Antiscalants Used in Seawater Desalination: Biodegradability and Effects on Microbial Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antiscalants
2.2. Enrichment Culture
2.3. Chemical Analysis
2.4. DNA Extraction, Library Preparation, and Sequencing
2.5. 16S rRNA Sequence Analysis
2.6. Statistical Analysis
3. Results and Discussion
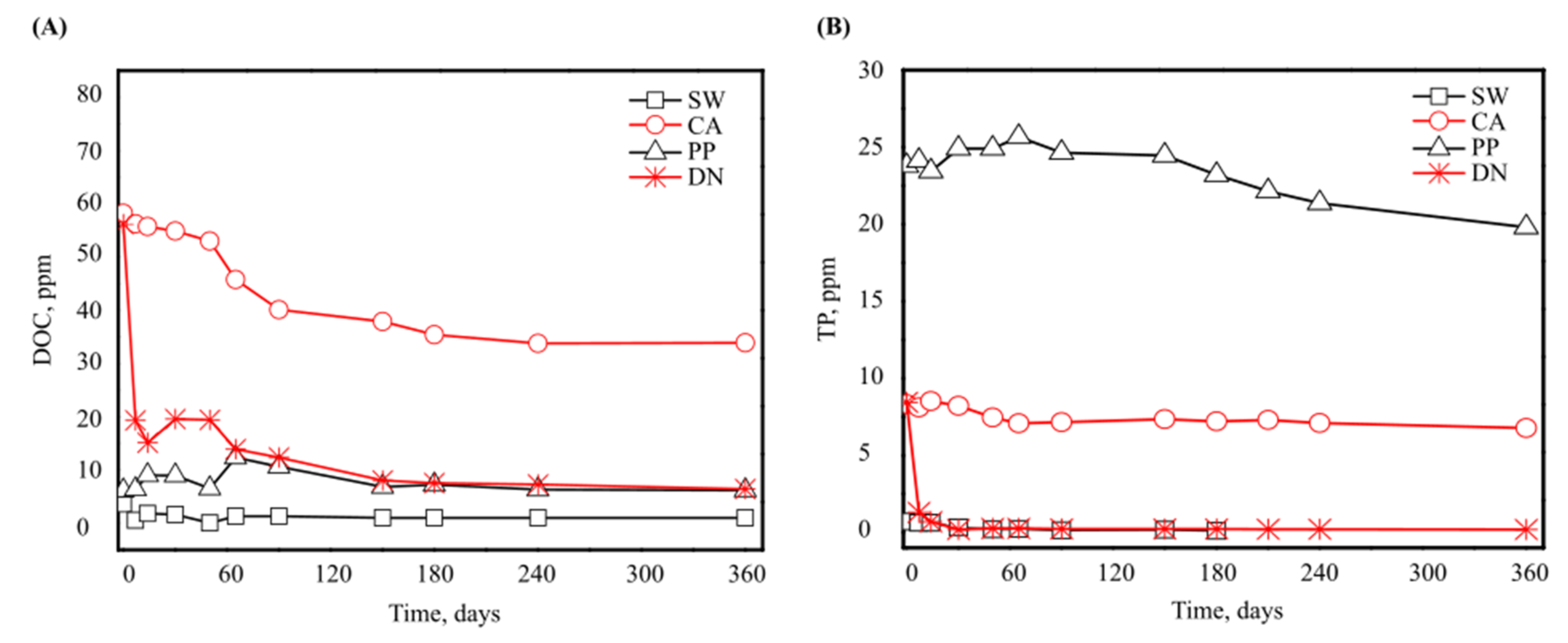
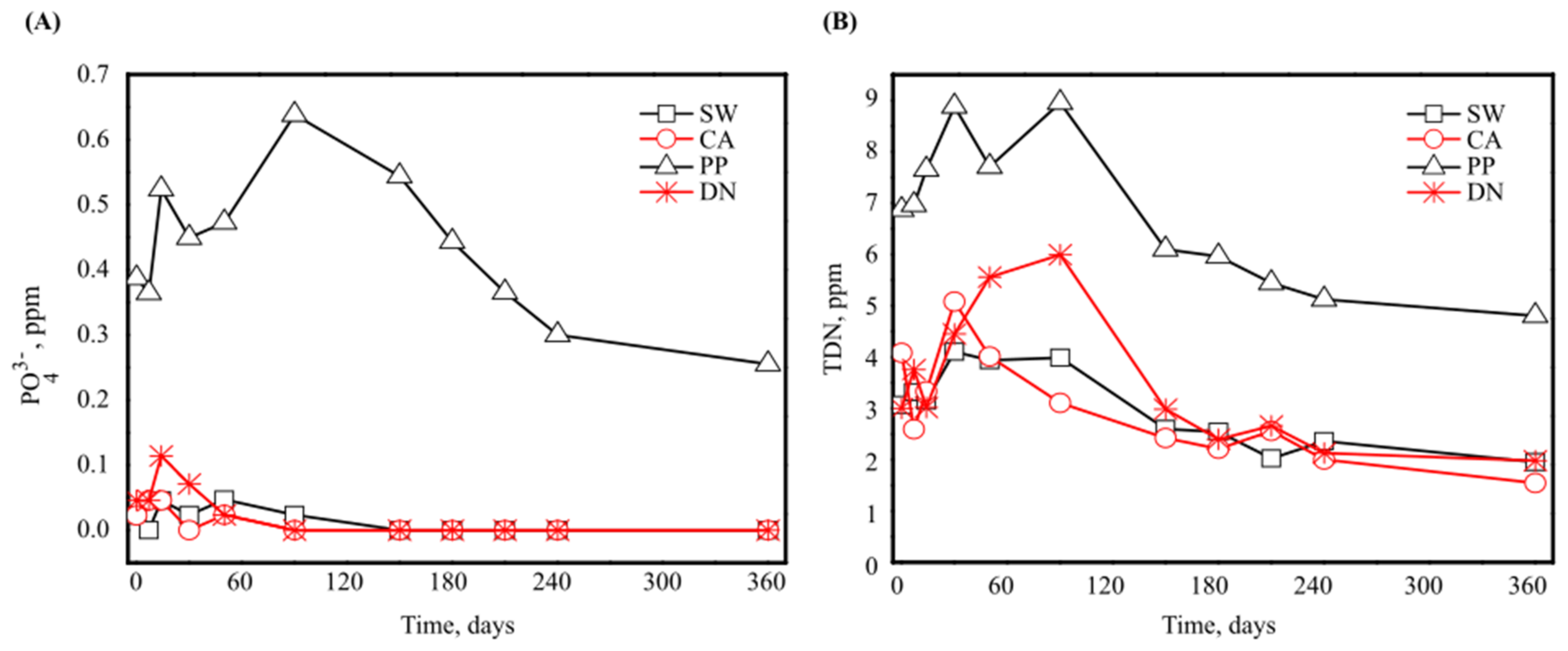
3.1. Antiscalant Biodegradability
3.2. Microbial Community Analysis
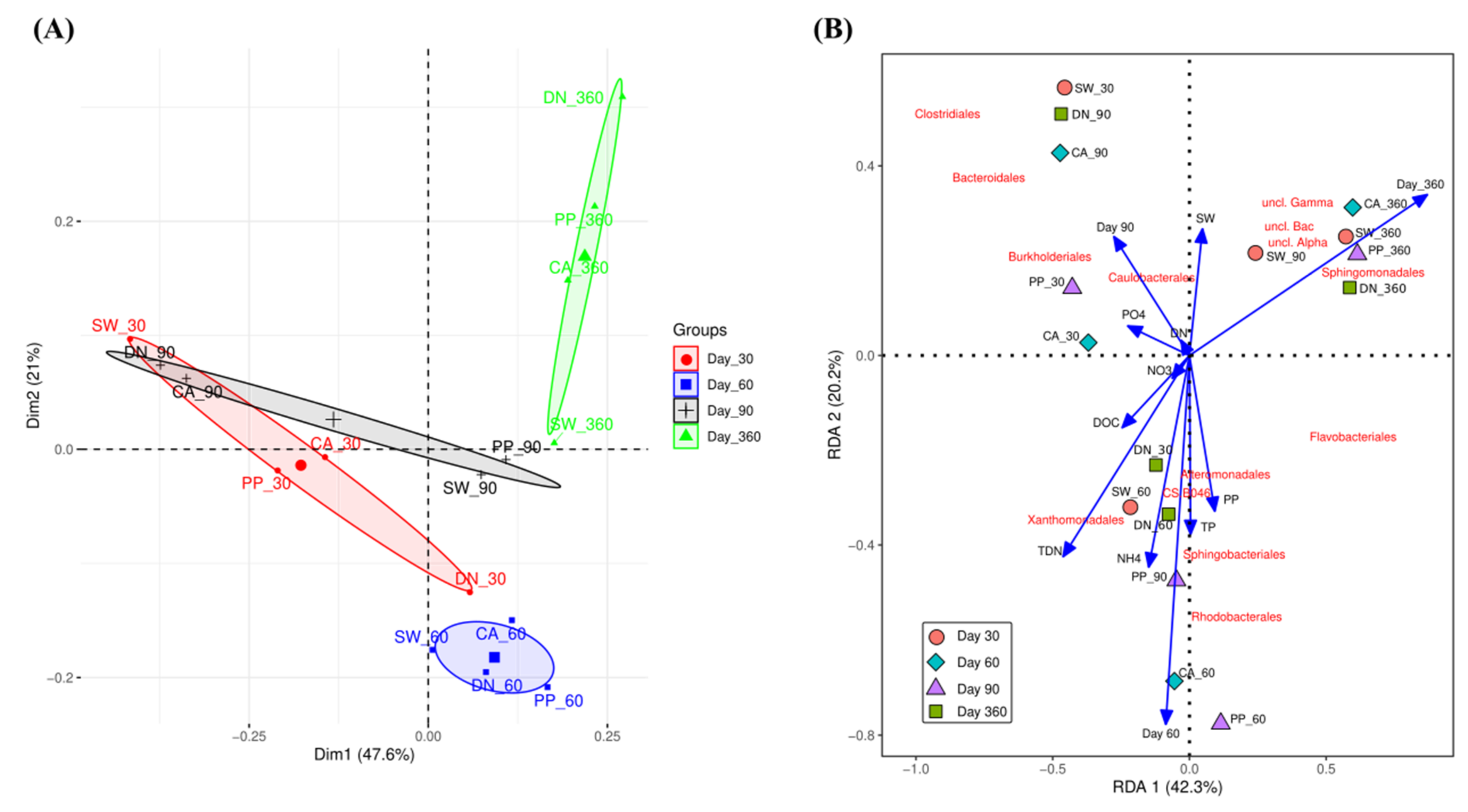
3.2.1. Bacterial Diversity
3.2.2. Effect of Different Types of Antiscalants and Incubation Time on the Bacterial Community Composition
3.2.3. Bacterial Community Composition
Bacterial Phylum Composition
Bacterial Order Compositions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amjad, Z. Applications of antiscalants to control calcium sulfate scaling in reverse osmosis systems. Desalination 1985, 54, 263–276. [Google Scholar] [CrossRef]
- Darton, E.G. Membrane chemical research: Centuries apart. Desalination 2000, 132, 121–131. [Google Scholar] [CrossRef]
- Fritzmann, C.; Löwenberg, J.; Wintgens, T.; Melin, T. State-of-the-art of reverse osmosis desalination. Desalination 2007, 216, 1–76. [Google Scholar] [CrossRef]
- Pearse, M.J. An overview of the use of chemical reagents in mineral processing. Miner. Eng. 2005, 18, 139–149. [Google Scholar] [CrossRef]
- Elsaid, K.; Kamil, M.; Sayed, E.T.; Abdelkareem, M.A.; Wilberforce, T.; Olabi, A. Environmental impact of desalination technologies: A review. Sci. Total Environ. 2020, 748, 141528. [Google Scholar] [CrossRef]
- Petersen, K.L.; Frank, H.; Paytan, A.; Bar-Zeev, E. Impacts of seawater desalination on coastal environments. In Sustainable Desalination Handbook; Elsevier: Amsterdam, The Netherlands, 2018; pp. 437–463. [Google Scholar]
- Petersen, K.L.; Paytan, A.; Rahav, E.; Levy, O.; Silverman, J.; Barzel, O.; Potts, D.; Bar-Zeev, E. Impact of brine and antiscalants on reef-building corals in the Gulf of Aqaba—Potential effects from desalination plants. Water Res. 2018, 144, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.A.; Johnston, E.L.; Knott, N.A. Impacts of desalination plant discharges on the marine environment: A critical review of published studies. Water Res. 2010, 44, 5117–5128. [Google Scholar] [CrossRef] [PubMed]
