; McLimans, C.J.; Tokarev, V.; Amaniera, I.; Baker, O.; Wong, H.T.; Brabec, J.; Drucker, R.; et al. Integrated Meta-omics Reveals a Fungus-Associated Bacteriome and Distinct Functional Pathways in Clostridioides difficile Infection. mSphere 2019, 4, e00454-19. [Google Scholar] [CrossRef]
- Wiegers, C.; van de Burgwal, L.H.M.; Larsen, O.F.A. Probiotics for the Management of Infectious Diseases: Reviewing the State of the Art. Front. Microbiol. 2022, 13, 877142. [Google Scholar] [CrossRef] [PubMed]
- Daquigan, N.; Seekatz, A.M.; Greathouse, K.L.; Young, V.B.; White, J.R. High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 2017, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiao, F.; Wang, X.; Ye, L.; Xiao, Y.; Li, D.; Zhang, T.; Wang, Y. Gut Microbial and Metabolic Features Associated With Clostridioides difficile Infection Recurrence in Children. Open Forum Infect. Dis. 2024, 11, ofae506. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Glendinning, L.; Wisedchanwet, T.; Watson, M. The Madness of Microbiome: Attempting To Find Consensus “Best Practice” for 16S Microbiome Studies. Appl. Environ. Microbiol. 2018, 84, e02627-17. [Google Scholar] [CrossRef]
- Beaudry, M.S.; Wang, J.; Kieran, T.J.; Thomas, J.; Bayona-Vasquez, N.J.; Gao, B.; Devault, A.; Brunelle, B.; Lu, K.; Wang, J.S.; et al. Improved Microbial Community Characterization of 16S rRNA via Metagenome Hybridization Capture Enrichment. Front. Microbiol. 2021, 12, 644662. [Google Scholar] [CrossRef] [PubMed]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. Author Correction: The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci. Data 2022, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.; Planche, T.; Eckert, C.; Barbut, F.; Terveer, E.M.; Dekkers, O.M.; Wilcox, M.H.; Kuijper, E.J. European Society of Clinical Microbiology and Infectious Diseases: Update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2016, 22 (Suppl. S4), S63–S81. [Google Scholar] [CrossRef]
- Posteraro, B.; De Maio, F.; Gasbarrini, A. Profiling the Gastrointestinal Microbiota. Methods Mol. Biol. 2021, 2283, 83–92. [Google Scholar] [CrossRef] [PubMed]
- De Maio, F.; Boru, C.E.; Avallone, M.; Velotti, N.; Bianco, D.M.; Capoccia, D.; Greco, F.; Guarisco, G.; Nogara, M.; Sanguinetti, M.; et al. Characterization of gut microbiota in patients with metabolic syndrome candidates for bariatric/metabolic surgery: Preliminary findings of a multi-center prospective study. Diabetes Res. Clin. Pract. 2021, 180, 109079. [Google Scholar] [CrossRef]
- Bianchetti, G.; De Maio, F.; Abeltino, A.; Serantoni, C.; Riente, A.; Santarelli, G.; Sanguinetti, M.; Delogu, G.; Martinoli, R.; Barbaresi, S.; et al. Unraveling the Gut Microbiome-Diet Connection: Exploring the Impact of Digital Precision and Personalized Nutrition on Microbiota Composition and Host Physiology. Nutrients 2023, 15, 3931. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Cascella, F.; Mellai, M.; Barizzone, N.; Mignone, F.; Massa, N.; Nobile, V.; Bona, E. Influence of Sex on the Microbiota of the Human Face. Microorganisms 2022, 10, 2470. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- De Maio, F.; Ianiro, G.; Coppola, G.; Santopaolo, F.; Abbate, V.; Bianco, D.M.; Del Zompo, F.; De Matteis, G.; Leo, M.; Nesci, A.; et al. Improved gut microbiota features after the resolution of SARS-CoV-2 infection. Gut Pathog. 2021, 13, 62. [Google Scholar] [CrossRef]
