Abstract
Quinoline is a typical nitrogen-heterocyclic compound with high toxicity and carcinogenicity which exists ubiquitously in industrial wastewater. In this study, a new quinoline-degrading bacterial strain Rhodococcus sp. JH145 was isolated from oil-contaminated soil. Strain JH145 could grow with quinoline as the sole carbon source. The optimum growth temperature, pH, and salt concentration were 30 °C, 8.0, and 1%, respectively. 100 mg/L quinoline could be completely removed within 28 h. Particularly, strain JH145 showed excellent quinoline biodegradation ability under a high-salt concentration of 7.5%. Two different quinoline degradation pathways, a typical 8-hydroxycoumarin pathway, and a unique anthranilate pathway were proposed based on the intermediates identified by liquid chromatography–time of flight mass spectrometry. Our present results provided new candidates for industrial application in quinoline-contaminated wastewater treatment even under high-salt conditions.
1. Introduction
Quinoline and its derivatives are common nitro-heterocyclic compounds in industry, which are refractory and widely distributed in industrial wastewater, such as coking wastewater and pulping wastewater [1,2,3,4,5,6]. Compared with other polycyclic aromatic hydrocarbons, quinoline more easily enters groundwater and soil due to its unique heterocyclic structure. Quinoline was identified as a toxic pollutant due to its carcinogenic and mutagenic properties. At present, the removal of quinoline pollutants from wastewater is of great interest.
Generally, quinoline has been removed by physical, chemical, or biological methods [7]. Among these methods, the physical methods have generally included adsorption, dissolution, and filtration [8]. A variety of chemical technologies have been applied for removing wastewater containing quinoline, such as wet oxidation [9,10] and ultraviolet (UV) photolysis [11,12,13]. Based on the present status of quinoline wastewater treatment, several methods have usually be combined to improve the removal efficiency. However, compared with physical or chemical methods, biological treatment has become a promising technology for removing various refractory pollutants, including quinoline, due to its merits that include less chemical agents, equipment, and secondary pollution [14,15,16,17].
Microorganisms play a major role in the biodegradation of quinoline in the environment. Several quinoline-degrading strains under aerobic or anaerobic conditions have been reported in recent years, such as Burkholderia sp. [18,19], Bacillus sp. [20], Brevundimonas sp. [21], Rhodococcus rhodochrous [22], Comamonas sp. [23,24], and Pseudomonas sp. [25,26]. However, most quinoline-degrading bacteria has shown relatively low adaptability and low degradation efficiency. Furthermore, many industrial wastewaters contain high concentrations of salt components, thus, the salt tolerance of bacterial strains plays an important role in wastewater treatment [6]. Nevertheless, high salt-tolerating quinoline-degrading bacteria have been investigated poorly in previous studies.
In this study, a new quinoline-degrading bacterial strain JH145 with high efficiency and halotolerance was isolated. The characteristics of quinoline degradation by strain JH145 were examined under different cultural conditions. The degrading intermediates were investigated and the pathways were proposed. The present findings will improve our understanding of quinoline biodegradation in a different environment.
2. Materials and Methods
2.1. Chemicals and Media
Quinoline (96% purity) and 2-hydroxyquinoline (97% purity) were purchased from Macklin Biochemical (Shanghai, China). These chemicals were dissolved in deionized water before use. Methanol used in high-performance liquid chromatography (HPLC) was purchased from Merck KGaA (Darmstadt, Germany). Luria-Bertani (LB) medium used to enrich bacteria contained: tryptone (10.0 g/L), yeast extract (5.0 g/L), and NaCl (10.0 g/L), at pH 7.0. The mineral salt medium (MSM) used for the bacterial growth and bacterial isolation contained (NH4)2SO4 (1.0 g/L), KH2PO4·2H2O (0.5 g/L), MgSO4·7H2O (0.2 g/L), K2HPO4·3H2O (1.5 g/L), NaCl (1.0 g/L), at pH 7.0. The corresponding solid media were prepared by adding 1.8% agar powder. All the above culture media needed to be sterilized in an autoclave (121 °C, 25 min) before being used.