- Oshchepkov, M.S.; Rudakova, G.Y.; Tkachenko, S.V.; Larchenko, V.E.; Popov, K.I.; Tusheva, M.A. Recent State-of-the-Art of Antiscalant-Driven Scale Inhibition Theory. Therm. Eng. 2021, 68, 370–380. [Google Scholar] [CrossRef]
- Jafar Mazumder, M.A. A review of green scale inhibitors: Process, types, mechanism and properties. Coatings 2020, 10, 928. [Google Scholar] [CrossRef]
- Hasson, D.; Shemer, H.; Sher, A. State of the art of friendly “green” scale control inhibitors: A review article. Ind. Eng. Chem. Res. 2011, 50, 7601–7607. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Al-Ghouti, M.A.; Qiblawey, H.; Rodrigues, D.F.; Hu, Y.; Zouari, N. Isolation, identification and biodiversity of antiscalant degrading seawater bacteria using MALDI-TOF-MS and multivariate analysis. Sci. Total Environ. 2019, 656, 910–920. [Google Scholar] [CrossRef]
- Larson, R.J.; Bookland, E.A.; Williams, R.T.; Yocom, K.M.; Saucy, D.A.; Freeman, M.B.; Swift, G. Biodegradation of acrylic acid polymers and oligomers by mixed microbial communities in activated sludge. J. Environ. Polym. Degrad. 1997, 5, 41–48. [Google Scholar]
- Campos, E.J.; Vieira, F.; Cavalcante, G.; Kjerfve, B.; Abouleish, M.; Shahriar, S.; Mohamed, R.; Gordon, A.L. Impacts of brine disposal from water desalination plants on the physical environment in the Persian/Arabian Gulf. Environ. Res. Commun. 2020, 2, 125003. [Google Scholar] [CrossRef]
- Hosseini, H.; Saadaoui, I.; Moheimani, N.; Al Saidi, M.; Al Jamali, F.; Al Jabri, H.; Hamadou, R.B. Marine health of the Arabian Gulf: Drivers of pollution and assessment approaches focusing on desalination activities. Mar. Pollut. Bull. 2021, 164, 112085. [Google Scholar] [CrossRef]
- Ihsanullah, I.; Atieh, M.A.; Sajid, M.; Nazal, M.K. Desalination and environment: A critical analysis of impacts, mitigation strategies, and greener desalination technologies. Sci. Total. Environ. 2021, 780, 146585. [Google Scholar] [CrossRef]
- Frank, H.; Fussmann, K.E.; Rahav, E.; Bar Zeev, E. Chronic effects of brine discharge form large-scale seawater reverse osmosis desalination facilities on benthic bacteria. Water Res. 2019, 151, 478–487. [Google Scholar] [CrossRef]
