Abstract
The Esx-1 family proteins of the Type VII secretion systems of Mycobacterium bovis and Mycobacterium tuberculosis have been assessed and are frequently used as candidates for tuberculosis (TB) diagnosis in both humans and animals. The presence of ESAT-6 and CFP 10 proteins, which are the most immunogenic proteins of the Esx-1 system and have been widely investigated for the immunodiagnosis of tuberculosis, in some Mycobacteriaceae and in Mycobacterium leprae, poses limitations for their use in specific diagnoses of TB. As such, to improve the specificity of the ESAT-6/CFP 10-based cell-mediated immunity (CMI) assays, other proteins encoded by genes within and outside the RD 1 region of the esx-1 locus have been evaluated as candidate antigens for CMI, as well as to investigate humoral responses in combination with ESAT-6 and or CFP 10, with varying specificity and sensitivity results. Hence, in this study, we evaluated various non-tuberculous mycobacteria (NTM), Mycolicibacterium, Mycolicibacter and Mycobacteroides species genomes available on the NCBI database for the presence and composition of the RD1 region of the esx-1 locus. In addition, we also assayed by polymerase chain reaction (PCR) and sequencing of Mycobacteriaceae available in our culture collection for the presence and sequence diversity of esxA and esxB genes encoding ESAT-6 and CFP 10, respectively. Whole genome sequence (WGS) data analysis revealed the presence of RD 1 gene orthologs in 70 of the over 100 published genomes of pathogenic and non- pathogenic Mycobcteriaceae other than tuberculosis. Among species evaluated from our culture collection, in addition to earlier reports of the presence of esxA and esxB in certain Mycolicibacterium, Mycolicibacterium septicum/peregrinum, Mycolicibacterium porcinum and Mycobacterium sp. N845T were also found to harbour orthologs of both genes. Orthologs of esxA only were detected in Mycobacterium brasiliensis, Mycolicibacterium elephantis and Mycolicibacterium flouroantheinivorans, whereas in Mycolicibacter engbackii, Mycolicibacterium mageritense and Mycobacterium paraffinicum, only esxB orthologs were detected. A phylogenetic analysis based on esxA and esxB sequences separated slow-growing from rapidly growing bacteria. These findings strengthen previous suggestions that esxA and esxB may be encoded in the majority of Mycobacteriaceae. The role of the Esx-1 system in both pathogenic and non-pathogenic Mycobacteriaceae needs further investigation, as these species may pose limitations to immunological assays for TB.
1. Introduction
Tuberculosis, caused by members of the Mycobacterium tuberculosis complex (MTBC) bacilli, is an important zoonotic disease in both humans and animals. The disease has long been recognized worldwide as a significant health and economic risk. The main etiological agent in humans is Mycobacterium tuberculosis, and in animals TB is mainly caused by Mycobacterium bovis [1]. The M. bovis and M. tuberculosis Esx-1 encodes 23 genes, namely esxA to esxW, which occur in pairs, and the singleton esxQ, related to the WXG100 family of proteins, which is characterized by a size of ~100 amino acids and the presence of a Trp-Xaa-Gly (W-X-G) motif [2]. The role of the Esx-1 secretion system in virulence as well as immunogenicity has been well described [3]. The WXG100 family of proteins are among the most immunodominant antigens recognised by the animal and human immune system [4]. Region of difference (RD1) encodes Esx-1 locus proteins, of which ESAT-6 encoded by esxA and CFP 10 encoded by esxB are the most immunodominant proteins and most widely investigated for use in cell-mediated immunity (CMI)-based diagnoses of TB in animals and humans [5,6]. ESAT-6 and CFP 10 have been reported to lack sequence homology with other Esx family proteins of M. bovis and M. tuberculosis; they are absent in the M. bovis BCG vaccine strain and M. microti, which lack the RD1 region. Consequently, they have been evaluated for use as diagnostic markers to differentiate between TB infection and M. bovis BCG vaccination [5,6]. Now, according to the new classification, mycobacteria are reclassified into the following five clades: Tuberculosis-Simiae, consisting of members of the MTBC as well as pathogenic slow-growing (SG) NTM; Terrae clade, consisting of most of the non-pathogenic slow-growing species belonging to the M. terrae complex; the Triviale clade consisting of slow-growing M. triviale and M. koreense; the Chelonae-abscessuss clade, consisting of the pathogenic rapidly growing (RG) species; and lastly the Fortuitum–vaccae clade, consisting of most of the non-pathogenic rapidly growing species. The above were proposed to be the following genera: an amended Mycobacterium, Mycolicibacter gen. nov., Mycolicibacillus gen. nov., Mycobacteroides gen. nov and Mycolicibacterium gen. nov, respectively [7]. Orthologs of esxA and esxB are found in a number of species of the genera Mycolicibacterium, Mycobacteroides, Mycobacterium and Mycolicibacter [7,8], but so far investigations into the immunodominance of the homologs of ESAT-6 and CFP 10 and others have mainly focused on pathogenic non-tuberculous mycobacteria (NTM) that are phylogenetically related to the Mycobacterium tuberculosis complex (MTBC), such as Mycobacterium kansasii and Mycobacterium marinum as well as Mycolicibacterium smegmatis [6]. This is despite the reported occurrence of these genes in other species of the different Mycobacteriaceae, including non-pathogenic spp., among others in addition to Mycolicibacterium smegmatis [7,8], Mycobacterium riyadhense [8], Mycobacterium gastri [9], Mycolicibacterium fortuitum, Mycolicibacterium malmesburyense, Mycolicibacterium komaniense, Mycolicibacterium. spp. JLS and Mycolicibacterium farcinogenes [10]. Homologs