2.2. Enrichment and Isolation of Bacteria
Oil-contaminated soils were collected to isolate quinoline-degrading bacteria. First, the oil-contaminated soil (35°21′–37°31′ N, 107°41′–110°31′ E) from an oil field was added into the sterile MSM (250 mL) containing 100 mg/L quinoline and then incubated in a shaker at 30 ℃ and 180 rpm. After cultivation for 7 days, the cell suspension (10 mL) was transferred and inoculated into another fresh MSM (250 mL) with 100 mg/L quinoline. The above cultures were repeated 5 times, then serially diluted and coated in MSM plates with 100 mg/L quinoline. The plates were incubated at 30 °C for several days. After visible colonies were formed on plates, individual colonies with different morphology were selected and strewn on MSM plates with 100 mg/L quinoline. The removal capacity of quinoline by the purified strain was verified. Finally, a strain of Rhodococcus sp. JH145 with high quinoline-degrading efficiency was selected for further study.
2.3. Identification of Strain JH145
The high-salt precipitation method was used to extract the genomic DNA of strain JH145. The 16S rRNA gene was amplified using the universal primers 27F and 1492R [27]. The amplified PCR product was purified and recovered with the gel recovery kit of Nanjing Novoz Biotech (Nanjing, China) Co., Ltd. The qualified product was further connected to pMD19-T and then was introduced into E. coli DH5α. Positive clones were screened on Luria-Bertani (LB) plates containing 100 mg/L ampicillin and then sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The obtained sequence was compared with those of the related microorganisms by BLAST and then identified as Rhodococcus sp. Finally, the phylogenetic tree of this strain was constructed with MEGA6 by using the neighbor-joining method.
2.4. Characteristics of Quinoline Biodegradation by Strain JH145
The strain JH145 was cultured in LB, collected, and washed twice with MSM. The cells were suspended at OD600 of 2.0. Then the degradation ability of strain under various environmental conditions was investigated in 250 mL Erlenmeyer flasks. First, the fresh bacterial solution was added in 100 mL MSM containing 100 mg/L quinoline with 1% inoculum. The biomass was measured by OD600nm and degradation of quinoline was monitored at the same time by sampling regularly.
Next, tests on the effects of temperature were examined at five levels 15, 20, 30, 37, and 45 °C, at pH 7.0. Then these were followed by tests on the effects of pH at six levels from pH 5.0 to 10.0 at the identified optimal temperature. Finally, at the optimal temperature and pH, the effects of salt concentration levels (1% to 10% [w/v]) on quinoline degradation were tested. All samples were tested in triplicate.
2.5. Detection of Quinoline Metabolites and Analysis Method
Samples at regular intervals of 4 h within 2 days were taken out from Erlenmeyer flasks, centrifuged for 5 min at 12,000 rpm, and then the supernatant was gathered. The concentrations of quinoline and its intermediates in the supernatant were analyzed by high-performance liquid chromatography (HPLC), using a Syncronis C18 column (4.6 μm × 250 mm × 5 μm). The methanol solution with (40:60, v/v) ultrapure water containing 1% formic acid was used as mobile phase at the flow rate of 1.0 mL min-1 by 20 μL injection volume. The UV detector was set at 275 nm.
To detect the metabolism intermediates of quinoline, the supernatant from the Erlenmeyer flask was lyophilized, dissolved by methanol, and then analyzed by liquid chromatography–time of flight mass spectrometry (LC–TOF/MS). By analyzing the positive and negative ion patterns, the metabolism intermediates of quinoline were determined by comparing literature and the standards [28].
3. Results and Discussion
3.1. Identification of Quinoline Degrading Bacteria
Quinoline was used as the sole carbon source for the enrichment culture. The fifth-generation enrichment solutions were applied for gradient dilution. Through continuous streaking and purification on the MSM plate with 100 mg/L quinoline as the sole carbon source, a single strain named JH145 was finally obtained. Strain JH145 is a Gram-positive bacteria, and its colony appears red, circular, slightly convex, and surface drying. Strain JH145 could degrade tyrosine but not casein, hypoxanthine, or xanthine. It hydrolysed aesculin weakly and isindole- and urease-negative. Meanwhile, strain JH145 was resistant to streptomycin and ampicillin, but susceptible to kanamycin and gentamicin. The strain was also positive for catalase and could grow in the presence of lysozyme, without production of H2S. At the same time, the phenotype and colony characteristics of this strain were very similar to those of Rhodococcus [20].
The BLAST search of the 16S rRNA gene sequence indicated that strain JH145 was closely related to the species in the genus of Rhodococcus, and also exhibited the highest similarity to Rhodococcus gordoniae (99.64%) and Rhodococcus pyridinivorans (98.79%) (Figure 1). Based on phenotypic and 16S rRNA gene phylogenetic analysis, the strain JH145 was identified as Rhodococcus sp. According to previous research reports, some strains in Rhodococcus can degrade quinoline under aerobic culture conditions, such as Rhodococcus pyridinivorans [29], Rhodococcus sp. Bl [30], and Rhodococcus rhodochrous [22]. The quinoline-degrading bacterial strains were diverse, and strain JH145 is the first strain of Rhodococcus gordoniae. reported to degrade quinoline.