- Studnik, H.; Liebsch, S.; Forlani, G.; Wieczorek, D.; Kafarski, P.; Lipok, J. Amino polyphosphonates—Chemical features and practical uses, environmental durability and biodegradation. New Biotechnol. 2015, 32, 1–6. [Google Scholar] [CrossRef]
- Hamed, O.A.; Al-Otaibi, H.A. Prospects of operation of MSF desalination plants at high TBT and low antiscalant dosing rate. Desalination 2010, 256, 181–189. [Google Scholar] [CrossRef]
- Ternan, N.G.; Mc Grath, J.W.; Mc Mullan, G.; Quinn, J.P. Review: Organophosphonates: Occurrence, synthesis and biodegradation by microorganisms. World J. Microbiol. Biotechnol. 1998, 14, 635–647. [Google Scholar] [CrossRef]
- Boels, L.; Witkamp, G.-J. Carboxymethyl inulin biopolymers: A green alternative for phosphonate calcium carbonate growth inhibitors. Cryst. Growth Des. 2011, 11, 4155–4165. [Google Scholar] [CrossRef]
- Nowack, B.; Stone, A.T. Degradation of nitrilotris(methylenephosphonic acid) and related (amino)phosphonate chelating agents in the presence of manganese and molecular oxygen. Environ. Sci. Technol. 2000, 34, 4759–4765. [Google Scholar] [CrossRef]
- Nowack, B. Aminopolyphosphonate removal during wastewater treatment. Water Res. 2002, 36, 4636–4642. [Google Scholar] [CrossRef]
- Steber, J.; Wierich, P. Properties of aminotris (methylenephosphonate) affecting its environmental fate: Degradability, sludge adsorption, mobility in soils, and bioconcentration. Chemosphere 1987, 16, 1323–1337. [Google Scholar] [CrossRef]
- Wang, X.-X.; Zhang, T.-Y.; Dao, G.-H.; Xu, Z.-B.; Wu, Y.-H.; Hu, H.-Y. Assessment and mechanisms of microalgae growth inhibition by phosphonates: Effects of intrinsic toxicity and complexation. Water Res. 2020, 186, 116333. [Google Scholar] [CrossRef]
- Belkin, N.; Kress, N.; Berman-Frank, I. Microbial communities in the process and effluents of seawater desalination plants. In Sustainable Desalination Handbook; Elsevier: Amsterdam, The Netherlands, 2018; pp. 465–488. [Google Scholar]
- Belkin, N.; Rahav, E.; Elifantz, H.; Kress, N.; Berman-Frank, I. The effect of coagulants and antiscalants discharged with seawater desalination brines on coastal microbial communities: A laboratory and in situ study from the southeastern Mediterranean. Water Res. 2017, 110, 321–331. [Google Scholar] [CrossRef]