- Dubberke, E.R.; McMullen, K.M.; Mayfield, J.L.; Reske, K.A.; Georgantopoulos, P.; Warren, D.K.; Fraser, V.J. Hospital-associated Clostridium difficile infection: Is it necessary to track community-onset disease? Infect. Control Hosp. Epidemiol. 2009, 30, 332–337. [Google Scholar] [CrossRef][Green Version]
- Johanesen, P.A.; Mackin, K.E.; Hutton, M.L.; Awad, M.M.; Larcombe, S.; Amy, J.M.; Lyras, D. Disruption of the Gut Microbiome: Clostridium difficile Infection and the Threat of Antibiotic Resistance. Genes 2015, 6, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Khanna, S. Community-acquired Clostridium difficile infection: An increasing public health threat. Infect. Drug Resist. 2014, 7, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ling, L.; Stratton, C.W.; Li, C.; Polage, C.R.; Wu, B.; Tang, Y.W. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg. Microbes Infect. 2018, 7, 15. [Google Scholar] [CrossRef]
- Herrera, G.; Vega, L.; Patarroyo, M.A.; Ramirez, J.D.; Munoz, M. Gut microbiota composition in health-care facility-and community-onset diarrheic patients with Clostridioides difficile infection. Sci. Rep. 2021, 11, 10849. [Google Scholar] [CrossRef]
- Martinez, E.; Taminiau, B.; Rodriguez, C.; Daube, G. Gut Microbiota Composition Associated with Clostridioides difficile Colonization and Infection. Pathogens 2022, 11, 781. [Google Scholar] [CrossRef]
- Perez-Cobas, A.E.; Artacho, A.; Ott, S.J.; Moya, A.; Gosalbes, M.J.; Latorre, A. Structural and functional changes in the gut microbiota associated to Clostridium difficile infection. Front. Microbiol. 2014, 5, 335. [Google Scholar] [CrossRef]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Vazquez-Cuesta, S.; Villar, L.; Garcia, N.L.; Fernandez, A.I.; Olmedo, M.; Alcala, L.; Marin, M.; Munoz, P.; Bouza, E.; Reigadas, E. Characterization of the gut microbiome of patients with Clostridioides difficile infection, patients with non-C. difficile diarrhea, and C. difficile-colonized patients. Front. Cell Infect. Microbiol. 2023, 13, 1130701. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Zhang, X.; Lu, H.; Lv, T.; Shen, P.; Lv, L.; Zheng, B.; Jiang, X.; Li, L. Identification of key taxa that favor intestinal colonization of Clostridium difficile in an adult Chinese population. Microbes Infect. 2016, 18, 30–38. [Google Scholar] [CrossRef]
- Elvers, K.T.; Wilson, V.J.; Hammond, A.; Duncan, L.; Huntley, A.L.; Hay, A.D.; van der Werf, E.T. Antibiotic-induced changes in the human gut microbiota for the most commonly prescribed antibiotics in primary care in the UK: A systematic review. BMJ Open 2020, 10, e035677. [Google Scholar] [CrossRef]