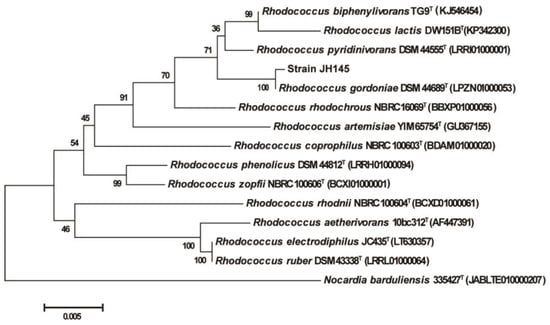
Figure 1.
Phylogenetic tree constructed from the 16S rRNA sequence of strain JH145 and some other strains by the neighbor-joining method.
3.2. Growth Status and Degradation Characteristics of Strain JH145
3.2.1. Study on the Growth Profile of Quinoline-Degrading Strain JH145
The bacterial suspension with 1% inoculum was added into the 100 mL sterile MSM with 100 mg/L quinoline, at pH 7.0, and then the flask was cultured in a shaker at 30 °C and 180 rpm. Figure 2a shows that 100 mg/L quinoline was degraded completely within 30 h. Meanwhile, the bacterial cells were grown up to OD600nm of 0.27 ± 0.02. The growth rate of strain JH145 was slow in the first 14 h due to the adaptation period to this medium. After 14 h, the growth rate of the strain increased significantly, indicating that strain JH145 entered the logarithmic growth phase and removed quinoline efficiently, and utilized it as the carbon source for cell growth.
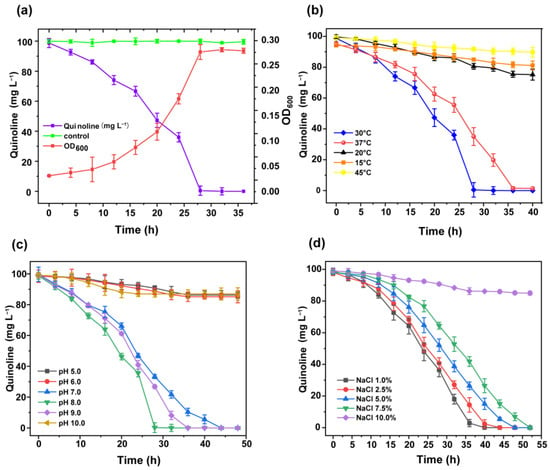
Figure 2.
Effect of JH145 on quinoline degradation under different conditions (a) The cell growth and degradation characteristics of strain JH145 with quinoline as the sole carbon source. (b) Effects of temperature on the quinoline biodegradation of strain JH145. (c) Effects of initial pH on the quinoline biodegradation of strain JH145. (d) Effects of NaCl on the quinoline biodegradation of strain JH145.
3.2.2. Study on the Degradability of Quinoline by Temperature
Temperature is a significant factor for quinoline degradation in the environment. As shown in Figure 2b, 100 mg/L quinoline was degraded completely by strain JH145 within 36 h at 30 °C and 37 °C, respectively. The quinoline degradation at 37 °C was a little slower than that at 30 °C. However, quinoline was hardly degraded by strain JH145 at 15 ℃, 20 °C, and 45 °C. The degradation rates were less than 20% of 100 mg/L quinoline. The order of quinoline degradation rates was as follows: 30 °C > 37 °C >> 20 °C > 45 °C > 15 °C. It might be due to the fact that strain JH145 is a mesophilic bacteria and the enzymes involved in quinoline biodegradation were temperature-sensitive.
3.2.3. Study on the Degradability of Quinoline by pH
The MSM with different pH values of 5.0, 6.0, 7.0, 8.0, 9.0. and 10.0 were adjusted by a 50 mM NaH2PO4-Na2HPO4 buffer system. The temperature used for degradation was 30 °C as investigated above. Figure 2c illustrates that 100 mg/L quinoline was degraded completely by strain JH145 at the initial pH of 7.0, 8.0, and 9.0. The optimum pH value for quinoline degradation was 8.0. The order of quinoline degradation rates was as follows: pH 7.0 > pH 8.0 > pH 9.0. Therefore, the strain JH145 could degrade quinoline well in neutral and light alkaline environments. In contrast, strain JH145 never degraded the added quinoline at the initial pH of 5.0, 6.0, and 10.0, indicating that extreme acidic or alkaline conditions were harmful to strain JH145. Since the actual growth environment of the bacteria was mostly a neutral environment, the following experimental conditions were set at pH 7.0.