- Kelaher, B.P.; Clark, G.F.; Johnston, E.L.; Coleman, M.A. Effect of desalination discharge on the abundance and diversity of reef fishes. Environ. Sci. Technol. 2020, 54, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Frank, H.; Rahav, E.; Bar-Zeev, E. Short-term effects of SWRO desalination brine on benthic heterotrophic microbial communities. Desalination 2017, 417, 52–59. [Google Scholar] [CrossRef]
- Sweity, A.; Oren, Y.; Ronen, Z.; Herzberg, M. The influence of antiscalants on biofouling of RO membranes in seawater desalination. Water Res. 2013, 47, 3389–3398. [Google Scholar] [CrossRef]
- Sweity, A.; Zere, T.R.; David, I.; Bason, S.; Oren, Y.; Ronen, Z.; Herzberg, M. Side effects of antiscalants on biofouling of reverse osmosis membranes in brackish water desalination. J. Memb. Sci. 2015, 481, 172–187. [Google Scholar] [CrossRef]
- Prihasto, N.; Liu, Q.-F.; Kim, S.-H. Pre-treatment strategies for seawater desalination by reverse osmosis system. Desalination 2009, 249, 308–316. [Google Scholar] [CrossRef]
- Hermony, A.; Sutzkover-Gutman, I.; Talmi, Y.; Fine, O. Palmachim Seawater desalination plant—Seven years of expansions with uninterrupted operation together with process improvements. Desalination Water Treat. 2015, 55, 2526–2535. [Google Scholar] [CrossRef]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater: 19th Edition Supplement; American Public Health Association: Washington, DC, USA, 1996. [Google Scholar]
- Dorsch, M.; Stackebrandt, E. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J Microbiol Methods 1992, 16, 271–279. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Al Ashhab, A.; Gillor, O.; Herzberg, M. Biofouling of reverse-osmosis membranes under different shear rates during tertiary wastewater desalination: Microbial community composition. Water Res. 2014, 67, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Huber, J.A.; Morrison, H.G.; Sogin, M.L.; Welch, D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007, 8, R143. [Google Scholar] [CrossRef]
- Keylock, C.J. Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos 2005, 109, 203–207. [Google Scholar]
- Hill, T.C.J.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Tsui, T.-H.; Zhang, L.; Zhang, J.; Dai, Y.; Tong, Y.W. Engineering interface between bioenergy recovery and biogas desulfurization: Sustainability interplays of biochar application. Renew. Sustain. Energy Rev. 2022, 157, 112053. [Google Scholar] [CrossRef]