of ESAT-6 (L-ESAT-6) and CFP 10 (L-CFP 10) in M. leprae have sequence similarities of 32% and 40%, respectively, to the M. bovis and M. tuberculosis proteins, and were reported to elicit adequate immune responses that may be used for the diagnosis of leprosy [11,12]. Despite these sequence differences, L-ESAT 6 and L-CFP 10 have also been shown to be recognised by T cells from TB patients [11,12]. Other proteins of the Esx-1 locus, including EspJ [13], and outside the Esx-1 locus, including Mb1992, Mb2031c, Mb2319, Mb2843, Mb2845c, Mb3212c, Rv0899 (OmpATB), EspC, EspF and others [13,14,15], either alone or in combination with ESAT 6/CFP 10, have also been investigated as candidate antigens for (animal) TB CMI assays with varying specificity and sensitivity results. Since (animal) TB diagnosis by CMI is complicated by the cross-reactivity of the immunodominant proteins with NTM orthologs, antibody-based assay development has therefore become a subject of research. Proteins such as MBP70 and MBP83, which are well-documented candidate antigens for humoral response; MBP63, which has been shown to induce both CMI and humoral response; and PE25 and PE41, which also form a dimer, and have been used as fusion antigens, have been evaluated as candidate antigens for recognition by Th2 lymphocytes in humoral immune response assays or in a combination of both CMI and humoral assays, including TB Enflerplex, Luminex technology and multi antigen print immune assay (MAPIA) [3]. Thus, there has been more of a focus on multiplex antigen assay development for antibody- and CMI-based diagnostics, as well as on the combination of both assays. A greater number of antigens may provide increased sensitivity, but specificity may be reduced due to cross reactions [3]. There has also been more of a focus on fusion of the ESAT 6/CFP 10 antigens with other antigens for improved sensitivity and specificity of CMI- and humoral-based assays, with these two antigens deemed important in (animal) TB diagnosis [16]
In the current study, we investigated the presence and arrangement of the RD1 region of Esx-1 in NTM, mycobacteroides, mycolicibacter and mycolicibacteria spp. This study is therefore believed to be able in the long run to assist in the development of TB diagnostic assays with improved specificity.
2. Materials and Methods
2.1. NCBI Database Search for RD1 Region Orthologs in Genomes of Mycobacteriaceae
Mycolicibacterium smegmatis (CP009494.1) sequences of esxA and esxB retrieved from the Smegmalist database (https://mycobrowser.epfl.ch/genes/; accessed on 20 January 2022), as well as Mycolicibacterium fortuitum (CP014258.1), Mycobacterium leprae (AL583917.1), Mycobacterium kansasii (CP006835.1), Mycobacterium szulgai (EU826486.1 and FJ014490.1, respectively), Mycobacterium marinum (CP024190.1), esxA and esxB sequences all retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/; accessed on 20 January 2022), were used as query reference sequences in an NCBI BLAST (blastn search). The maximum number of 100 hits for sequences producing significant alignments was selected in all the analyses. The BLAST results and the location/arrangement of the two genes, as well as translated gene orthologs of the RD1 region, were investigated using an NCBI nucleotide search (https://www.ncbi.nlm.nih.gov/nuccore/; accessed on 20 January 2022).
2.2. PCR Primer Design for esxA and esxB Genes’ Evaluation
To cover both slow growing (SG) as well as rapid growing (RG) Mycobacteriaceae, we used primer sequences designed from M. bovis and sequences derived from a rapidly growing, well-studied species, M. smegmatis, respectively. The first sets of primers were designed manually from the M. smegmatis MC2155 nucleotide sequences using NCBI primer BLAST. The M. smegmatis MC2155 sequences of esxA and esxB were derived from the Smegmalist database (https://mycobrowser.epfl.ch/genes/; accessed on 20 January 2022). The second sets of primers were designed manually from M. bovis AF2122/97 nucleotide sequences using NCBI primer BLAST. The M. bovis sequences of esxA and esxB were derived from Bovilist database (http://genolist.pasteur.fr/BoviList/genome.cgi?; accessed on 20 January 2022). The M. bovis esxA and esxB nucleotide sequences were also compared to M. smegmatis sequences by pairwise alignment using the Molecular Evolutionary Genetic Analysis (MEGA-X) platform [17]. The oligonucleotide sequences of the two genes are captured in Table 1. The M. smegmatis-derived primers were evaluated on M. smegmatis ATCC 14468 using PCR and sequencing, whereas M. bovis-derived primers were evaluated on M. bovis field isolates that were previously identified and characterised by Hlokwe et al. [1]. Each of the primer sequences was also evaluated for sequence match on NCBI BLAST, the using blastn algorithm.

Table 1.
Primer sequences designed and used for the amplification of esxA and esxB.
2.3. Bacterial Species Subjected to Assessment for the Presence of esxA and esxB Using PCR-Sequencing
Forty-one isolates belonging to twenty-one bacterial species available in the NTM, mycolicibacteria and mycolicibacter culture collection of the ARC-OVI (Agricultural Research Council—Onderstepoort Veterinary Institute, Pretoria, South Africa) were included in this study. All isolates were derived from different sources, including soil, water, bovine nasal swabs and animal tissue, and had previously been identified to species level by Gcebe et al. [18]; Gcebe and Hlokwe [19]; and Gcebe et al. [20]. The M. bovis strains used for primer verification were previously identified by Hlokwe et al. [1]. In addition, American Type Culture Collection (ATCC) strains were used. Bacterial species used in this study, except for M. bovis, are presented in Table 2.

Table 2.
The origin of Mycobacteriaceae used in the study.