- Wang, L.; Guo, G.; Xu, Y.; Li, L.; Yang, B.; Zhao, D.; Tian, H.; Ye, C.; Lin, Z.; Cui, J.; et al. The effect of fecal microbiota transplantation on antibiotic-associated diarrhea and its impact on gut microbiota. BMC Microbiol. 2024, 24, 160. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Porchas, M.; Villalpando-Canchola, E.; Ortiz Suarez, L.E.; Vargas-Albores, F. How conserved are the conserved 16S-rRNA regions? PeerJ 2017, 5, e3036. [Google Scholar] [CrossRef]
- Pinna, N.K.; Dutta, A.; Monzoorul Haque, M.; Mande, S.S. Can Targeting Non-Contiguous V-Regions With Paired-End Sequencing Improve 16S rRNA-Based Taxonomic Resolution of Microbiomes?: An In Silico Evaluation. Front. Genet. 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Ticinesi, A.; Gerritsen, J.; Nouvenne, A.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Duranti, S.; Mangifesta, M.; Viappiani, A.; et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: A metagenomic study. Sci. Rep. 2016, 6, 25945. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, Q.; Gu, S.; Chen, Y.; Lv, L.; Zheng, B.; Wang, Q.; Wang, K.; Wang, S.; Xia, J.; et al. Akkermansia muciniphila Ameliorates Clostridioides difficile Infection in Mice by Modulating the Intestinal Microbiome and Metabolites. Front. Microbiol. 2022, 13, 841920. [Google Scholar] [CrossRef]
- Kameoka, S.; Motooka, D.; Watanabe, S.; Kubo, R.; Jung, N.; Midorikawa, Y.; Shinozaki, N.O.; Sawai, Y.; Takeda, A.K.; Nakamura, S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1-V2 and V3-V4 primer sets. BMC Genom. 2021, 22, 527. [Google Scholar] [CrossRef]
- Palkova, L.; Tomova, A.; Repiska, G.; Babinska, K.; Bokor, B.; Mikula, I.; Minarik, G.; Ostatnikova, D.; Soltys, K. Evaluation of 16S rRNA primer sets for characterisation of microbiota in paediatric patients with autism spectrum disorder. Sci. Rep. 2021, 11, 6781. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Liu, Y.; Wang, X.; Dong, X.; Zou, X. Akkermansia muciniphila as a Next-Generation Probiotic in Modulating Human Metabolic Homeostasis and Disease Progression: A Role Mediated by Gut-Liver-Brain Axes? Int. J. Mol. Sci. 2023, 24, 3900. [Google Scholar] [CrossRef] [PubMed]
- Quaranta, G.; Ianiro, G.; De Maio, F.; Guarnaccia, A.; Fancello, G.; Agrillo, C.; Iannarelli, F.; Bibbo, S.; Amedei, A.; Sanguinetti, M.; et al. “Bacterial Consortium”: A Potential Evolution of Fecal Microbiota Transplantation for the Treatment of Clostridioides difficile Infection. Biomed. Res. Int. 2022, 2022, 5787373. [Google Scholar] [CrossRef]
- Pellegrino, A.; Coppola, G.; Santopaolo, F.; Gasbarrini, A.; Ponziani, F.R. Role of Akkermansia in Human Diseases: From Causation to Therapeutic Properties. Nutrients 2023, 15, 1815. [Google Scholar] [CrossRef]
- Chiantera, V.; Lagana, A.S.; Basciani, S.; Nordio, M.; Bizzarri, M. A Critical Perspective on the Supplementation of Akkermansia muciniphila: Benefits and Harms. Life 2023, 13, 1247. [Google Scholar] [CrossRef] [PubMed]