- Paramasivam, S.; Alva, A.K.; Prakash, O.; Cui, S.L. Denitrification in the vadose zone and in surficial groundwater of a sandy entisol with citrus production. Plant Soil 1999, 208, 307–319. [Google Scholar] [CrossRef]
- Mavredaki, E.; Stathoulopoulou, A.; Neofotistou, E.; Demadis, K.D. Environmentally benign chemical additives in the treatment and chemical cleaning of process water systems: Implications for green chemical technology. Desalination 2007, 210, 257–265. [Google Scholar] [CrossRef]
- Nowack, B. Environmental chemistry of phosphonates. Water Res. 2003, 37, 2533–2546. [Google Scholar] [CrossRef]
- Ménesguen, A.; Desmit, X.; Dulière, V.; Lacroix, G.; Thouvenin, B.; Thieu, V.; Dussauze, M. How to avoid eutrophication in coastal seas? A new approach to derive river-specific combined nitrate and phosphate maximum concentrations. Sci. Total Environ. 2018, 628, 400–414. [Google Scholar] [CrossRef]
- Farmer, A.M. Phosphate pollution: A global overview of the problem. In Phosphorus: Polluter and Resource of the Future—Removal and Recovery from Wastewater; Schaum, C., Ed.; International Water Association: London, UK, 2018; pp. 35–55. [Google Scholar]
- Patey, M.D.; Rijkenberg, M.J.A.; Statham, P.J.; Stinchcombe, M.C.; Achterberg, E.P.; Mowlem, M. Determination of nitrate and phosphate in seawater at nanomolar concentrations. TrAC Trends Anal. Chem. 2008, 27, 169–182. [Google Scholar] [CrossRef]
- Jackson, G.A.; Williams, P.M. Importance of dissolved organic nitrogen and phosphorus to biological nutrient cycling. Deep. Sea Res. Part A Oceanogr. Res. Pap. 1985, 32, 223–235. [Google Scholar] [CrossRef]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237. [Google Scholar] [CrossRef]
- Smith, E.M.; Prairie, Y.T. Bacterial metabolism and growth efficiency in lakes: The importance of phosphorus availability. Limnol. Oceanogr. 2004, 49, 137–147. [Google Scholar] [CrossRef]
- Dyhrman, S.; Ammerman, J.; Van Mooy, B. Microbes and the marine phosphorus cycle. Oceanography 2007, 20, 110–116. [Google Scholar] [CrossRef]
- Carlsson, P.; Caron, D.A. Seasonal variation of phosphorus limitation of bacterial growth in a small lake. Limnol. Oceanogr. 2001, 46, 108–120. [Google Scholar] [CrossRef]