2.4. Polymerase Chain Reaction (PCR) for the Detection of esxA and esxB Gene Orthologs
Crude DNA was prepared from colonies and used as a template in polymerase chain reactions, as described by Gcebe et al. [18]. Briefly, individual colonies were suspended in PCR water and heated for 25 min at 100 °C. The supernatant was used as template DNA in the PCR protocols. The two primer pairs mentioned in Section 2.2, above, were used in the PCR assays (Table 1). The PCR conditions used for the amplification of the two gene fragments in separate PCR reactions are as follows: a 50 µL PCR reaction mixture was prepared, containing 28.5 µL de-ionised water, 3 µL MgCl2 (25 mM), 1 µL dNTP mix (10 mM), 4.75 µL of 10× PCR buffer (160 mM) (Tris -HCl, MgCl2, Tween 20, (NH4)2, SO4), 0.75 µL Taq DNA Polymerase (5 U/µL) (Supertherm TM), 1 µL of each forward and reverse primers (50 pmol) and 10 µL of DNA template. The PCR cycling parameters were as follows: 45 cycles of denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, and elongation at 72 °C for 1 min and final extension at 72 °C for 10 min. The PCR amplification products were electrophoresed on ethidium-stained 1.5% agarose gel and visualised under ultraviolet light (UV). The integrity of DNA of all the isolates that yielded negative results in all PCR assays for both esxA and esxB was tested by PCR targeting the hsp65 gene, as described by Gcebe et al. [21].
2.5. Extraction and Purification of DNA from the Agarose Gel
For sequencing and to avoid the inclusion of non-specific PCR products if present, selected amplification products were excised as accurately as possible from the gel using a clean scalpel under transillumination (Spectroline UV Transulliminator, Model T312A). The weight of every excised DNA gel sample was recorded. DNA was extracted from the gel using the Qiagen gel extraction kit following the manufacturer’s Quick-Start protocol (QIAquick® Gel Extraction Kit, Qiagen, Hilden, Germany). An aliquot of the extracted DNA was electrophoresed on agarose gel (1.5%) to confirm the success of DNA extraction.
2.6. Sequencing and Subsequent BLAST Analysis of the Orthologs of esxA and esxB Genes
Sequencing of the PCR products was performed at Inqaba Biotechnical Industries (Pty) Ltd. (Pretoria, South Africa) using an ABI sequencer. The PCR products were sequenced in both directions using the forward and reverse primer sequences that were initially used for amplification. Sequences from both strands were manually edited and pairwise alignments undertaken using the BioEdit Sequence alignment editor (version 7.1.9) and Molecular Evolutionary Genetics Analysis (MEGA X) platform [17]. The resulting consensus sequences were analysed on the NCBI platform for gene sequence identity/similarity using the Basic local alignment tool (BLAST) (www.blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 20 January 2022).
2.7. Phylogenetic Analysis
The phylogenetic analysis of 58 species of Mycobacterium, Mycolicibacterium and Mycobacteriodes combined was performed based on esxA and esxB gene sequences retrieved from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide; accessed on 20 January 2022). Multiple sequence alignments were performed using the MEGA-X platform [22]. Sequences were first trimmed manually at the 5′ and 3′ ends, so that they all began and ended at the same nucleotide position. Phylogenetic trees were created using the neighbour-joining method [23]. The neighbour-joining trees were verified using the maximum composite likelihood method, and one thousand bootstrap repeats were performed [24]. M. bovis AF2122/97 (LT 708304.1) and Mycobacterium shinjukeunse JCM14233 (AP.022575.1) were used as outgroup sequences.
3. Results
3.1. Evaluation of the Designed Primers for PCR
The amplification of the esxA and esxB gene fragments was shown in the M. smegmatis ATCC 14468 strain, resulting in products of approximately 250 bp and 270 bp for esxB and esxA, respectively, as well as in M. bovis field isolates, resulting in products of approximately 290 bp and 300 bp for esxA and esxB, respectively. Sequence data BLAST search results for the respective gene fragment sequences indicated that the amplified sequence using esxA primers was 100% identical to the M. smegmatis INHR2 strain (CP009496.1), position 87348–87617 in the genome. The amplified sequence when esxB primers were used was also identical to the M. smegmatis INHR2 strain (CP009496.1), at position 87041–87295 in the genome. Likewise, the verification of M. bovis-derived primers on M. bovis field isolates from our culture collection [1] revealed 100% sequence identity to esxA and esxB of the M. bovis AF2122/97 genome, at position 4288929–4289216 and position 4288594–42888896, respectively. The alignment of the M. bovis AF2122/97 esxA and esxB and M. smegmatis MC2155 esxA ortholog (Msmeg 0066) and esxB ortholog (Msmeg 0065) revealed 28% and 33% sequence divergence (Figure 1A,B). The combined results of the NCBI BLAST using M. smegmatis-derived esxA forward and reverse primers revealed sequence identities to esxA/WXG100 family gene fragments of the following species, i.e., different strains of M. smegmatis, Mycolicibacterium hassiacum, Mycolicibacterium goodii, Mycolicibacterium thermoresistible, Mycolicibacterium septicum, Mycolicibacterium boeneckei, Mycolicibacterium farcinogenes, Mycolicibacterium sp. VKM Ac-1817D, Mycolicibacterium fortuitum and Mycolicibacterium senegalense. Similarly, the NCBI BLAST of M. smegmatis-derived esxB primers revealed sequence identities to different strains of M. smegmatis, M. goodii, M. boenickei, M. farcinogenes, Mycolicibacterium lentiflavum, Mycolicibacterium malmoense, Mycolicibacterium parakoreensis and Mycobacterium subspecies paratuberculosis esxB/WXG 100 family gene fragments. On the other hand, the NCBI BLAST of the M. bovis-derived esxA primers revealed matches to M. tuberculosis complex (MTBC) species, M. kansasii and Mycobacterium ostraviense esxA fragments. The M. bovis-derived esxB primer NCBI BLAST showed sequence matches to those in the MTBC species, as well as in Mycolicibacterium novocastrense, Mycobacterium pseudoshotsii, Mycobacterium shotsii and M. kansasii.

