Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives
Abstract
1. Introduction
2. A Gut Feeling: The Microbiome as a Complementary Brain
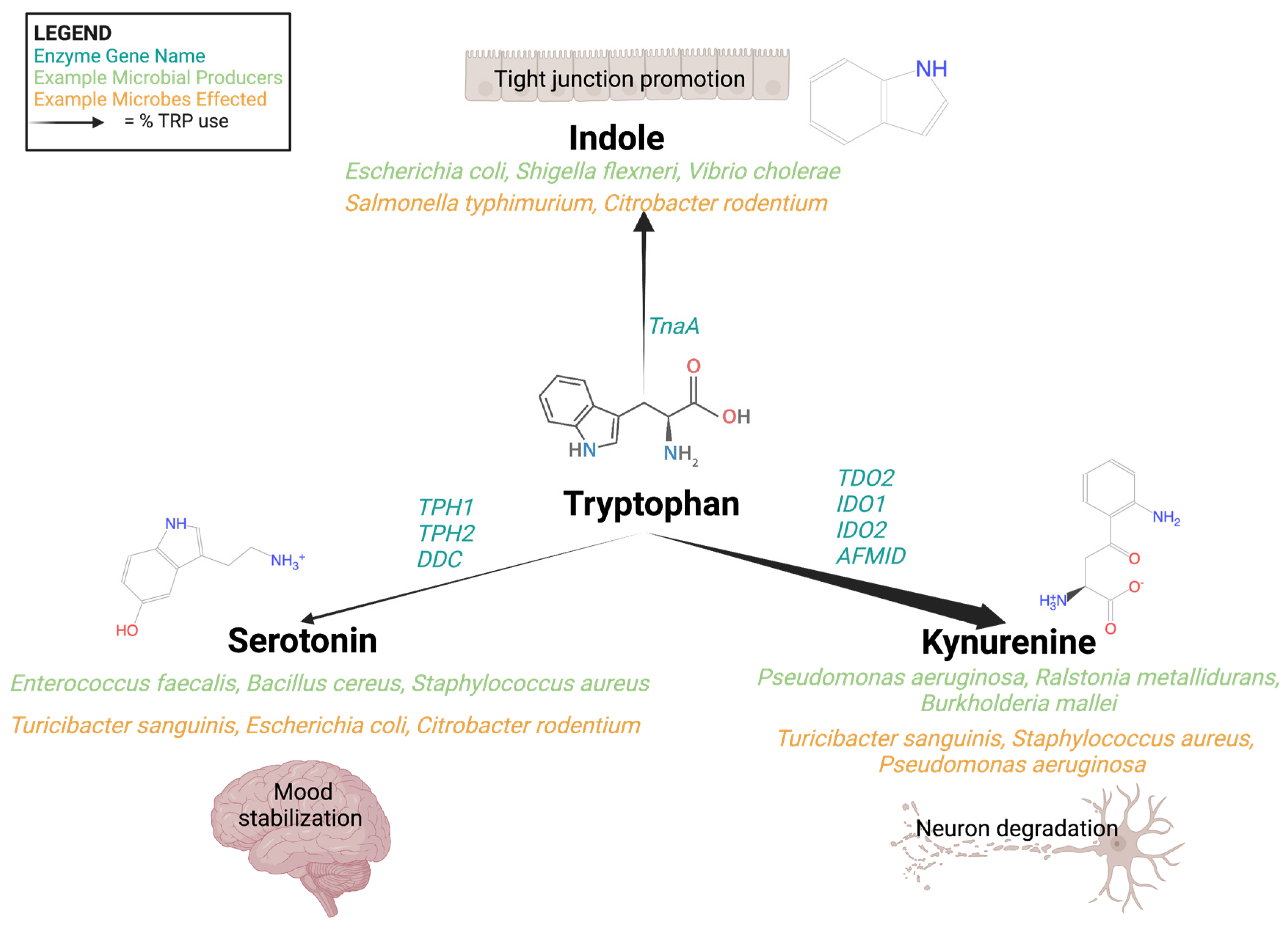
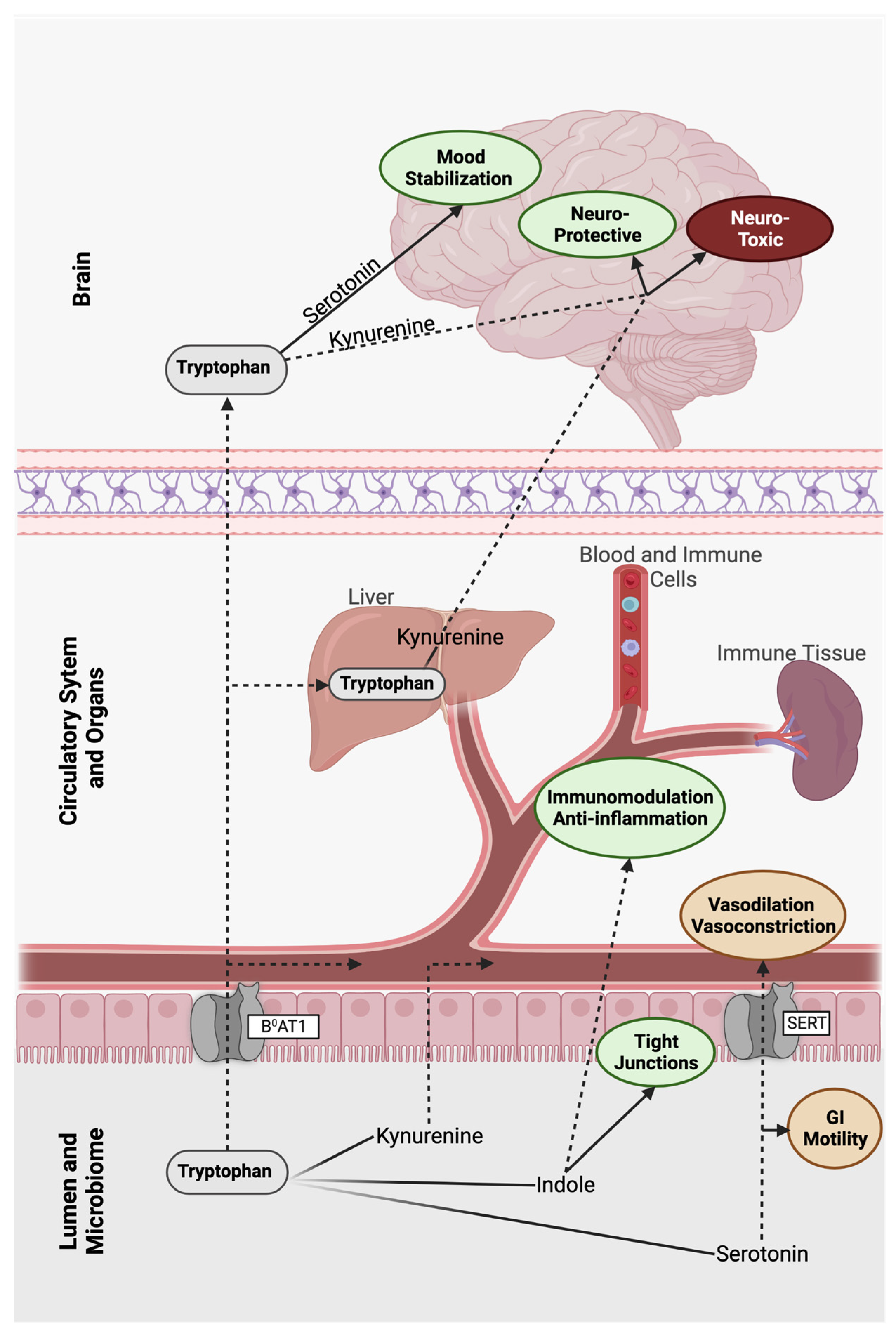
3. The Tryptophan Pathway: Microbial-Derived Metabolic Mind Control
3.1. Indole
3.2. Serotonin
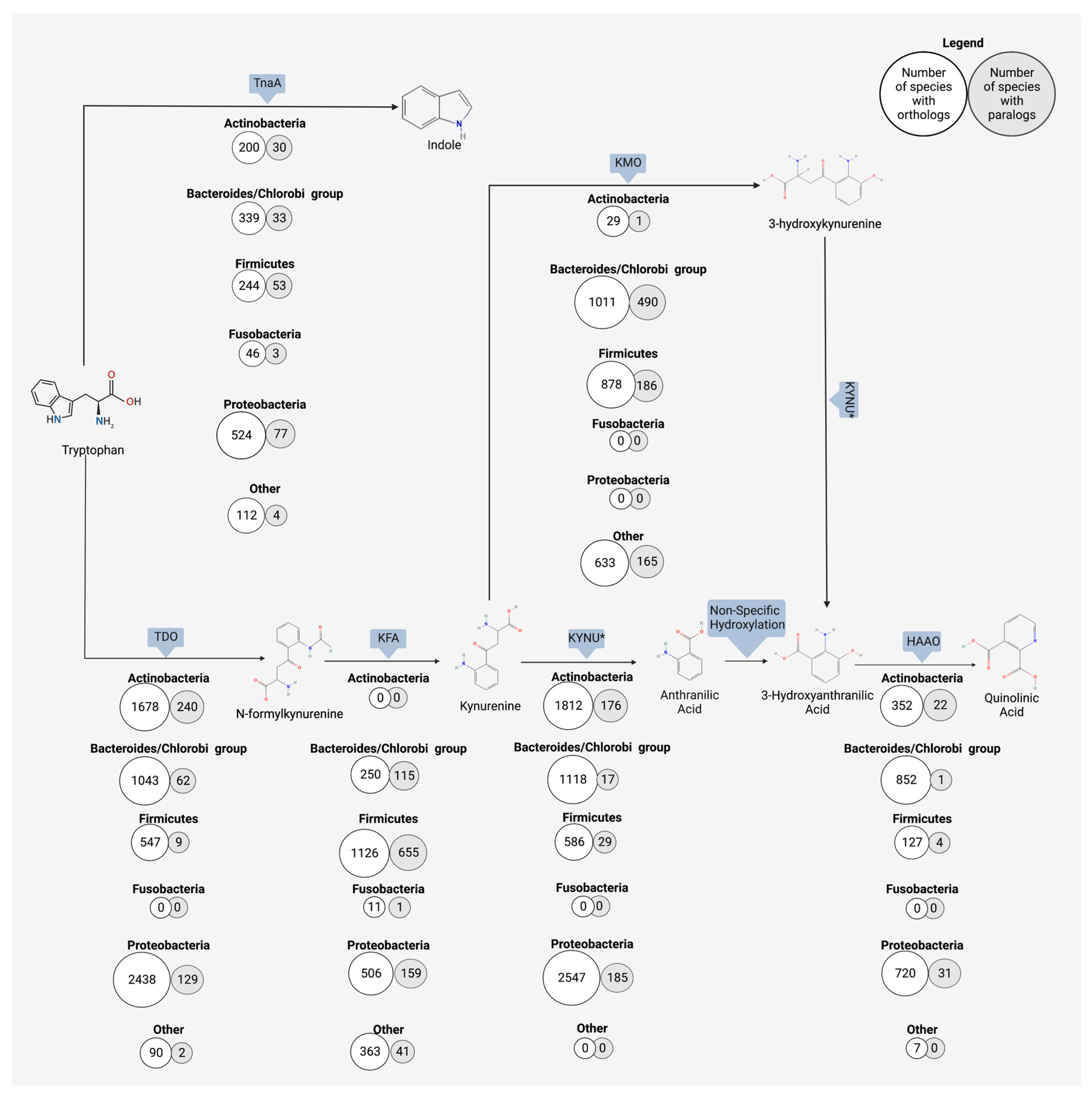
3.3. Kynurenine
4. Anthranilic Acid and Beyond
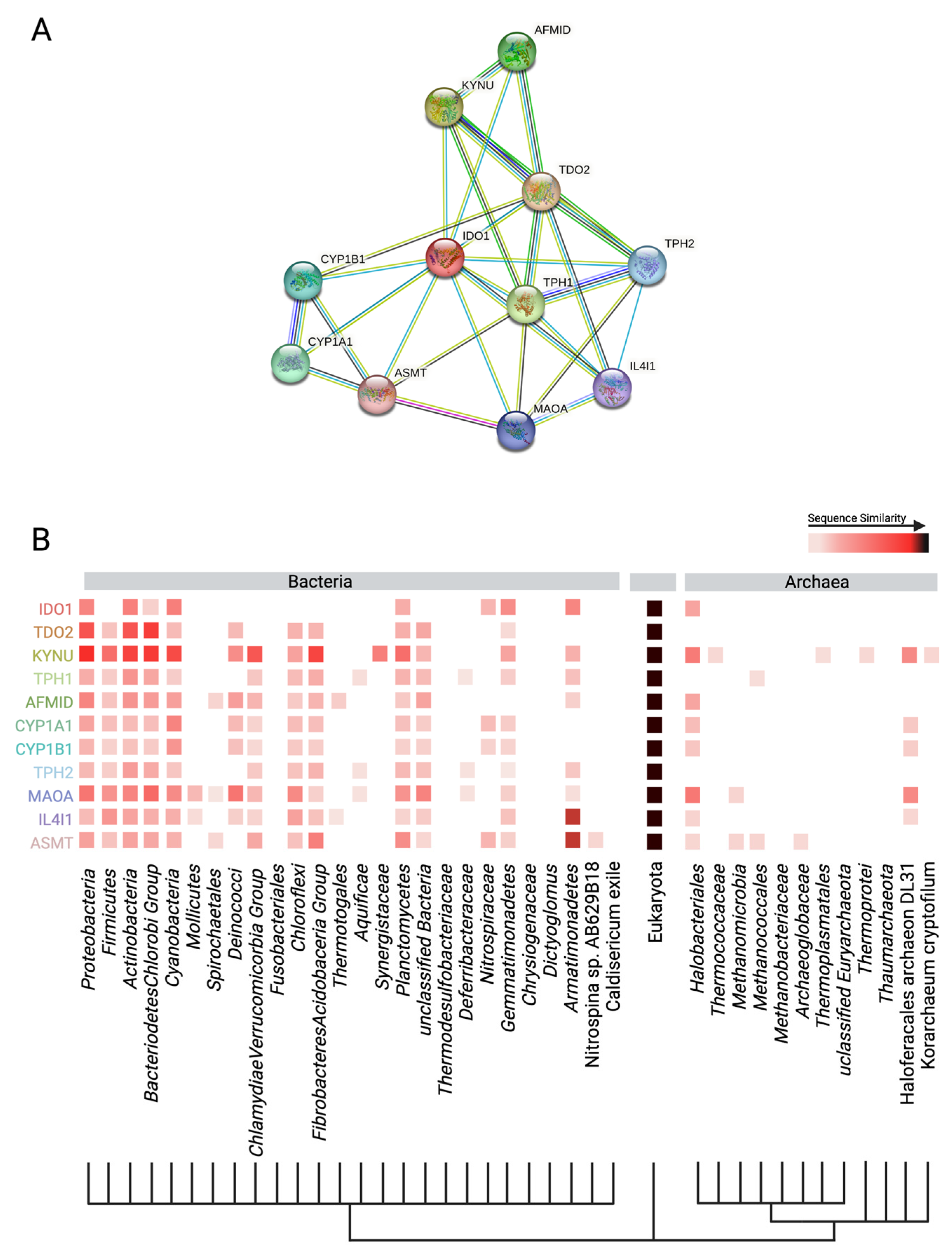
4.1. Human Production of Anthranilic Acid and Derivatives
4.2. Bacterial Production of Anthranilic Acid and Derivatives
4.3. Neurological Outcomes of Anthranilic Acid
5. Balancing the Microbes and Metabolites
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 4505. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Mande, S.S. Host-microbiome interactions: Gut-Liver axis and its connection with other organs. NPJ Biofilms Microbiomes 2022, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Reparaz, J.; Kasper, L.H. The Second Brain: Is the Gut Microbiota a Link between Obesity and Central Nervous System Disorders? Curr. Obes. Rep. 2016, 5, 51–64. [Google Scholar] [CrossRef]
- Kastl, A.J., Jr.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Morowitz, M.J.; Carlisle, E.M.; Alverdy, J.C. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surg. Clin. N. Am. 2011, 91, 771–785. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Peng, L.; He, Z.; Chen, W.; Holzman, I.R.; Lin, J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 2007, 61, 37–41. [Google Scholar] [CrossRef]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Kim, C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef]
- Neis, E.P.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Allison, C.; Gibson, S.A.; Cummings, J.H. Contribution of the microflora to proteolysis in the human large intestine. J. Appl. Bacteriol. 1988, 64, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Rogan, C.J.; Pang, Y.Y.; Davis, E.W., 2nd; Anderson, J.C. Ancient co-option of an amino acid ABC transporter locus in Pseudomonas syringae for host signal-dependent virulence gene regulation. PLoS Pathog. 2020, 16, e1008680. [Google Scholar] [CrossRef]
- Ma, J.; Zhu, Y.; Wang, Z.; Yu, X.; Hu, R.; Wang, X.; Cao, G.; Zou, H.; Shah, A.M.; Peng, Q.; et al. Glutamine supplementation affected the gut bacterial community and fermentation leading to improved nutrient digestibility in growth-retarded yaks. FEMS Microbiol. Ecol. 2021, 97, fiab084. [Google Scholar] [CrossRef]
- Roth, W.; Zadeh, K.; Vekariya, R.; Ge, Y.; Mohamadzadeh, M. Tryptophan Metabolism and Gut-Brain Homeostasis. Int. J. Mol. Sci. 2021, 22, 2973. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Mu, C.L.; Farzi, A.; Zhu, W.Y. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef]
- Kumar, A.; Sperandio, V. Indole Signaling at the Host-Microbiota-Pathogen Interface. mBio 2019, 10, e01031-19. [Google Scholar] [CrossRef]
- Boon, N.; Kaur, M.; Aziz, A.; Bradnick, M.; Shibayama, K.; Eguchi, Y.; Lund, P.A. The Signaling Molecule Indole Inhibits Induction of the AR2 Acid Resistance System in Escherichia coli. Front. Microbiol. 2020, 11, 474. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Brew, B.J. Implications of the kynurenine pathway and quinolinic acid in Alzheimer’s disease. Redox Rep. 2002, 7, 199–206. [Google Scholar] [CrossRef]
- Heyes, M.P.; Saito, K.; Crowley, J.S.; Davis, L.E.; Demitrack, M.A.; Der, M.; Dilling, L.A.; Elia, J.; Kruesi, M.J.; Lackner, A.; et al. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain 1992, 115 Pt 5, 1249–1273. [Google Scholar] [CrossRef]
- Pappolla, M.A.; Perry, G.; Fang, X.; Zagorski, M.; Sambamurti, K.; Poeggeler, B. Indoles as essential mediators in the gut-brain axis. Their role in Alzheimer’s disease. Neurobiol. Dis. 2021, 156, 105403. [Google Scholar] [CrossRef]
- Ting, K.K.; Brew, B.J.; Guillemin, G.J. Effect of quinolinic acid on human astrocytes morphology and functions: Implications in Alzheimer’s disease. J. Neuroinflamm. 2009, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Wiedlocha, M.; Marcinowicz, P.; Janoska-Jazdzik, M.; Szulc, A. Gut microbiota, kynurenine pathway and mental disorders—Review. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 106, 110145. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Ma, Z.S.; Li, W.; Shi, P. Microbiome-host-phylogeny relationships in animal gastrointestinal tract microbiomes. FEMS Microbiol. Ecol. 2022, 98, fiac021. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Longhi, G.; Tarracchini, C.; Mancabelli, L.; Lugli, G.A.; Alessandri, G.; Turroni, F.; Milani, C.; Ventura, M. The human gut microbiome of athletes: Metagenomic and metabolic insights. Microbiome 2023, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Morgan, X.C.; Segata, N.; Waldron, L.; Reyes, J.; Earl, A.M.; Giannoukos, G.; Boylan, M.R.; Ciulla, D.; Gevers, D.; et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA 2014, 111, E2329–E2338. [Google Scholar] [CrossRef] [PubMed]
- Madi, N.; Chen, D.; Wolff, R.; Shapiro, B.J.; Garud, N.R. Community diversity is associated with intra-species genetic diversity and gene loss in the human gut microbiome. eLife 2023, 12, e78530. [Google Scholar] [CrossRef]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 2017, 5, 98. [Google Scholar] [CrossRef]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Braidy, N.; Guillemin, G.J.; Mansour, H.; Chan-Ling, T.; Grant, R. Changes in kynurenine pathway metabolism in the brain, liver and kidney of aged female Wistar rats. FEBS J. 2011, 278, 4425–4434. [Google Scholar] [CrossRef]
- Gourbeyre, P.; Desbuards, N.; Gremy, G.; Tranquet, O.; Champ, M.; Denery-Papini, S.; Bodinier, M. Perinatal and postweaning exposure to galactooligosaccharides/inulin prebiotics induced biomarkers linked to tolerance mechanism in a mouse model of strong allergic sensitization. J. Agric. Food Chem. 2013, 61, 6311–6320. [Google Scholar] [CrossRef]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef]
- Kountouras, J.; Chatzopoulos, D.; Zavos, C. Reactive oxygen metabolites and upper gastrointestinal diseases. Hepatogastroenterology 2001, 48, 743–751. [Google Scholar]