3.2.4. Study on Salt Tolerance of Strain JH145
Figure 2d shows the quinoline removal by strain JH145 at different initial concentrations (1.0%, 2.5%, 5.0%, 7.5%, and 10%) of NaCl. 100 mg/L quinoline was degraded well with 1.0~7.5% NaCl within 50 h by strain JH145. With the increase of NaCl concentration from 1.0% to 7.5%, the removal rate of quinoline slightly decreased gradually. Nevertheless, only approximately 15% of the initial quinoline was consumed at 50 h when NaCl reached 10.0%, probably due to the inhibition effects of NaCl concentration on biodegradability.
In the current wastewater treatment by bacteria, high salt concentration conditions are a common problem [30]. Bacteria does not degrade pollutions in those high salt concentration conditions probably due to the high osmotic pressure of salt. Our present results showed that strain JH145 could remove quinoline efficiently under NaCl concentration of up to 7.5%, indicating the superiority of strain JH145 in industrial treatment high-salt concentration quinoline wastewater.
3.3. Identification of Metabolites and Proposal of Quinoline Aerobic Catabolism Pathway
HPLC was used to detect the possible catabolic intermediates during quinoline degradation. As shown in Figure 3, a new peak around 4.86 min appeared when quinoline (6.82 min) degraded and finally disappeared. LC-MS results of this new peak showed a molecular ion peak at m/z 144.0463 [M-H] and fragment ion peak at 116.0518 [M-H], indicating the adding of the oxygen atom. Generally, the first step of the aerobic degradation of quinoline was 2-hydroxyquinoline as shown in Figure 4B [23,29], reported in almost all quinoline biodegradation pathways. HPLC results showed that the retention time of the new peak was equal to that of the commercial 2-hydroxyquinoline standard. Thus, the new peak was identified as 2-hydroxyquinoline.
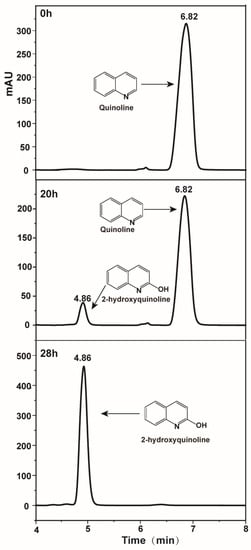
Figure 3.
HPLC chromatograms of quinoline and 2-hydroxyquinoline.
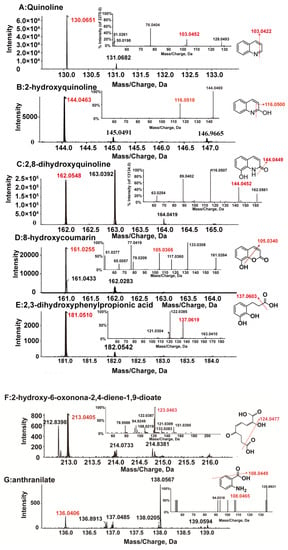
Figure 4.
HPLC–MS mass spectrograms and proposed structures of quinoline and its metabolites.
Although only one visible peak emerged in HPLC analyses, several trace amount intermediates were detected by HPLC–MS. As shown in Figure 4, five more metabolites were detected. According to their molecular and fragment ion peaks, these intermediates were proposed as 2-hydroxyquinoline (Figure 4B), 2,8-dihydroxyquinoline (Figure 4C), 8-hydroxycoumarin (Figure 4D), 2,3-dihydroxyphenylpropionic acids (Figure 4E), 2-hydroxy-6-oxonona-2,4-diene-1,9-dioate (Figure 4F), and anthranilate (Figure 4G). Based on these intermediates, two different quinoline biodegradation pathways in strain JH145 were proposed (Figure 5). In the first path, 2-hydroxyquinoline was converted to 2,8-dihydroxyquinoline. Afterward, 2,8-dihydroxyquinoline was converted to 8-hydroxycoumarin and accompanied by NH4+, which underwent ring-opening to form 2,3-dihydroxyphenylpropionic acids. Thereafter, the catabolic compound was meta-cleaved to yield 2-hydroxy-6-oxonona-2,4-diene-1,9-dioate, followed by downstream metabolism ending up in the citrate cycle (TCA). In another pathway, the quinoline cleaved the ring to form an anthranilate. As no other intermediates were observed in the experiments, subsequent metabolites of anthranilate were unclear.