Figure 1.
(A) Alignment of M. bovis esxA and M. smegmatis msmeg_0066 gene sequences encoding for Esat-6. (B) Alignment of M. bovis esxB and M. smegmatis msmeg_0065 gene sequences encoding for CFP 10. Sequence differences are highlighted in yellow, and positions of oligonucleotides are underlined.
3.2. The Presence of esxA and esxB in NTM, Mycolicibacteria, Mycolicibacter and Mycobacteroides Isolates as Determined by PCR and Sequencing
Among the reference strains shown in Table 3, both esxA and esxB genes were amplified from the DNA of M. fortuitum (ATCC 6481) and M. smegmatis (ATCC 14468) using M. smegmatis-derived primers. In M. moriokaense (ATCC 43059), neither of the genes were detected using either M. smegmatis- or M. bovis-derived primers for amplification. Field isolates belonging to M. fortuitum, M. mageritense, Mycolicibacterium sp. N845T, the Mycolicibacterium septicum/M. peregrinum group, Mycolicibacterium paraffinicum and Mycolicibacterium porcinum were also found to harbour both esxA and esxB genes. Only the esxA gene was detected in DNA of isolates belonging to Mycolicibacterium flouroanthenivorans, Mycolicibacterium elephantis and Mycolicibacterium brasiliensis using M. smegmatis-derived primers. Only the esxB gene was identified in DNA of Mycolicibacter engbaeckii using M. bovis-derived primers. The amplification of neither esxA nor esxB genes was observed in isolates belonging to Mycolicibacterium acapulcensis, Mycolicibacterium chitae, Mycolicibacterium confluentis, Mycolicibacterium vaccae/M. vanbaalenii, Mycolicibacterium parafortuitum, Mycolicibacterium austroafricanum, Mycolicibacterium madagascariense, Mycolicibacterium komaniense, Mycolicibacterium malmesburyense, Mycolicibacterium neoaurum and Mycolicibacterium moriokaense. Figures S1 and S2 show examples of amplified esxA and esxB gene fragments, respectively, using M. smegmatis-derived primers, while Figures S3 and S4 are examples of gel electrophoresis images for amplified esxA and esxB, respectively, using M. bovis-derived primers.

Table 3.
Assessment of Mycobacteriaceae for the presence of esxA and esxB by PCR.
3.3. The Presence of RD1 Region Orthologs in NTM, Mycolicibacteria and Mycobacteroides Genomes: An NCBI Database Search
Using NCBI BLAST searches employing Mycolicibacterium smegmatis, Mycolicibacterium fortuitum, Mycobacterium leprae, Mycobacterium kansasii, Mycobacterium szulgai and Mycobacterium marinum esxA and esxB as reference query sequences, we observed that these two genes and orthologs were harboured in the genomes of at least 70 Mycobacteriaceae, including NTM (n = 27), Mycolicibacterium (n = 42) and Mycobacteroides (n = 1) species, as shown in Table 4. The two genes were also detected as individual gene coding sequences in M. szulgai, as no data for whole genome sequences for this species were available. The NCBI nucleotide data search revealed the occurrence of the RD1 region in all the 70 Mycobactericeae species except for Mycolicibacterium branderi, where only esxA was found and no ortholog of esxB. Predicted protein products of the genes within the RD1 region for different species are captured in Table 4. The WGS of Mycobacterium riyadhense was only available on the NCBI nucleotide database as a summary, and thus the RD1 region analysis could not be carried out for this bacillus.

Table 4.
Region in NTM, mycolicibacteria and mycobacteroides.
3.4. The esxA- and esxB-Based Phylogenetic Analysis
Phylogenetic analyses of mycobacteria, mycolicibacteria and mycobacteroides, based on esxA and esxB sequences, respectively, revealed a clear separation of slow-growing Mycobacterium species and rapidly growing Mycolicibacterium and Mycobacteroides species, supported by up to 100% bootstrap values (Figure 2 and Figure 3). M. chelonae, the only species of the Mycobacteroides genus included in the analysis, clustered with Mycolicibacterium spp. and was found to be closer to Mycolicibacterium phlei. This clustering was supported by 99% and 100% bootstraps in esxA and esxB sequence-based trees, respectively. Sequences that were too divergent (≤50% to any of the included sequences) were excluded from this analysis.
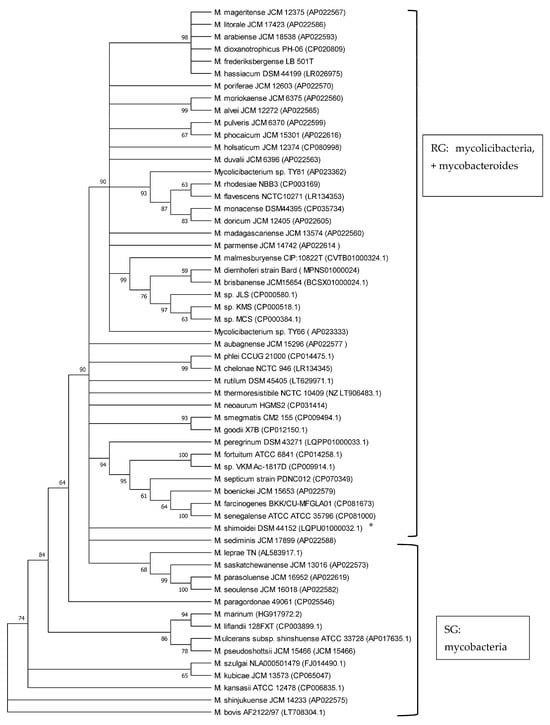
Figure 2.