- Xu, M.; Cen, M.; Shen, Y.; Zhu, Y.; Cheng, F.; Tang, L.; Hu, W.; Dai, N. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig. Dis. Sci. 2021, 66, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Genestet, C.; Le Gouellec, A.; Chaker, H.; Polack, B.; Guery, B.; Toussaint, B.; Stasia, M.J. Scavenging of reactive oxygen species by tryptophan metabolites helps Pseudomonas aeruginosa escape neutrophil killing. Free Radic. Biol. Med. 2014, 73, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Kurnasov, O.; Jablonski, L.; Polanuyer, B.; Dorrestein, P.; Begley, T.; Osterman, A. Aerobic tryptophan degradation pathway in bacteria: Novel kynurenine formamidase. FEMS Microbiol. Lett. 2003, 227, 219–227. [Google Scholar] [CrossRef]
- Ulvik, A.; Theofylaktopoulou, D.; Midttun, O.; Nygard, O.; Eussen, S.J.; Ueland, P.M. Substrate product ratios of enzymes in the kynurenine pathway measured in plasma as indicators of functional vitamin B-6 status. Am. J. Clin. Nutr. 2013, 98, 934–940. [Google Scholar] [CrossRef]
- Tennoune, N.; Andriamihaja, M.; Blachier, F. Production of Indole and Indole-Related Compounds by the Intestinal Microbiota and Consequences for the Host: The Good, the Bad, and the Ugly. Microorganisms 2022, 10, 930. [Google Scholar] [CrossRef]
- Nelp, M.T.; Kates, P.A.; Hunt, J.T.; Newitt, J.A.; Balog, A.; Maley, D.; Zhu, X.; Abell, L.; Allentoff, A.; Borzilleri, R.; et al. Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form. Proc. Natl. Acad. Sci. USA 2018, 115, 3249–3254. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, S.; Nunes-Costa, D.; Cardoso, S.M.; Empadinhas, N.; Marugg, J.D. Enzyme Promiscuity in Serotonin Biosynthesis, From Bacteria to Plants and Humans. Front. Microbiol. 2022, 13, 873555. [Google Scholar] [CrossRef]
- Zhu, J.; Hua, X.; Yang, T.; Guo, M.; Li, Q.; Xiao, L.; Li, L.; Chen, J.; Li, T. Alterations in Gut Vitamin and Amino Acid Metabolism are Associated with Symptoms and Neurodevelopment in Children with Autism Spectrum Disorder. J. Autism Dev. Disord. 2022, 52, 3116–3128. [Google Scholar] [CrossRef]
- Bortolotti, P.; Hennart, B.; Thieffry, C.; Jausions, G.; Faure, E.; Grandjean, T.; Thepaut, M.; Dessein, R.; Allorge, D.; Guery, B.P.; et al. Tryptophan catabolism in Pseudomonas aeruginosa and potential for inter-kingdom relationship. BMC Microbiol. 2016, 16, 137. [Google Scholar] [CrossRef]
- Chobot, V.; Hadacek, F.; Weckwerth, W.; Kubicova, L. Iron chelation and redox chemistry of anthranilic acid and 3-hydroxyanthranilic acid: A comparison of two structurally related kynurenine pathway metabolites to obtain improved insights into their potential role in neurological disease development. J. Organomet. Chem. 2015, 782, 103–110. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Dunham, R.M.; Iwai, S.; Maher, M.C.; Albright, R.G.; Broadhurst, M.J.; Hernandez, R.D.; Lederman, M.M.; Huang, Y.; Somsouk, M.; et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013, 5, 193ra191. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.S. Structure and mechanism of kynureninase. Arch. Biochem. Biophys. 2014, 544, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kuc, D.; Zgrajka, W.; Parada-Turska, J.; Urbanik-Sypniewska, T.; Turski, W.A. Micromolar concentration of kynurenic acid in rat small intestine. Amino Acids 2008, 35, 503–505. [Google Scholar] [CrossRef] [PubMed]
- Kurnasov, O.; Goral, V.; Colabroy, K.; Gerdes, S.; Anantha, S.; Osterman, A.; Begley, T.P. NAD biosynthesis: Identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 2003, 10, 1195–1204. [Google Scholar] [CrossRef]
- Darlington, L.G.; Forrest, C.M.; Mackay, G.M.; Smith, R.A.; Smith, A.J.; Stoy, N.; Stone, T.W. On the Biological Importance of the 3-hydroxyanthranilic Acid: Anthranilic Acid Ratio. Int. J. Tryptophan Res. 2010, 3, 51–59. [Google Scholar] [CrossRef]
- Chirino, B.; Strahsburger, E.; Agullo, L.; Gonzalez, M.; Seeger, M. Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS ONE 2013, 8, e75746. [Google Scholar] [CrossRef]
- Zhang, P.; Jin, T.; Kumar Sahu, S.; Xu, J.; Shi, Q.; Liu, H.; Wang, Y. The Distribution of Tryptophan-Dependent Indole-3-Acetic Acid Synthesis Pathways in Bacteria Unraveled by Large-Scale Genomic Analysis. Molecules 2019, 24, 1411. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Jin, U.H.; Lee, S.O.; Sridharan, G.; Lee, K.; Davidson, L.A.; Jayaraman, A.; Chapkin, R.S.; Alaniz, R.; Safe, S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol. Pharmacol. 2014, 85, 777–788. [Google Scholar] [CrossRef]
- Miyagi, M.; Wilson, R.; Saigusa, D.; Umeda, K.; Saijo, R.; Hager, C.L.; Li, Y.; McCormick, T.; Ghannoum, M.A. Indole-3-acetic acid synthesized through the indole-3-pyruvate pathway promotes Candida tropicalis biofilm formation. PLoS ONE 2020, 15, e0244246. [Google Scholar] [CrossRef]
- Li, Y.; Xu, W.; Zhang, F.; Zhong, S.; Sun, Y.; Huo, J.; Zhu, J.; Wu, C. The Gut Microbiota-Produced Indole-3-Propionic Acid Confers the Antihyperlipidemic Effect of Mulberry-Derived 1-Deoxynojirimycin. mSystems 2020, 5, e00313-20. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.L.; Eckstrom, K.; Lile, K.H.; Caldwell, S.; Heney, E.R.; Lahue, K.G.; D’Alessandro, A.; Wargo, M.J.; Krementsov, D.N. Lactobacillus reuteri tryptophan metabolism promotes host susceptibility to CNS autoimmunity. Microbiome 2022, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Konopelski, P.; Mogilnicka, I. Biological Effects of Indole-3-Propionic Acid, a Gut Microbiota-Derived Metabolite, and Its Precursor Tryptophan in Mammals’ Health and Disease. Int. J. Mol. Sci. 2022, 23, 1222. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Rothrock, M.J., Jr.; Loughrin, J.H.; Doerner, K.C. Characterization of skatole-producing microbial populations in enriched swine lagoon slurry. FEMS Microbiol. Ecol. 2007, 60, 329–340. [Google Scholar] [CrossRef]
- Song, L.; He, M.; Sun, Q.; Wang, Y.; Zhang, J.; Fang, Y.; Liu, S.; Duan, L. Roseburia hominis Increases Intestinal Melatonin Level by Activating p-CREB-AANAT Pathway. Nutrients 2021, 14, 117. [Google Scholar] [CrossRef]