- Pinhassi, J.; Gómez-Consarnau, L.; Alonso-Sáez, L.; Sala, M.M.; Vidal, M.; Pedrós-Alió, C.; Gasol, J.M. Seasonal changes in bacterioplankton nutrient limitation and their effects on bacterial community composition in the NW Mediterranean Sea. Aquat. Microb. Ecol. 2006, 44, 241–252. [Google Scholar]
- Bohn, K.; Pavlick, R.; Reu, B.; Kleidon, A. The strengths of r- and K-selection shape diversity-disturbance relationships. PLoS ONE 2014, 9, e95659. [Google Scholar] [CrossRef]
- Boersma, A.S.; Kallscheuer, N.; Wiegand, S.; Rast, P.; Peeters, S.H.; Mesman, R.J.; Heuer, A.; Boedeker, C.; Jetten, M.S.; Rohde, M.; et al. Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie Van Leeuwenhoek, 2020; 113, 1751–1766. [Google Scholar]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Pinto, D.; Vollmers, J.; Rivas-Marín, E.; Kohn, T.; Peeters, S.H.; Heuer, A.; Rast, P.; et al. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 2020, 5, 126–140. [Google Scholar] [CrossRef]
- Lage, O.M.; van Niftrik, L.; Jogler, C.; Devos, D.P. Planctomycetes; Reference module in life sciences; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Rabus, R.; Gade, D.; Helbig, R.; Bauer, M.; Glöckner, F.O.; Kube, M.; Schlesner, H.; Reinhardt, R.; Amann, R. Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2002, 2, 649–655. [Google Scholar] [PubMed]
- Schlesner, H. The Development of Media Suitable for the Microorganisms Morphologically Resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from Various Aquatic Habitats Using Dilute Media. Syst. Appl. Microbiol. 1994, 17, 135–145. [Google Scholar] [CrossRef]
- Wecker, P.; Klockow, C.; Ellrott, A.; Quast, C.; Langhammer, P.; Harder, J.; Glöckner, F.O. Transcriptional response of the model planctomycete Rhodopirellula baltica SH1T to changing environmental conditions. BMC Genom. 2009, 10, 410. [Google Scholar] [CrossRef]
- Tremblay, J.; Greer, C.W. Metagenomic data mining in oil spill studies. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 211–223. [Google Scholar]
- Bhushan, B.; Halasz, A.; Thiboutot, S.; Ampleman, G.; Hawari, J. Chemotaxis-mediated biodegradation of cyclic nitramine explosives RDX, HMX, and CL-20 by Clostridium sp. EDB2. Biochem. Biophys. Res. Commun. 2004, 316, 816–821. [Google Scholar] [CrossRef]
- Ekambaram, S.P.; Perumal, S.S.; Annamalai, U. Decolorization and biodegradation of remazol reactive dyes by Clostridium species. 3 Biotech 2016, 6, 20. [Google Scholar] [CrossRef]