Phylogenetic tree illustrating the evolutionary relationships of Mycobacteriaceae based on esxA gene sequences constructed using the neighbour-joining method. All the sequences were retrieved from Genbank. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. * is SG bacterium. All RG are Mycolicibacterium species and Mycobacteroides chelonae; all SG are Mycobacterium species.
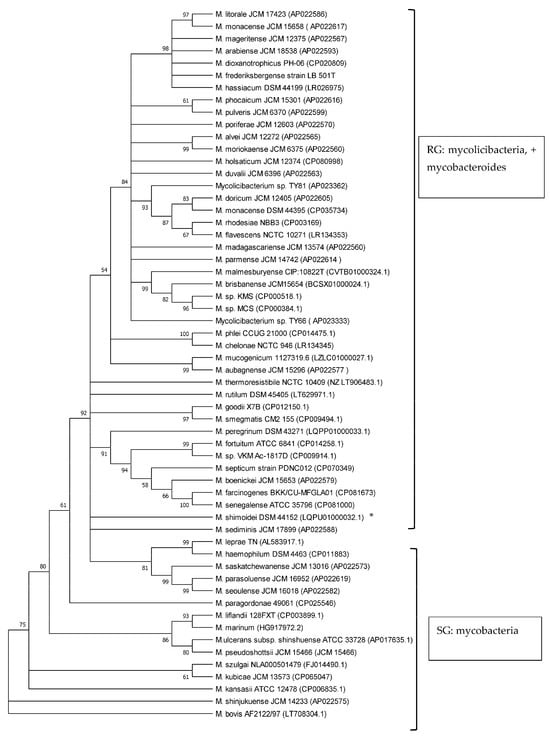
Figure 3.
Phylogenetic tree illustrating the evolutionary relationship of Mycobacteriaceae based on esxB gene sequences constructed using the neighbour-joining method. All the sequences were retrieved from Genbank. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. * is SG bacterium. All RG are Mycolicibacterium species and Mycobacteroides chelonae; all SG are Mycobacterium species.
4. Discussion
The presence of esxA and esxB genes and their protein products in NTM has been a subject of research, following the detection of these two genes and immunogenic epitopes of their protein products in some pathogenic NTMs closely related to MTBC, such as M. kansasii and M. marinum. This has been a great concern, as these protein products have been broadly used as markers in the cell-mediated immunodiagnosis of both human and animal TB, since they were thought to be specific to members of the MTBC [6,10,25]. As such, other proteins have been investigated for use as markers for the detection of CMI and humoral responses or as a combination of both for the diagnosis of (animal) TB [3,16].
To further understand the role of NTM, Mycobacteroides, Mycolicibacterium and Mycolicibacter spp. in host immune responsiveness, we set up and screened isolates available in our biobank for esxA and esxB using PCR and sequence analysis of these two genes. We also analysed genomes in the NCBI database for the presence of the RD1 region orthologs of the esx-1 locus using esxA and esxB as query sequences in the NCBI BLAST analysis. Previous studies that reported NTM orthologs of esxA and esxB used PCR with primers designed from M. tuberculosis/M. bovis to screen SG as well as RG species [8,24,25,26]. Disparities in findings from some of these studies were seen as some reported the presence of these genes and others the absence in similar species, probably due to differences in primers used for amplification or PCR failures [8,24,25,26]. We therefore used primer sequences derived from a rapidly growing species, M. smegmatis, as well as primers designed from M. bovis for the amplification of the two genes for improved sensitivity. Employing the PCR-sequencing approach using primers derived from M. smegmatis, we confirmed the occurrence of esxA and esxB in M. smegmatis and M. fortuitum reference ATCC strains as well as field isolates, as reported previously [8,24]. We also showed the presence of the two genes in field isolates belonging to rapidly growing (RG) species, including, Mycobacterium sp. N845T, the M. septicum/M. peregrinum group and M. porcinum. It should be noted that Mycobacterium sp. N845T is not a validly published species; however, it is phylogenetically closer to RG species, as previously determined by 16S rRNA gene analysis [20]. Therefore, the amplification of both the esxA and esxB genes confirmed that some non-pathogenic RG species also harbour esxA and esxB gene orthologs.