- Xie, X.; Ding, D.; Bai, D.; Zhu, Y.; Sun, W.; Sun, Y.; Zhang, D. Melatonin biosynthesis pathways in nature and its production in engineered microorganisms. Synth. Syst. Biotechnol. 2022, 7, 544–553. [Google Scholar] [CrossRef]
- Broekhuizen, M.; Danser, A.H.J.; Reiss, I.K.M.; Merkus, D. The Function of the Kynurenine Pathway in the Placenta: A Novel Pharmacotherapeutic Target? Int. J. Environ. Res. Public Health 2021, 18, 11545. [Google Scholar] [CrossRef]
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut microbes and the brain: Paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef]
- Ye, L.; Bae, M.; Cassilly, C.D.; Jabba, S.V.; Thorpe, D.W.; Martin, A.M.; Lu, H.Y.; Wang, J.; Thompson, J.D.; Lickwar, C.R.; et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe 2021, 29, 179–196.e9. [Google Scholar] [CrossRef]
- Yu, C.D.; Xu, Q.J.; Chang, R.B. Vagal sensory neurons and gut-brain signaling. Curr. Opin. Neurobiol. 2020, 62, 133–140. [Google Scholar] [CrossRef]
- Prescott, S.L.; Liberles, S.D. Internal senses of the vagus nerve. Neuron 2022, 110, 579–599. [Google Scholar] [CrossRef]
- Kaelberer, M.M.; Buchanan, K.L.; Klein, M.E.; Barth, B.B.; Montoya, M.M.; Shen, X.; Bohorquez, D.V. A gut-brain neural circuit for nutrient sensory transduction. Science 2018, 361, eaat5236. [Google Scholar] [CrossRef]
- Jacobson, A.; Yang, D.; Vella, M.; Chiu, I.M. The intestinal neuro-immune axis: Crosstalk between neurons, immune cells, and microbes. Mucosal Immunol. 2021, 14, 555–565. [Google Scholar] [CrossRef]
- Walker, J.R.; Ediger, J.P.; Graff, L.A.; Greenfeld, J.M.; Clara, I.; Lix, L.; Rawsthorne, P.; Miller, N.; Rogala, L.; McPhail, C.M.; et al. The Manitoba IBD cohort study: A population-based study of the prevalence of lifetime and 12-month anxiety and mood disorders. Am. J. Gastroenterol. 2008, 103, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N. Psychological Stress and Depression: Risk Factors for IBD? Dig. Dis. 2016, 34, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Limbana, T.; Khan, F.; Eskander, N. Gut Microbiome and Depression: How Microbes Affect the Way We Think. Cureus 2020, 12, e9966. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Martin, P.; Becker, J.A.J.; Caramello, N.; Fernandez, S.P.; Costa-Campos, R.; Canaguier, J.; Barbosa, S.; Martinez-Gili, L.; Myridakis, A.; Dumas, M.E.; et al. The microbial metabolite p-Cresol induces autistic-like behaviors in mice by remodeling the gut microbiota. Microbiome 2021, 9, 157. [Google Scholar] [CrossRef]
- Zheng, Y.; Prince, N.Z.; Peralta Marzal, L.N.; Ahmed, S.; Garssen, J.; Perez Pardo, P.; Kraneveld, A.D. The Autism Spectrum Disorder-Associated Bacterial Metabolite p-Cresol Derails the Neuroimmune Response of Microglial Cells Partially via Reduction of ADAM17 and ADAM10. Int. J. Mol. Sci. 2022, 23, 11013. [Google Scholar] [CrossRef]
- Pascucci, T.; Colamartino, M.; Fiori, E.; Sacco, R.; Coviello, A.; Ventura, R.; Puglisi-Allegra, S.; Turriziani, L.; Persico, A.M. P-cresol Alters Brain Dopamine Metabolism and Exacerbates Autism-Like Behaviors in the BTBR Mouse. Brain Sci. 2020, 10, 233. [Google Scholar] [CrossRef]
- Guzman-Salas, S.; Weber, A.; Malci, A.; Lin, X.; Herrera-Molina, R.; Cerpa, W.; Dorador, C.; Signorelli, J.; Zamorano, P. The metabolite p-cresol impairs dendritic development, synaptogenesis, and synapse function in hippocampal neurons: Implications for autism spectrum disorder. J. Neurochem. 2022, 161, 335–349. [Google Scholar] [CrossRef]
- Altieri, L.; Neri, C.; Sacco, R.; Curatolo, P.; Benvenuto, A.; Muratori, F.; Santocchi, E.; Bravaccio, C.; Lenti, C.; Saccani, M.; et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers 2011, 16, 252–260. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Cerullo, S.; Neri, C.; Urbani, A.; Tripi, G.; Malvy, J.; Barthelemy, C.; Bonnet-Brihault, F.; Persico, A.M. Urinary p-cresol is elevated in young French children with autism spectrum disorder: A replication study. Biomarkers 2014, 19, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Bosi, A.; Banfi, D.; Bistoletti, M.; Giaroni, C.; Baj, A. Tryptophan Metabolites Along the Microbiota-Gut-Brain Axis: An Interkingdom Communication System Influencing the Gut in Health and Disease. Int. J. Tryptophan Res. 2020, 13, 1178646920928984. [Google Scholar] [CrossRef] [PubMed]
- Karayol, R.; Medrihan, L.; Warner-Schmidt, J.L.; Fait, B.W.; Rao, M.N.; Holzner, E.B.; Greengard, P.; Heintz, N.; Schmidt, E.F. Serotonin receptor 4 in the hippocampus modulates mood and anxiety. Mol. Psychiatry 2021, 26, 2334–2349. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Ghia, J.E.; Khan, W.I. Serotonin in the gut: Blessing or a curse. Biochimie 2019, 161, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Toth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef]
- Banerjee, A.; Herring, C.A.; Chen, B.; Kim, H.; Simmons, A.J.; Southard-Smith, A.N.; Allaman, M.M.; White, J.R.; Macedonia, M.C.; McKinley, E.T.; et al. Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 2020, 159, 2101–2115.e5. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016, 24, 151–157. [Google Scholar] [CrossRef]
- Kawasoe, J.; Uchida, Y.; Kawamoto, H.; Miyauchi, T.; Watanabe, T.; Saga, K.; Tanaka, K.; Ueda, S.; Terajima, H.; Taura, K.; et al. Propionic Acid, Induced in Gut by an Inulin Diet, Suppresses Inflammation and Ameliorates Liver Ischemia and Reperfusion Injury in Mice. Front. Immunol. 2022, 13, 862503. [Google Scholar] [CrossRef]
- Yoshida, H.; Ishii, M.; Akagawa, M. Propionate suppresses hepatic gluconeogenesis via GPR43/AMPK signaling pathway. Arch. Biochem. Biophys. 2019, 672, 108057. [Google Scholar] [CrossRef]
- Bellono, N.W.; Bayrer, J.R.; Leitch, D.B.; Castro, J.; Zhang, C.; O’Donnell, T.A.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell 2017, 170, 185–198.e16. [Google Scholar] [CrossRef]
- Ashniev, G.A.; Petrov, S.N.; Iablokov, S.N.; Rodionov, D.A. Genomics-Based Reconstruction and Predictive Profiling of Amino Acid Biosynthesis in the Human Gut Microbiome. Microorganisms 2022, 10, 740. [Google Scholar] [CrossRef]