- Berkell, M.; Mysara, M.; Xavier, B.B.; van Werkhoven, C.H.; Monsieurs, P.; Lammens, C.; Ducher, A.; Vehreschild, M.; Goossens, H.; de Gunzburg, J.; et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 2021, 12, 2241. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ufondu, A.; Lee, K.; Jayaraman, A. Emerging computational tools and models for studying gut microbiota composition and function. Curr. Opin. Biotechnol. 2020, 66, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Abeltino, A.; Hatem, D.; Serantoni, C.; Riente, A.; De Giulio, M.M.; De Spirito, M.; De Maio, F.; Maulucci, G. Unraveling the Gut Microbiota: Implications for Precision Nutrition and Personalized Medicine. Nutrients 2024, 16, 3806. [Google Scholar] [CrossRef] [PubMed]
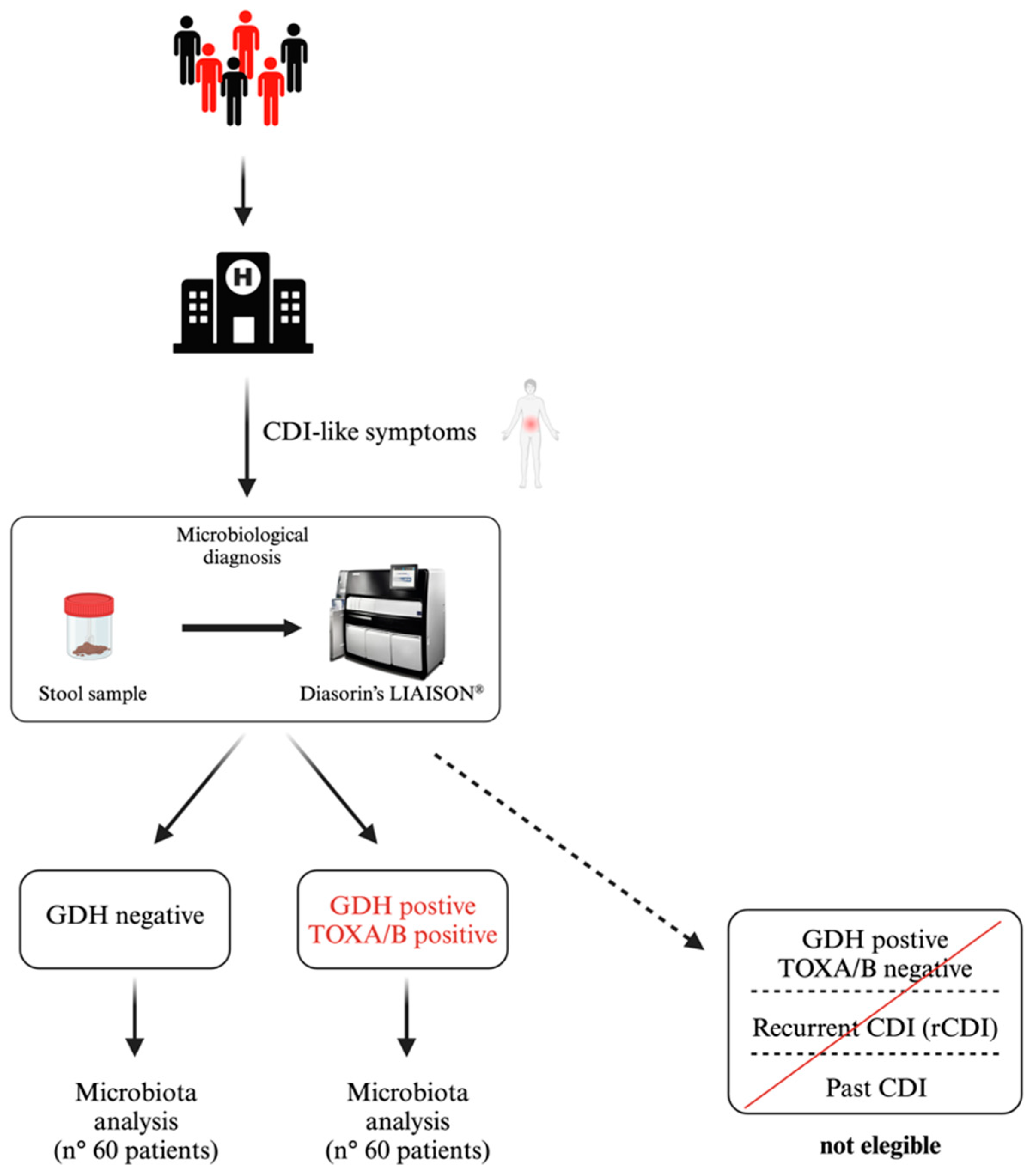
| Parameter | CDI-Positive (n. 60) | CDI-Negative (n. 60) | |
|---|---|---|---|
| Gender, n (%) Female | 26 (43.3) | 33 (55.0) | |
| Age (years, median [Q1–Q3]) | 75.50; [62.00–82.00] | 62 [48.50–76.00] | |
| Hospitalization (days, median [Q1–Q3]) | 9.50; [2.00–24.75] | 4.50 [1.00–10.75] | |
| Community-acquired infections, n (%) | 23 (38.3) | - | |
| Dead patients, n (%) | 5 (74.0) | 5 (74.0) | |
| GDH (median [Q1–Q3]) | 95.00; [95.00–95.00] | 0.26 [0.23–0.29] | |
| Toxins A/B (median [Q1–Q3]) | 10.1; [3.17–71.00] | - | |
| Number of antibiotics and antibiotic classes received before onset of CDI, n (%) | |||
| 0 | 32 (53.33) | 57 (95.00) | |
1
| 9 (15.00) | 1
| 3 (5.00) |
2
| 6 (10.00) | - | |
3
| 5 (8.33) | - | |
4
| 6 (10.00) | - | |
5
| 2 (3.33) | - |
| 16s rRNA Regions | CDI-Positive Mean; Median [Q1–Q3] | CDI-Negative Mean; Median [Q1–Q3] | Wilcoxon | |
|---|---|---|---|---|
| Shannon diversity | V1-V3 | 3.424; 3.468 [3.221–3.697]; | 3.330; 3.637 [2.844–3.955] | p = 0.565 |
| V3-V4-V6 | 3.432; 3.495 [3.227–3.778] | 3.391; 3.699 [2.964–3.946] | p = 0.317 | |
| Pielou’s evenness | V1-V3 | 0.581; 0.586 [0.550–0.632]; | 0.564; 0.617 [0.500–0.656] | p = 0.626 |
| V3-V4-V6 | 0.609; 0.622 [0.580–0.633] | 0.596; 0.636 [0.543–0.670] | p = 0.637 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosato, R.; Quaranta, G.; Santarelli, G.; Fancello, G.; Bianco, D.M.; Monzo, F.R.; Bibbò, S.; Cammarota, G.; Sanguinetti, M.; Masucci, L.; et al. Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset. Microorganisms 2025, 13, 16. https://doi.org/10.3390/microorganisms13010016
Rosato R, Quaranta G, Santarelli G, Fancello G, Bianco DM, Monzo FR, Bibbò S, Cammarota G, Sanguinetti M, Masucci L, et al. Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset. Microorganisms. 2025; 13(1):16. https://doi.org/10.3390/microorganisms13010016
Chicago/Turabian StyleRosato, Roberto, Gianluca Quaranta, Giulia Santarelli, Giovanni Fancello, Delia Mercedes Bianco, Francesca Romana Monzo, Stefano Bibbò, Giovanni Cammarota, Maurizio Sanguinetti, Luca Masucci, and et al. 2025. "Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset" Microorganisms 13, no. 1: 16. https://doi.org/10.3390/microorganisms13010016
APA StyleRosato, R., Quaranta, G., Santarelli, G., Fancello, G., Bianco, D. M., Monzo, F. R., Bibbò, S., Cammarota, G., Sanguinetti, M., Masucci, L., & De Maio, F. (2025). Can Gut Microbiota Analysis Reveal Clostridioides difficile Infection? Evidence from an Italian Cohort at Disease Onset. Microorganisms, 13(1), 16. https://doi.org/10.3390/microorganisms13010016