Only esxB orthologs were detected in M. paraffinicum, M. engbaeckii and M. mageritense and only esxA orthologs were detected in Mycolicibacterium elephantis, M. brasiliensis and Mycolicibacterium flouroantheinivorans. To obtain a better understanding of RD1 region orthologs in Mycobacteriaceae other than tuberculosis, we analysed whole genome sequences of NTM, mycolicibacteria, mycolicibacter and mycobacteroides available in the public databases for the presence of genes of the RD1 region. Currently, there are more than 230 NTM, Mycolicibacterium and Mycobacteroides species combined on the List of Prokaryotic names with Standing in Nomenclature (LPSN) [27], and more than 100 genomes of these genera are available in the NCBI genome (http://ncbi.nhlm.gov/genome; accessed on 20 January 2022) and the NCBI Bio-project (http://ncbi.nhlm.gov/bio-project; accessed on 20 January 2022) databases. In this study, the RD 1 region orthologs were found to be harboured in genomes of more than 70 species. These include genomes of slow-growing (SG) pathogenic NTM, which are known to harbour RD1 region orthologs, among others M. marinum and M. kansasii; and genomes of non-pathogenic, at most opportunistic pathogenic RG spp., including, among others, M. fortuitum, M. smegmatis, Mycolicibacterium spp. JLS and Mycolicibacterium spp. MCS, as well as genomes of species not previously reported to harbour the RD 1 region [6,7,10,20]. Mycolicibacterium pulveris, Mycobacteroides chelonae, M. moriokaense, Mycolicibacterium. boenickei, Mycobacterium seoulense and Mycobacterium paraseoulense are among those genomes of species not previously reported to harbour the RD 1 region, but in this current study, RD 1 orthologs are reported. All these genomes were sequenced in other studies and submitted to the NCBI databases between the years 2013 and 2021 (http://ncbi.nhlm.gov/genome; http://ncbi.nhlm.gov/bio-project; accessed on 20 January 2022) (Table 4). The approach employed in the current study, using NCBI BLAST to investigate the presence of the RD 1 region, may not have been exhaustive of all NTM, mycolicibacteria and mycobactoides that may harbour these genes, due to the possible large sequence diversity of esxA and esxB, which were used as reference markers for the investigations.
The identification of the RD 1 region of the Esx-1 locus in more than 70 sequenced genomes of Mycobacteriaceae, including NTM, mycolicibacteria and mycobacteroides, confirms that this locus may be typical of the Mycobacteriaceae family [8,10]. The location of esxA and esxB genes next to each other in the genomes of the analysed species, as well as the presence of neighbouring gene orthologs of the RD1 region, including the recently investigated espJ as an additional marker for the immunodiagnosis of (animal) TB, calls for further investigation regarding their expression and the secretion of their protein products. Should these immunogenic protein homologs be expressed, secreted and recognised by T-cells, they may impact on the specificity of the diagnosis of (animal) TB by CMI assays that use either purified protein derivatives (PPD) or specific antigens such as ESAT-6, CFP10 and other antigens encoded in the RD 1 region, as previously experienced [28,29].
The use of 16S rRNA gene sequence analysis, as well as the analysis of other Mycobacterium housekeeping genes like rpoB, hsp65, ITS and sodA, has long been proven to be a robust tool to study phylogenetic relationships and the classification of the now-amended genus Mycobacterium, Mycolicibacterium and Mycolicibacter, Mycobacteroides and Mycolicibacillus spp. by growth rate, as well as to investigate their pathogenic potential [7,20,30,31,32,33]. Previous studies have shown that esxA and esxB sequences have the potential to be employed in phylogenetic analyses of the genera and their classification according to growth rate, i.e., as SG and RG species [8,20]. In the current study, the phylogenetic analysis of 58 Mycobacteriaceae including Mycobacterium, Mycobacteroides and Mycolicibacterium using esxA and esxB sequences grouped these into slow-growing and rapidly growing organisms. These findings are in line with results from other studies employing markers such as 16S rRNA, hsp65, ITS and sodA that demonstrated a robust phylogenetic classification of Mycobacteriaceae by growth rate [20,30,31,32,33]. Recently, core genomes were employed for the phylogenetic classification and reclassification of mycobacteria into five new genera [7]. Our analysis separated mycolicibacteria from mycobacteria. However, in contrast to the classification using core genomes, as described by Gupta et al. [7], in our study, using esxA and esxB as phylogenetic markers, Mycobacteroides chelonae could not be separated from the Mycolicibacterium clade and was closer to Mycolicibacterium phlei in both trees. The separation of RG from SG using esxA and esxB further strengthens earlier suggestions about the use of these two genes as potential phylogenetic markers for mycobacteria and now-amended mycobacteria, mycolicibacteria, mycobacteroides, mycolicibacilli and mycolicibacter [8,20].
In conclusion, we have demonstrated the presence of esxA and esxB orthologs as well as the presence of gene orthologs of the RD1 region in 70 Mycobacteriaceae including NTM, Mycolicibacterium and Mycobacteroides species’ full genomes using an in silico genome search approach. We also demonstrated the presence of these genes in species of the three genera and in Mycolicibacter engbaekii using PCR and sequencing techniques. In addition, whole genome sequence analysis provided a better understanding of the presence of esxA and esxB as well as the RD1 orthologs in Mycobacteriaceae other than tuberculosis compared to the PCR amplification. The characterization of Mycobacteriaceae for the presence as well as sequences of genes such as esxA and esxB and other genes of the esx-1 locus that play an important role in immune responses should form part of future studies describing new species. This warrants further investigations of the actual expression and secretion of these antigen homologs, as well as their role in cross-reactive immune responses. Lastly, esxA and esxB may be useful tools for the phylogenetic classification of species of Mycobacteriaceae.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12061151/s1, Figure S1: Gel electrophoresis image illustrating amplification of esxA gene fragment using M. smegmatis derived primers. Lane ‘M’ is 100 bp Molecular weight markers (O’ gene ruler from Thermofisher Scientific), Lanes 6, 7 and 8 are positive samples, Lanes 2, 3, 4, 9, 10,11,12,13,14, and 15 are negative samples, and lane PC is M. smegmatis positive control and lane NC1 and NC2 are negative controls. Figure S2: Gel electrophoresis image illustrating amplification of esxB gene fragment using M. smegmatis derived primers. Lane 1 is a positive sample, lane NC is a negative control, while lane PC is M. smegmatis positive control, and lane M is a 100 bp Molecular weight marker (100 bp O’gene Ruler from Thermo Fisher Scientific); Figure S3: Gel electrophoresis image illustrating amplification of esxA gene fragment using M. bovis derived primers. Lanes 1 and 4 are negative samples, Lane 2, is a positive sample and lane 3 is M. bovis control while lane 5 is a negative control, and lane M, a 100 bp Molecular weight marker (100 bp O’gene Ruler from Thermo Fisher Scientific); Figure S4: Gel electrophoresis image illustrating amplification of esxB gene fragment using M. bovis derived primers. Lanes 1 is M. bovis control, Lane 2, is a negative sample and lane 3 is a positive sample while lane 4 is a negative control, and lane M, a 100 bp Molecular weight marker (100 bp O’gene Ruler from Thermo Fisher Scientific).