- Romeo, E.; Dave, M.H.; Bacic, D.; Ristic, Z.; Camargo, S.M.; Loffing, J.; Wagner, C.A.; Verrey, F. Luminal kidney and intestine SLC6 amino acid transporters of B0AT-cluster and their tissue distribution in Mus musculus. Am. J. Physiol. Renal Physiol. 2006, 290, F376–F383. [Google Scholar] [CrossRef] [PubMed]
- Pramod, A.B.; Foster, J.; Carvelli, L.; Henry, L.K. SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Aspects Med. 2013, 34, 197–219. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, N.; Ge, Y.; Yang, Y.; Ren, F.; Wu, Z. Tryptophan and the innate intestinal immunity: Crosstalk between metabolites, host innate immune cells, and microbiota. Eur. J. Immunol. 2022, 52, 856–868. [Google Scholar] [CrossRef]
- Pardridge, W.M.; Fierer, G. Transport of tryptophan into brain from the circulating, albumin-bound pool in rats and in rabbits. J. Neurochem. 1990, 54, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Chojnacki, C.; Konrad, P.; Blonska, A.; Medrek-Socha, M.; Przybylowska-Sygut, K.; Chojnacki, J.; Poplawski, T. Altered Tryptophan Metabolism on the Kynurenine Pathway in Depressive Patients with Small Intestinal Bacterial Overgrowth. Nutrients 2022, 14, 3217. [Google Scholar] [CrossRef]
- Singhal, M.; Turturice, B.A.; Manzella, C.R.; Ranjan, R.; Metwally, A.A.; Theorell, J.; Huang, Y.; Alrefai, W.A.; Dudeja, P.K.; Finn, P.W.; et al. Serotonin Transporter Deficiency is Associated with Dysbiosis and Changes in Metabolic Function of the Mouse Intestinal Microbiome. Sci. Rep. 2019, 9, 2138. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, C.W.; Yang, Y.; Hsiao, Y.C.; Ru, H.; Lu, K. High-coverage metabolomics uncovers microbiota-driven biochemical landscape of interorgan transport and gut-brain communication in mice. Nat. Commun. 2021, 12, 6000. [Google Scholar] [CrossRef]
- Van der Wielen, N.; Moughan, P.J.; Mensink, M. Amino Acid Absorption in the Large Intestine of Humans and Porcine Models. J. Nutr. 2017, 147, 1493–1498. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, H.; Anjum, K.; Zhong, X.; Miao, S.; Zheng, G.; Liu, W.; Li, L. Dual Role of Indoles Derived from Intestinal Microbiota on Human Health. Front. Immunol. 2022, 13, 903526. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Lee, J.H.; Wood, T.K.; Lee, J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Santana Machado, T.; Poitevin, S.; Paul, P.; McKay, N.; Jourde-Chiche, N.; Legris, T.; Mouly-Bandini, A.; Dignat-George, F.; Brunet, P.; Masereeuw, R.; et al. Indoxyl Sulfate Upregulates Liver P-Glycoprotein Expression and Activity through Aryl Hydrocarbon Receptor Signaling. J. Am. Soc. Nephrol. 2018, 29, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.N.; Swimm, A.; Sonowal, R.; Bretin, A.; Gewirtz, A.T.; Jones, R.M.; Kalman, D. Indoles from the commensal microbiota act via the AHR and IL-10 to tune the cellular composition of the colonic epithelium during aging. Proc. Natl. Acad. Sci. USA 2020, 117, 21519–21526. [Google Scholar] [CrossRef] [PubMed]
- Rattanaphan, P.; Mittraparp-Arthorn, P.; Srinoun, K.; Vuddhakul, V.; Tansila, N. Indole signaling decreases biofilm formation and related virulence of Listeria monocytogenes. FEMS Microbiol. Lett. 2020, 367, fnaa116. [Google Scholar] [CrossRef] [PubMed]
- Domka, J.; Lee, J.; Wood, T.K. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 2006, 72, 2449–2459. [Google Scholar] [CrossRef]
- Hirakawa, H.; Kodama, T.; Takumi-Kobayashi, A.; Honda, T.; Yamaguchi, A. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology (Reading) 2009, 155 Pt 2, 541–550. [Google Scholar] [CrossRef]
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425. [Google Scholar] [CrossRef]
- Darkoh, C.; Chappell, C.; Gonzales, C.; Okhuysen, P. A rapid and specific method for the detection of indole in complex biological samples. Appl. Environ. Microbiol. 2015, 81, 8093–8097. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Sawrey-Kubicek, L.; Beals, E.; Rhodes, C.H.; Houts, H.E.; Sacchi, R.; Zivkovic, A.M. Human gut microbiome composition and tryptophan metabolites were changed differently by fast food and Mediterranean diet in 4 days: A pilot study. Nutr. Res. 2020, 77, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.D.; Murray, I.A.; Bisson, W.H.; Lahoti, T.S.; Gowda, K.; Amin, S.G.; Patterson, A.D.; Perdew, G.H. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015, 5, 12689. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.L.; Curran, M.A.; Marconi, S.A.; Carpenter, C.D.; Lubbers, L.S.; McAbee, M.D. Distribution of mRNAs encoding the arylhydrocarbon receptor, arylhydrocarbon receptor nuclear translocator, and arylhydrocarbon receptor nuclear translocator-2 in the rat brain and brainstem. J. Comp. Neurol. 2000, 427, 428–439. [Google Scholar] [CrossRef]
- Sheu, M.L.; Pan, L.Y.; Sheehan, J.; Yang, M.Y.; Pan, H.C. Modulation of Aryl Hydrocarbon Receptor Expression Alleviated Neuropathic Pain in a Chronic Constriction Nerve Injury Animal Model. Int. J. Mol. Sci. 2022, 23, 1255. [Google Scholar] [CrossRef]
- Kou, Z.; Dai, W. Aryl hydrocarbon receptor: Its roles in physiology. Biochem. Pharmacol. 2021, 185, 114428. [Google Scholar] [CrossRef]
- Denison, M.S.; Faber, S.C. And Now for Something Completely Different: Diversity in Ligand-Dependent Activation of Ah Receptor Responses. Curr. Opin. Toxicol. 2017, 2, 124–131. [Google Scholar] [CrossRef]
- Dong, F.; Perdew, G.H. The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut Microbes 2020, 12, 1859812. [Google Scholar] [CrossRef]
- Stockinger, B.; Di Meglio, P.; Gialitakis, M.; Duarte, J.H. The aryl hydrocarbon receptor: Multitasking in the immune system. Annu. Rev. Immunol. 2014, 32, 403–432. [Google Scholar] [CrossRef]
- Buckley, M.M.; O’Brien, R.; Brosnan, E.; Ross, R.P.; Stanton, C.; Buckley, J.M.; O’Malley, D. Glucagon-Like Peptide-1 Secreting L-Cells Coupled to Sensory Nerves Translate Microbial Signals to the Host Rat Nervous System. Front. Cell Neurosci. 2020, 14, 95. [Google Scholar] [CrossRef]