- Kutty, R.; Bennett, G.N. Role of clostridial nitroreductases in bioremediation. In Optimization and Applicability of Bioprocesses; Purohit, H.J., Kalia, V.C., Vaidya, A.N., Khardenavis, A.A., Eds.; Springer: Singapore, 2017; pp. 175–186. [Google Scholar]
- Hashlamon, A.; Ahmad, A.; Choonhong, L. Pre-treatment Methods for Seawater Desalination and Industrial Wastewater Treatment: A Brief Review. Int. J. Sci. Res. Sci. Eng. Technol. 2015, 1, 2394–4099. [Google Scholar]
- Ventresque, C.; Gisclon, V.; Bablon, G.; Chagneau, G. An outstanding feat of modern technology: The Mery-sur-Oise nanofiltration Treatment plant (340,000 m3/d). Desalination 2000, 131, 1–16. [Google Scholar] [CrossRef]
- Hafsi, M. Analysis of Boujdour desalination plant performance. Desalination 2001, 134, 93–104. [Google Scholar] [CrossRef]
- Ansari, A.; Peña-Bahamonde, J.; Fanourakis, S.K.; Hu, Y.; Rodrigues, D.F. Microbially-induced mineral scaling in desalination conditions: Mechanisms and effects of commercial antiscalants. Water Res. 2020, 179, 115863. [Google Scholar] [CrossRef]
- Martínez, A.; Ventouras, L.-A.; Wilson, S.T.; Karl, D.M.; Delong, E.F. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front. Microbiol. 2013, 4, 340. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Ashhab, A.; Sweity, A.; Al-Hadidi, L.; Herzberg, M.; Ronen, Z. Antiscalants Used in Seawater Desalination: Biodegradability and Effects on Microbial Diversity. Microorganisms 2022, 10, 1580. https://doi.org/10.3390/microorganisms10081580
Al-Ashhab A, Sweity A, Al-Hadidi L, Herzberg M, Ronen Z. Antiscalants Used in Seawater Desalination: Biodegradability and Effects on Microbial Diversity. Microorganisms. 2022; 10(8):1580. https://doi.org/10.3390/microorganisms10081580
Chicago/Turabian StyleAl-Ashhab, Ashraf, Amer Sweity, Luna Al-Hadidi, Moshe Herzberg, and Zeev Ronen. 2022. "Antiscalants Used in Seawater Desalination: Biodegradability and Effects on Microbial Diversity" Microorganisms 10, no. 8: 1580. https://doi.org/10.3390/microorganisms10081580
APA StyleAl-Ashhab, A., Sweity, A., Al-Hadidi, L., Herzberg, M., & Ronen, Z. (2022). Antiscalants Used in Seawater Desalination: Biodegradability and Effects on Microbial Diversity. Microorganisms, 10(8), 1580. https://doi.org/10.3390/microorganisms10081580