Author Contributions
Conceptualization, N.G.; Formal analysis, T.M.H., A.M. and V.P.M.G.R.; Funding acquisition, A.M. and V.P.M.G.R.; Investigation, N.G., T.M.H., A.B., A.M. and V.P.M.G.R.; Methodology, N.G., T.M.H., A.B., A.M. and V.P.M.G.R.; Writing—original draft, N.G. and A.B.; Writing—review and editing, N.G., T.M.H., A.M. and V.P.M.G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by WOTRO Science for Global Development; grant number W01.65.321.00, and we received no funding for APC publication fees.
Data Availability Statement
In this study, we used publicly available data which were accessed from NCBI (https://www.ncbi.nlm.nih.gov/), Smegmlist (https://mycobrowser.epfl.ch/genes/) and Bovilist (http://genolist.pasteur.fr/BoviList/genome.cgi?; accessed on 20 January 2022) databases. All other data generated or analysed in the current study are available upon request.
Acknowledgments
We would like to thank I-Chang Lee of the ARC-OVI for assisting with the PCR work. We would also like to thank Ernest Ngoepe of the ARC-OVI for critically reviewing this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hlokwe, T.M.; Van Helden, P.; Michel, A.L. Evidence of increasing intra and inter-species transmission of Mycobacterium bovis in South Africa: Are we losing the battle? Prev. Vet. Med. 2014, 115, 10–17. [Google Scholar] [CrossRef]
- Pallen, M.J. The ESAT-6/WXG100 superfamily–and a new Gram-positive secretion system? Trends Microbiol. 2002, 10, 209–212. [Google Scholar] [CrossRef]
- Moens, C.; Filée, P.; Boes, A.; Alie, C.; Dufrasne, F.; André, E.; Marché, S.; Fretin, D. Identification of New Mycobacterium bovis antigens and development of a multiplexed serological bead-immunoassay for the diagnosis of bovine tuberculosis in cattle. PLoS ONE 2023, 18, e0292590. [Google Scholar] [CrossRef]
- Brodin, P.; de Jonge, M.I.; Majlessi, L.; Leclerc, C.; Nilges, M.; Cole, S.T.; Brosch, R. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J. Biol. Chem. 2005, 280, 33953–33959. [Google Scholar] [CrossRef]
- Ganguly, N.; Siddiqui, I.; Sharma, P. Role of M. tuberculosis RD-1 region encoded secretory proteins in protective response and virulence. Tuberculosis 2008, 88, 510–517. [Google Scholar] [CrossRef]
- Vordermeier, H.M.; Brown, J.; Cockle, P.J.; Franken, W.P.; Arend, S.M.; Ottenhoff, T.H.; Jahans, K.; Hewinson, R.G. Assessment of cross-reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-cell epitope level. Clin. Vaccine Immunol. 2007, 14, 1203–1209. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Gey van Pittius, N.C.; Gamieldien, J.; Hide, W.; Brown, G.D.; Siezen, R.J.; Beyers, A.D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+ C Gram-positive bacteria. Genome Biol. 2001, 2, research0044.1. [Google Scholar] [CrossRef]
- van Ingen, J.; de Zwaan, R.; Dekhuijzen, R.; Boeree, M.; van Soolingen, D. Region of difference 1 in nontuberculous Mycobacterium species adds a phylogenetic and taxonomical character. J. Bacteriol. 2009, 191, 5865–5867. [Google Scholar] [CrossRef]
- Colangeli, R.; Spencer, J.S.; Bifani, P.; Williams, A.; Lyashchenko, K.; Keen, M.A.; Hill, P.J.; Belisle, J.; Gennaro, M.L. MTSA-10, the product of the Rv3874 gene of Mycobacterium tuberculosis, elicits tuberculosis-specific, delayed-type hypersensitivity in guinea pigs. Infect. Immun. 2000, 68, 990–993. [Google Scholar] [CrossRef][Green Version]
- Gcebe, N.; Michel, A.; Gey van Pittius, N.C.; Rutten, V. Comparative genomics and proteomic analysis of four non-tuberculous Mycobacterium species and Mycobacterium tuberculosis complex: Occurrence of shared immunogenic proteins. Front. Microbiol. 2016, 7, 160883. [Google Scholar] [CrossRef]
- Geluk, A.; Van Meijgaarden, K.E.; Franken, K.L.; Wieles, B.; Arend, S.M.; Faber, W.R.; Naafs, B.; Ottenhoff, T.H. Immunological crossreactivity of the Mycobacterium leprae CFP-10 with its homologue in Mycobacterium tuberculosis. Scand. J. Immunol. 2004, 59, 66–70. [Google Scholar] [CrossRef]
- Geluk, A.; van Meijgaarden, K.E.; Franken, K.L.; Subronto, Y.W.; Wieles, B.; Arend, S.M.; Sampaio, E.P.; de Boer, T.; Faber, W.R.; Naafs, B.; et al. Identification and characterization of the ESAT-6 homologue of Mycobacterium leprae and T-cell cross-reactivity with Mycobacterium tuberculosis. Infect. Immun. 2002, 70, 2544–2548. [Google Scholar] [CrossRef]