- Young, S.N.; Anderson, G.M.; Gauthier, S.; Purdy, W.C. The origin of indoleacetic acid and indolepropionic acid in rat and human cerebrospinal fluid. J. Neurochem. 1980, 34, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Jaglin, M.; Rhimi, M.; Philippe, C.; Pons, N.; Bruneau, A.; Goustard, B.; Dauge, V.; Maguin, E.; Naudon, L.; Rabot, S. Indole, a Signaling Molecule Produced by the Gut Microbiota, Negatively Impacts Emotional Behaviors in Rats. Front. Neurosci. 2018, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.Z.; Martin, K.A.; Xing, P.Y.; Agrawal, R.; Whiley, L.; Wood, T.K.; Hejndorf, S.; Ng, Y.Z.; Low, J.Z.Y.; Rossant, J.; et al. Tryptophan-metabolizing gut microbes regulate adult neurogenesis via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2021, 118, e2021091118. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Semak, I.; Pisarchik, A.; Sweatman, T.; Szczesniewski, A.; Wortsman, J. Conversion of L-tryptophan to serotonin and melatonin in human melanoma cells. FEBS Lett. 2002, 511, 102–106. [Google Scholar] [CrossRef]
- Hata, T.; Asano, Y.; Yoshihara, K.; Kimura-Todani, T.; Miyata, N.; Zhang, X.T.; Takakura, S.; Aiba, Y.; Koga, Y.; Sudo, N. Regulation of gut luminal serotonin by commensal microbiota in mice. PLoS ONE 2017, 12, e0180745. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef]
- Ge, X.; Ding, C.; Zhao, W.; Xu, L.; Tian, H.; Gong, J.; Zhu, M.; Li, J.; Li, N. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J. Transl. Med. 2017, 15, 13. [Google Scholar] [CrossRef]
- De Vadder, F.; Grasset, E.; Manneras Holm, L.; Karsenty, G.; Macpherson, A.J.; Olofsson, L.E.; Backhed, F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. USA 2018, 115, 6458–6463. [Google Scholar] [CrossRef]
- Shah, P.A.; Park, C.J.; Shaughnessy, M.P.; Cowles, R.A. Serotonin as a Mitogen in the Gastrointestinal Tract: Revisiting a Familiar Molecule in a New Role. Cell Mol. Gastroenterol. Hepatol. 2021, 12, 1093–1104. [Google Scholar] [CrossRef]
- Zhao, Y.; Gan, L.; Ren, L.; Lin, Y.; Ma, C.; Lin, X. Factors influencing the blood-brain barrier permeability. Brain Res. 2022, 1788, 147937. [Google Scholar] [CrossRef]
- Fu, J.; Li, L.; Huo, D.; Yang, R.; Yang, B.; Xu, B.; Yang, X.; Dai, M.; Tan, C.; Chen, H.; et al. Meningitic Escherichia coli alpha-hemolysin aggravates blood-brain barrier disruption via targeting TGFbeta1-triggered hedgehog signaling. Mol. Brain 2021, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Pirina, P.; Zinellu, E.; Paliogiannis, P.; Fois, A.G.; Marras, V.; Sotgia, S.; Carru, C.; Zinellu, A. Circulating serotonin levels in COPD patients: A pilot study. BMC Pulm. Med. 2018, 18, 167. [Google Scholar] [CrossRef] [PubMed]
- Herr, N.; Bode, C.; Duerschmied, D. The Effects of Serotonin in Immune Cells. Front. Cardiovasc. Med. 2017, 4, 48. [Google Scholar] [CrossRef]
- Mann, D.A.; Oakley, F. Serotonin paracrine signaling in tissue fibrosis. Biochim. Biophys. Acta 2013, 1832, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Brzezinski, A. Melatonin in humans. N. Engl. J. Med. 1997, 336, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Huether, G. Melatonin synthesis in the gastrointestinal tract and the impact of nutritional factors on circulating melatonin. Ann. N. Y. Acad. Sci. 1994, 719, 146–158. [Google Scholar] [CrossRef]
- Velarde, E.; Alonso-Gomez, A.L.; Azpeleta, C.; Isorna, E.; De Pedro, N.; Delgado, M.J. Melatonin effects on gut motility are independent of the relaxation mediated by the nitrergic system in the goldfish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011, 159, 367–371. [Google Scholar] [CrossRef]
- Song, G.H.; Leng, P.H.; Gwee, K.A.; Moochhala, S.M.; Ho, K.Y. Melatonin improves abdominal pain in irritable bowel syndrome patients who have sleep disturbances: A randomised, double blind, placebo controlled study. Gut 2005, 54, 1402–1407. [Google Scholar] [CrossRef]
- Xu, P.; Wang, J.; Hong, F.; Wang, S.; Jin, X.; Xue, T.; Jia, L.; Zhai, Y. Melatonin prevents obesity through modulation of gut microbiota in mice. J. Pineal Res. 2017, 62, e12399. [Google Scholar] [CrossRef]
- Escribano, B.M.; Colin-Gonzalez, A.L.; Santamaria, A.; Tunez, I. The role of melatonin in multiple sclerosis, Huntington’s disease and cerebral ischemia. CNS Neurol. Disord. Drug Targets 2014, 13, 1096–1119. [Google Scholar] [CrossRef]
- Bentley, G.E. Unraveling the enigma: The role of melatonin in seasonal processes in birds. Microsc. Res. Tech. 2001, 53, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Filadelfi, A.M.; Castrucci, A.M. Comparative aspects of the pineal/melatonin system of poikilothermic vertebrates. J. Pineal Res. 1996, 20, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human Gut Bacteria Are Sensitive to Melatonin and Express Endogenous Circadian Rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef] [PubMed]
- Graniczkowska, K.B.; Shaffer, C.L.; Cassone, V.M. Transcriptional effects of melatonin on the gut commensal bacterium Klebsiella aerogenes. Genomics 2022, 114, 110321. [Google Scholar] [CrossRef]
- Chen, L.M.; Bao, C.H.; Wu, Y.; Liang, S.H.; Wang, D.; Wu, L.Y.; Huang, Y.; Liu, H.R.; Wu, H.G. Tryptophan-kynurenine metabolism: A link between the gut and brain for depression in inflammatory bowel disease. J. Neuroinflamm. 2021, 18, 135. [Google Scholar] [CrossRef]
- Tan, V.X.; Guillemin, G.J. Kynurenine Pathway Metabolites as Biomarkers for Amyotrophic Lateral Sclerosis. Front. Neurosci. 2019, 13, 1013. [Google Scholar] [CrossRef]
- Venkatesan, D.; Iyer, M.; Narayanasamy, A.; Siva, K.; Vellingiri, B. Kynurenine pathway in Parkinson’s disease-An update. eNeurologicalSci 2020, 21, 100270. [Google Scholar] [CrossRef]
- Groven, N.; Reitan, S.K.; Fors, E.A.; Guzey, I.C. Kynurenine metabolites and ratios differ between Chronic Fatigue Syndrome, Fibromyalgia, and healthy controls. Psychoneuroendocrinology 2021, 131, 105287. [Google Scholar] [CrossRef]
- Solvang, S.H.; Nordrehaug, J.E.; Aarsland, D.; Lange, J.; Ueland, P.M.; McCann, A.; Midttun, O.; Tell, G.S.; Giil, L.M. Kynurenines, Neuropsychiatric Symptoms, and Cognitive Prognosis in Patients with Mild Dementia. Int. J. Tryptophan Res. 2019, 12, 1178646919877883. [Google Scholar] [CrossRef]
- Bartoli, F.; Cioni, R.M.; Cavaleri, D.; Callovini, T.; Crocamo, C.; Misiak, B.; Savitz, J.B.; Carra, G. The association of kynurenine pathway metabolites with symptom severity and clinical features of bipolar disorder: An overview. Eur. Psychiatry 2022, 65, e82. [Google Scholar] [CrossRef]
- Bartoli, F.; Misiak, B.; Callovini, T.; Cavaleri, D.; Cioni, R.M.; Crocamo, C.; Savitz, J.B.; Carra, G. The kynurenine pathway in bipolar disorder: A meta-analysis on the peripheral blood levels of tryptophan and related metabolites. Mol. Psychiatry 2021, 26, 3419–3429. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 2017, 112 Pt B, 399–412. [Google Scholar] [CrossRef]
- Savitz, J. The kynurenine pathway: A finger in every pie. Mol. Psychiatry 2020, 25, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Huang, X.F.; Newell, K.A. The kynurenine pathway in major depression: What we know and where to next. Neurosci. Biobehav. Rev. 2021, 127, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, D.; Song, P.; Zou, M.H. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front. Biosci. (Landmark Ed.) 2015, 20, 1116–1143. [Google Scholar] [CrossRef]
- Joisten, N.; Ruas, J.L.; Braidy, N.; Guillemin, G.J.; Zimmer, P. The kynurenine pathway in chronic diseases: A compensatory mechanism or a driving force? Trends Mol. Med. 2021, 27, 946–954. [Google Scholar] [CrossRef]
- Huang, Y.S.; Ogbechi, J.; Clanchy, F.I.; Williams, R.O.; Stone, T.W. IDO and Kynurenine Metabolites in Peripheral and CNS Disorders. Front. Immunol. 2020, 11, 388. [Google Scholar] [CrossRef] [PubMed]
- Prasher, P.; Sharma, M. Medicinal chemistry of anthranilic acid derivatives: A mini review. Drug Dev. Res. 2021, 82, 945–958. [Google Scholar] [CrossRef]
- Nittoli, T.; Curran, K.; Insaf, S.; DiGrandi, M.; Orlowski, M.; Chopra, R.; Agarwal, A.; Howe, A.Y.; Prashad, A.; Floyd, M.B.; et al. Identification of anthranilic acid derivatives as a novel class of allosteric inhibitors of hepatitis C NS5B polymerase. J. Med. Chem. 2007, 50, 2108–2116. [Google Scholar] [CrossRef]
- Jahan, H.; Choudhary, M.I.; Atta, A.; Khan, K.M.; Ur-Rahman, A. Anthranilic Acid Derivatives: Novel Inhibitors of Protein Glycation and the Associated Oxidative Stress in the Hepatocytes. Med. Chem. 2018, 14, 516–523. [Google Scholar] [CrossRef]
- Guillemin, G.J.; Cullen, K.M.; Lim, C.K.; Smythe, G.A.; Garner, B.; Kapoor, V.; Takikawa, O.; Brew, B.J. Characterization of the kynurenine pathway in human neurons. J. Neurosci. 2007, 27, 12884–12892. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Kerr, S.J.; Smythe, G.A.; Smith, D.G.; Kapoor, V.; Armati, P.J.; Croitoru, J.; Brew, B.J. Kynurenine pathway metabolism in human astrocytes: A paradox for neuronal protection. J. Neurochem. 2001, 78, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Smith, D.G.; Smythe, G.A.; Armati, P.J.; Brew, B.J. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Adv. Exp. Med. Biol. 2003, 527, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Werner-Felmayer, G.; Werner, E.R.; Fuchs, D.; Hausen, A.; Reibnegger, G.; Wachter, H. Characteristics of interferon induced tryptophan metabolism in human cells in vitro. Biochim. Biophys. Acta 1989, 1012, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Sorgdrager, F.J.H.; Naude, P.J.W.; Kema, I.P.; Nollen, E.A.; Deyn, P.P. Tryptophan Metabolism in Inflammaging: From Biomarker to Therapeutic Target. Front. Immunol. 2019, 10, 2565. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, H.; Yang, G.; Yang, Y.; Li, W.; Song, M.; Shao, M.; Su, X.; Lv, L. Associations between expression of indoleamine 2, 3-dioxygenase enzyme and inflammatory cytokines in patients with first-episode drug-naive Schizophrenia. Transl. Psychiatry 2021, 11, 595. [Google Scholar] [CrossRef]
- Litzenburger, U.M.; Opitz, C.A.; Sahm, F.; Rauschenbach, K.J.; Trump, S.; Winter, M.; Ott, M.; Ochs, K.; Lutz, C.; Liu, X.; et al. Constitutive IDO expression in human cancer is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Oncotarget 2014, 5, 1038–1051. [Google Scholar] [CrossRef]
- Wolf, A.M.; Wolf, D.; Rumpold, H.; Moschen, A.R.; Kaser, A.; Obrist, P.; Fuchs, D.; Brandacher, G.; Winkler, C.; Geboes, K.; et al. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin. Immunol. 2004, 113, 47–55. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, H.; Wen, Q.; Zhang, Y. Indoleamine 2,3-dioxygenase expression in human inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 2012, 24, 695–701. [Google Scholar] [CrossRef]
- Gibney, S.M.; Fagan, E.M.; Waldron, A.M.; O’Byrne, J.; Connor, T.J.; Harkin, A. Inhibition of stress-induced hepatic tryptophan 2,3-dioxygenase exhibits antidepressant activity in an animal model of depressive behaviour. Int. J. Neuropsychopharmacol. 2014, 17, 917–928. [Google Scholar] [CrossRef]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y.; Hirayama, C. Serum indole and skatole in patients with various liver diseases. Clin. Chim. Acta 1988, 176, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Hong, Y.; Lee, M.; Keum, B.R.; Kim, G.H. Natural Product Skatole Ameliorates Lipotoxicity-Induced Multiple Hepatic Damage under Hyperlipidemic Conditions in Hepatocytes. Nutrients 2023, 15, 1490. [Google Scholar] [CrossRef]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Teunis, C.; Nieuwdorp, M.; Hanssen, N. Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcoholic Fatty Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 2022, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Cho, J.S.; Lee, S.Y. Microbial production of methyl anthranilate, a grape flavor compound. Proc. Natl. Acad. Sci. USA 2019, 116, 10749–10756. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Kazemi Shariat Panahi, H.; Guillemin, G.J. Microorganisms, Tryptophan Metabolism, and Kynurenine Pathway: A Complex Interconnected Loop Influencing Human Health Status. Int. J. Tryptophan Res. 2019, 12, 1178646919852996. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Berkeley, M.; Kriventseva, E.V. OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Res 2021, 49, D389–D393. [Google Scholar] [CrossRef]