- Ruhwald, M.; de Thurah, L.; Kuchaka, D.; Zaher, M.R.; Salman, A.M.; Abdel-Ghaffar, A.R.; Shoukry, F.A.; Michelsen, S.W.; Soborg, B.; Blauenfeldt, T.; et al. Introducing the ESAT-6 free IGRA, a companion diagnostic for TB vaccines based on ESAT-6. Sci. Rep. 2017, 7, 45969. [Google Scholar] [CrossRef]
- Eirin, M.E.; Macias, A.; Magnano, G.; Morsella, C.; Mendez, L.; Blanco, F.C.; Bianco, M.V.; Severina, W.; Alito, A.; de los Angeles Pando, M.; et al. Identification and evaluation of new Mycobacterium bovis antigens in the in vitro interferon gamma release assay for bovine tuberculosis diagnosis. Tuberculosis 2015, 95, 795–801. [Google Scholar] [CrossRef]
- Schiller, I.; Vordermeier, H.M.; Waters, W.R.; Palmer, M.; Thacker, T.; Whelan, A.; Hardegger, R.; Marg-Haufe, B.; Raeber, A.; Oesch, B. Assessment of Mycobacterium tuberculosis OmpATb as a novel antigen for the diagnosis of bovine tuberculosis. Clin. Vaccine Immunol. 2009, 16, 1314–1321. [Google Scholar] [CrossRef]
- Gcebe, N.; Rutten, V.P.; van Pittius, N.G.; Naicker, B.; Michel, A.L. Mycobacterium komaniense sp. nov., a rapidly growing non-tuberculous Mycobacterium species detected in South Africa. Int. J. Syst. Evol. Microbiol. 2018, 68, 1526–1532. [Google Scholar] [CrossRef]
- Lyashchenko, K.P.; Sridhara, A.A.; Johnathan-Lee, A.; Sikar-Gang, A.; Lambotte, P.; Esfandiari, J.; Bernitz, N.; Kerr, T.J.; Miller, M.A.; Waters, W.R. Differential antigen recognition by serum antibodies from three bovid hosts of Mycobacterium bovis infection. Comp. Immunol. Microbiol. Infect. Dis. 2020, 69, 101424. [Google Scholar] [CrossRef]
- Gcebe, N.; Rutten, V.; Gey van Pittius, N.C.; Michel, A. Prevalence and Distribution of Non-Tuberculous Mycobacteria (NTM) in Cattle, African Buffaloes (Syncerus caffer) and their Environments in South Africa. Transbound. Emerg. Dis. 2013, 60, 74–84. [Google Scholar] [CrossRef]
- Gcebe, N.; Hlokwe, T.M. Non-tuberculous mycobacteria in South African wildlife: Neglected pathogens and potential impediments for bovine tuberculosis diagnosis. Front. Cell. Infect. Microbiol. 2017, 7, 15. [Google Scholar] [CrossRef]
- Gcebe, N.; Michel, A.L.; Hlokwe, T.M. Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa. BMC Microbiol. 2018, 18, 32. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Scherrer, S.; Landolt, P.; Friedel, U.; Stephan, R. Distribution and expression of esat-6 and cfp-10 in non-tuberculous mycobacteria isolated from lymph nodes of slaughtered cattle in Switzerland. J. Vet. Diagn. Investig. 2019, 31, 217–221. [Google Scholar] [CrossRef]
- Arend, S.M.; de Haas, P.; Leyten, E.; Rosenkrands, I.; Rigouts, L.; Andersen, P.; Mijs, W.; van Dissel, J.T.; van Soolingen, D. ESAT-6 and CFP-10 in clinical versus environmental isolates of Mycobacterium kansasii. J. Infect. Dis. 2005, 191, 1301–1310. [Google Scholar] [CrossRef][Green Version]
- Hughes, M.S.; Ball, N.W.; McCarroll, J.; Erskine, M.; Taylor, M.J.; Pollock, J.M.; Skuce, R.A.; Neill, S.D. Molecular analyses of mycobacteria other than the M. tuberculosis complex isolated from Northern Ireland cattle. Vet. Microbiol. 2005, 108, 101–112. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607. [Google Scholar] [CrossRef]
- Jenkins, A.O.; Gormley, E.; Gcebe, N.; Fosgate, G.T.; Conan, A.; Aagaard, C.; Michel, A.L.; Rutten, V.P. Cross reactive immune responses in cattle arising from exposure to Mycobacterium bovis and non-tuberculous mycobacteria. Prev. Vet. Med. 2018, 152, 16–22. [Google Scholar] [CrossRef]
- Michel, A.L. Mycobacterium fortuitum infection interference with Mycobacterium bovis diagnostics: Natural infection cases and a pilot experimental infection. J. Vet. Diagn. Investig. 2008, 20, 501–503. [Google Scholar] [CrossRef]
- Drancourt, M.; Raoult, D. Sequence-based identification of new bacteria: A proposition for creation of an orphan bacterium repository. J. Clin. Microbiol. 2005, 43, 4311–4315. [Google Scholar] [CrossRef][Green Version]
- Kim, H.; Kim, S.H.; Shim, T.S.; Kim, M.N.; Bai, G.H.; Park, Y.G.; Lee, S.H.; Chae, G.T.; Cha, C.Y.; Kook, Y.H.; et al. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 2005, 55, 1649–1656. [Google Scholar] [CrossRef]
- Roth, A.; Fischer, M.; Hamid, M.E.; Michalke, S.; Ludwig, W.; Mauch, H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 1998, 36, 139–147. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).