- Karcher, N.; Nigro, E.; Puncochar, M.; Blanco-Miguez, A.; Ciciani, M.; Manghi, P.; Zolfo, M.; Cumbo, F.; Manara, S.; Golzato, D.; et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol. 2021, 22, 209. [Google Scholar] [CrossRef]
- Tierney, B.T.; Yang, Z.; Luber, J.M.; Beaudin, M.; Wibowo, M.C.; Baek, C.; Mehlenbacher, E.; Patel, C.J.; Kostic, A.D. The Landscape of Genetic Content in the Gut and Oral Human Microbiome. Cell Host Microbe 2019, 26, 283–295.e8. [Google Scholar] [CrossRef]
- Xiao, L.; Yan, J.; Yang, T.; Zhu, J.; Li, T.; Wei, H.; Chen, J. Fecal Microbiome Transplantation from Children with Autism Spectrum Disorder Modulates Tryptophan and Serotonergic Synapse Metabolism and Induces Altered Behaviors in Germ-Free Mice. mSystems 2021, 6, e01343-20. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, L.; Zhu, R.; Zhang, S.; Liu, S.; Wang, Y.; Wu, Y.; Xing, S.; Liao, X.; Mi, J. Porcine gut microbiota in mediating host metabolic adaptation to cold stress. NPJ Biofilms Microbiomes 2022, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Oxenkrug, G.; van der Hart, M.; Roeser, J.; Summergrad, P. Anthranilic Acid: A Potential Biomarker and Treatment Target for Schizophrenia. Ann. Psychiatry Ment. Health 2016, 4, 1059. [Google Scholar] [PubMed]
- Krause, D.; Suh, H.S.; Tarassishin, L.; Cui, Q.L.; Durafourt, B.A.; Choi, N.; Bauman, A.; Cosenza-Nashat, M.; Antel, J.P.; Zhao, M.L.; et al. The tryptophan metabolite 3-hydroxyanthranilic acid plays anti-inflammatory and neuroprotective roles during inflammation: Role of hemeoxygenase-1. Am. J. Pathol. 2011, 179, 1360–1372. [Google Scholar] [CrossRef]
- Coplan, J.D.; George, R.; Syed, S.A.; Rozenboym, A.V.; Tang, J.E.; Fulton, S.L.; Perera, T.D. Early Life Stress and the Fate of Kynurenine Pathway Metabolites. Front. Hum. Neurosci. 2021, 15, 636144. [Google Scholar] [CrossRef]
- Parrott, J.M.; Redus, L.; Santana-Coelho, D.; Morales, J.; Gao, X.; O’Connor, J.C. Neurotoxic kynurenine metabolism is increased in the dorsal hippocampus and drives distinct depressive behaviors during inflammation. Transl. Psychiatry 2016, 6, e918. [Google Scholar] [CrossRef]
- Birner, A.; Platzer, M.; Bengesser, S.A.; Dalkner, N.; Fellendorf, F.T.; Queissner, R.; Pilz, R.; Rauch, P.; Maget, A.; Hamm, C.; et al. Increased breakdown of kynurenine towards its neurotoxic branch in bipolar disorder. PLoS ONE 2017, 12, e0172699. [Google Scholar] [CrossRef]
- Steiner, J.; Dobrowolny, H.; Guest, P.C.; Bernstein, H.G.; Fuchs, D.; Roeser, J.; Summergrad, P.; Oxenkrug, G. Gender-specific elevation of plasma anthranilic acid in schizophrenia: Protection against glutamatergic hypofunction? Schizophr. Res. 2022, 243, 483–485. [Google Scholar] [CrossRef]
- Oxenkrug, G.; van der Hart, M.; Roeser, J.; Summergrad, P. Peripheral Tryptophan—Kynurenine Metabolism Associated with Metabolic Syndrome is Different in Parkinson’s and Alzheimer’s Diseases. Endocrinol. Diabetes Metab. J. 2017, 1. [Google Scholar] [CrossRef]
- Giorgini, F.; Huang, S.Y.; Sathyasaikumar, K.V.; Notarangelo, F.M.; Thomas, M.A.; Tararina, M.; Wu, H.Q.; Schwarcz, R.; Muchowski, P.J. Targeted deletion of kynurenine 3-monooxygenase in mice: A new tool for studying kynurenine pathway metabolism in periphery and brain. J. Biol. Chem. 2013, 288, 36554–36566. [Google Scholar] [CrossRef] [PubMed]
- Jayawickrama, G.S.; Nematollahi, A.; Sun, G.; Gorrell, M.D.; Church, W.B. Inhibition of human kynurenine aminotransferase isozymes by estrogen and its derivatives. Sci. Rep. 2017, 7, 17559. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, E.; D’Esposito, M. Estrogen shapes dopamine-dependent cognitive processes: Implications for women’s health. J. Neurosci. 2011, 31, 5286–5293. [Google Scholar] [CrossRef]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693 Pt B, 128–133. [Google Scholar] [CrossRef]
- Curto, M.; Lionetto, L.; Negro, A.; Capi, M.; Fazio, F.; Giamberardino, M.A.; Simmaco, M.; Nicoletti, F.; Martelletti, P. Altered kynurenine pathway metabolites in serum of chronic migraine patients. J. Headache Pain. 2015, 17, 47. [Google Scholar] [CrossRef]
- Curto, M.; Lionetto, L.; Negro, A.; Capi, M.; Perugino, F.; Fazio, F.; Giamberardino, M.A.; Simmaco, M.; Nicoletti, F.; Martelletti, P. Altered serum levels of kynurenine metabolites in patients affected by cluster headache. J. Headache Pain. 2015, 17, 27. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.; Dobrowolny, H.; Guest, P.C.; Bernstein, H.G.; Fuchs, D.; Roeser, J.; Summergrad, P.; Oxenkrug, G.F. Plasma Anthranilic Acid and Leptin Levels Predict HAM-D Scores in Depressed Women. Int. J. Tryptophan Res. 2021, 14, 11786469211016474. [Google Scholar] [CrossRef] [PubMed]
- Colle, R.; Masson, P.; Verstuyft, C.; Feve, B.; Werner, E.; Boursier-Neyret, C.; Walther, B.; David, D.J.; Boniface, B.; Falissard, B.; et al. Peripheral tryptophan, serotonin, kynurenine, and their metabolites in major depression: A case-control study. Psychiatry Clin. Neurosci. 2020, 74, 112–117. [Google Scholar] [CrossRef]
- Pawlowski, T.; Pawlak, D.; Inglot, M.; Zalewska, M.; Marciniak, D.; Bugajska, J.; Janocha-Litwin, J.; Malyszczak, K. The role of anthranilic acid in the increase of depressive symptoms and major depressive disorder during treatment for hepatitis C with pegylated interferon-alpha2a and oral ribavirin. J. Psychiatry Neurosci. 2021, 46, E166–E175. [Google Scholar] [CrossRef]
- Pompili, M.; Lionetto, L.; Curto, M.; Forte, A.; Erbuto, D.; Montebovi, F.; Seretti, M.E.; Berardelli, I.; Serafini, G.; Innamorati, M.; et al. Tryptophan and Kynurenine Metabolites: Are They Related to Depression? Neuropsychobiology 2019, 77, 23–28. [Google Scholar] [CrossRef]
- Darlington, L.G.; Mackay, G.M.; Forrest, C.M.; Stoy, N.; George, C.; Stone, T.W. Altered kynurenine metabolism correlates with infarct volume in stroke. Eur. J. Neurosci. 2007, 26, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kwak, J.H.; Pyo, S. Inhibition of LPS-induced inflammatory mediators by 3-hydroxyanthranilic acid in macrophages through suppression of PI3K/NF-kappaB signaling pathways. Food Funct. 2016, 7, 3073–3082. [Google Scholar] [CrossRef] [PubMed]
- Cervellati, C.; Trentini, A.; Pecorelli, A.; Valacchi, G. Inflammation in Neurological Disorders: The Thin Boundary between Brain and Periphery. Antioxid. Redox Signal 2020, 33, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, B.; Bhargava, P.; Tremlett, H.; Hart, J.; Graves, J.; Waubant, E. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann. Clin. Transl. Neurol. 2018, 5, 1211–1221. [Google Scholar] [CrossRef]
- Rodrigues, F.B.; Byrne, L.M.; Lowe, A.J.; Tortelli, R.; Heins, M.; Flik, G.; Johnson, E.B.; De Vita, E.; Scahill, R.I.; Giorgini, F.; et al. Kynurenine pathway metabolites in cerebrospinal fluid and blood as potential biomarkers in Huntington’s disease. J. Neurochem. 2021, 158, 539–553. [Google Scholar] [CrossRef]
- Fathi, M.; Vakili, K.; Yaghoobpoor, S.; Tavasol, A.; Jazi, K.; Hajibeygi, R.; Shool, S.; Sodeifian, F.; Klegeris, A.; McElhinney, A.; et al. Dynamic changes in metabolites of the kynurenine pathway in Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease: A systematic Review and meta-analysis. Front. Immunol. 2022, 13, 997240. [Google Scholar] [CrossRef]
- Inglis, J.J.; Criado, G.; Andrews, M.; Feldmann, M.; Williams, R.O.; Selley, M.L. The anti-allergic drug, N-(3′,4′-dimethoxycinnamonyl) anthranilic acid, exhibits potent anti-inflammatory and analgesic properties in arthritis. Rheumatology 2007, 46, 1428–1432. [Google Scholar] [CrossRef]
- Maes, M.; Leonard, B.E.; Myint, A.M.; Kubera, M.; Verkerk, R. The new ‘5-HT’ hypothesis of depression: Cell-mediated immune activation induces indoleamine 2,3-dioxygenase, which leads to lower plasma tryptophan and an increased synthesis of detrimental tryptophan catabolites (TRYCATs), both of which contribute to the onset of depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2011, 35, 702–721. [Google Scholar] [CrossRef]
- Traylor, M.; Rutten-Jacobs, L.C.; Holliday, E.G.; Malik, R.; Sudlow, C.; Rothwell, P.M.; Maguire, J.M.; Koblar, S.A.; Bevan, S.; Boncoraglio, G.; et al. Differences in Common Genetic Predisposition to Ischemic Stroke by Age and Sex. Stroke 2015, 46, 3042–3047. [Google Scholar] [CrossRef]
- Caspi, A.; Sugden, K.; Moffitt, T.E.; Taylor, A.; Craig, I.W.; Harrington, H.; McClay, J.; Mill, J.; Martin, J.; Braithwaite, A.; et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science 2003, 301, 386–389. [Google Scholar] [CrossRef]
- Qin, Y.; Havulinna, A.S.; Liu, Y.; Jousilahti, P.; Ritchie, S.C.; Tokolyi, A.; Sanders, J.G.; Valsta, L.; Brozynska, M.; Zhu, Q.; et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 2022, 54, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.H.; Foster, C.M.; Vishnivetskaya, T.; Campbell, A.G.; Yang, Z.K.; Wymore, A.; Palumbo, A.V.; Chesler, E.J.; Podar, M. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012, 6, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
| Substrate | Metabolite | Known Microbial Producer (s) | Bioactive Function | Cofactors |
|---|---|---|---|---|
| Tryptophan | Kynurenine | Pseudomonas aeruginosa [42], Ralstonia metallidurans [43] | Neuronal damage mediator | INF-gamma [44] |
| Indole | Escherichia coli, Proteus vulgaris, Clostridium sp., Bacteroides sp. [45] | Anti-inflammatory, epithelial tight-junction regulation | Iron [46] | |
| Serotonin | Pseudomonas putida KT2440 [47] | Mood regulation | Vitamin B2 [48] | |
| Kynurenine | Anthranilic acid | Pseudomonas aeruginosa [49], Burkholderia fungorum, Bacillus cereus, Ralstonia solanacearum, and Ralstonia metallidurans [43] | Inflammation activation | Iron [50] |
| 3-Hydroxyanthranilic acid | Proteobacteria sp., Actinobacteria sp., Firmicutes sp. Bacteroidetes sp., Chloroflexi sp., Cyanobacteria sp., Euryarchaeota sp. [51] | Free radical generation, apoptotic regulation | Pyridoxal-5′-phosphate [52] | |
| Kynurenic acid | Escherichia coli [53] | NMDA agonist, anticonvulsant | ||
| 3-hydroxykynurenine | Proteobacteria sp., Actinobacteria sp., Firmicutes sp. Bacteroidetes sp., Chloroflexi sp., Cyanobacteria sp., Euryarchaeota sp. [51] | Oxidative damage, apoptotic regulation | ||
| Quinolinic acid | Streptomyces antibioticus, Cyanidium caldarium, Karlingia rosea [54] | NMDA activator | Anthranilic Acid [55] | |
| Picolinic acid | Burkholderia xenovorans, [56] | Iron and zinc chelator, antiviral, antifungal | Anthranilic Acid [55] | |
| Indole | Indole-3-acetamide | Acidobacteria sp., Bacteroidetes sp., Firmicutes sp. [57] | Antioxidant, anti-hyperglycemic | |
| Tryptamine | Ruminococcus gnavus and Clostridium sporogenes [58] | Neuroinflammatory modulation | ||
| Indole-3-acetic acid | Clostridium sp., Bacteroides sp., Bifidobacterium sp. [59] | Anti-angiogenic | ||
| Indole-3-pyruvic acid | Pseudomonas fluorescens and Candida tropicalis [60] | Anti-inflammatory, anti-angiogenic | ||
| Indole-3-acetate | Clostridium sporogenes [61] | Anti-inflammatory | ||
| Indole-3-aldehyde | Lactobacillus sp. [62] | Epithelial cell cycle regulation, goblet cell differentiation regulation | ||
| Indole-3-propionic acid | Lactobacillus reuteri, Akkermansia sp., Clostridium sp., Peptostreptococi sp. [63] | Anti-inflammatory, neuronal apoptosis regulation | ||
| Skatole | Clostridium sp. and Bacteroides sp. [64] | Induces colonocyte apoptosis | ||
| Serotonin | Melatonin | Roseburia hominis [65] | Circadian rhythm regulation, antioxidant | Tetrahydrobiop-terin and O2 [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, C.; Hess, M.; Weimer, B.C. Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives. Microorganisms 2023, 11, 1825. https://doi.org/10.3390/microorganisms11071825
Shaw C, Hess M, Weimer BC. Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives. Microorganisms. 2023; 11(7):1825. https://doi.org/10.3390/microorganisms11071825
Chicago/Turabian StyleShaw, Claire, Matthias Hess, and Bart C. Weimer. 2023. "Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives" Microorganisms 11, no. 7: 1825. https://doi.org/10.3390/microorganisms11071825
APA StyleShaw, C., Hess, M., & Weimer, B. C. (2023). Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives. Microorganisms, 11(7), 1825. https://doi.org/10.3390/microorganisms11071825