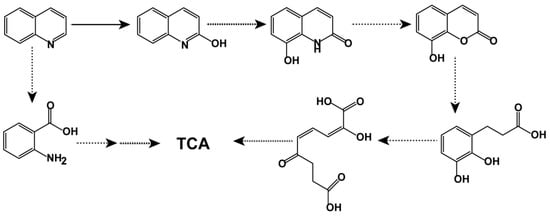
Figure 5.
The proposed quinoline biodegradation pathway of strain JH145.
As shown in previous studies, two common quinoline degradation pathways named the 5,6-dihydroxy-2(1H) quinolinone pathway and the 8-hydroxycoumarin pathway, exist in quinoline-degrading microorganisms [31,32]. Bacteria usually employ one quinoline degradation pathway, for example, Comamonas with the 8-hydroxycoumarin pathway [23]. Recently, quinoline was found to be removed by two pathways in one strain, such as Rhodococcus [28] and Pseudomonas putida [33]. In this study, we found that the 8-hydroxycoumarin pathway and, interestingly, a unique anthranilic acid pathway were used by Rhodococcus sp. JH145. This unique anthranilic acid degradation pathway did not appear in Gram-positive bacteria but was reported in a Gram-negative bacterium Comamonas sp. recently [34]. Considering the significant differences of Gram-positive and Gram-negative bacteria, more studies of Rhodococcus sp. JH145 are expected in our future research.
4. Conclusions
In this study, a new quinoline-degrading strain JH145 was isolated from oil-contaminated soil. The strain was identified as Rhodococcus sp. based on its morphology and 16S rRNA gene sequence. Strain JH145 could grow with quinoline as the sole carbon source. The optimum growth temperature, pH, and salt concentration were 30 °C, 8.0 and 1%, respectively, and 100 mg/L quinoline could be completely degraded within 28 h. Particularly, strain JH145 showed excellent quinoline biodegradation ability under a high-salt concentration of 7.5%. Two different quinoline degradation pathways, a typical 8-hydroxycoumarin pathway, and a unique anthranilate pathway were proposed based on the intermediates identified by HPLC–MS. Our present results provided new candidates for industrial application in quinoline-contaminated wastewater treatment even under high-salt conditions.
Author Contributions
Conceptualization, C.C. and J.H.; methodology, S.X. and P.Y.; software, S.X. and X.W.; validation, Q.H. and C.C.; formal analysis, Y.J., X.Z., Q.H. and J.H.; investigation, Y.J., F.Z., S.X. and P.Y.; resources, J.Q., C.C. and J.H.; data curation, Y.J. and F.Z.; writing—original draft preparation, Y.J.; writing—review and editing, Q.H., J.Q., C.C. and J.H.; visualization, Y.J., X.W. and X.Z.; supervision, J.Q. and J.H.; project administration, Q.H., J.Q. and J.H.; funding acquisition, C.C., J.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Cuiwei Chu, grant number [Nos. 31800097 and 32070092].
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 31800097 and 32070092).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Felczak, A.; Zawadzka, K.; Lisowska, K. Efficient biodegradation of quinolone-Factors determining the process. Int. Biodeter. Biodegr. 2014, 96, 127–134. [Google Scholar] [CrossRef]
- Shi, J.; Han, H.; Xu, C. A novel enhanced anaerobic biodegradation method using biochar and Fe (OH)3@biochar for the removal of nitrogen heterocyclic compounds from coal gasification wastewater. Sci. Total Environ. 2019, 697, 134052. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, R.; Liu, C.; Chen, L. Bioaugmentation with isolated strains for the removal of toxic and refractory organics from coking wastewater in a membrane bioreactor. Biodegradation 2015, 26, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Chen, Q.; Parales, R.E.; Lu, Z.; Hong, Q.; He, J.; Qiu, J.; Jiang, J. Bacterial catabolism of nicotine: Catabolic strains, pathways and modules. Environ. Res. 2020, 183, 109258. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Meng, N.; Li, Y.; Wang, J. Occurrence, impacts, and microbial transformation of 3-methylindole (skatole): A critical review. J. Hazard. Mater. 2021, 416, 126181. [Google Scholar] [CrossRef]
- Shaikhulova, S.; Fakhrullina, G.; Nigamatzyanova, L.; Akhatova, F.; Fakhrullin, R. Worms eat oil: Alcanivorax borkumensis hydrocarbonoclastic bacteria colonise Caenorhabditis elegans nematodes intestines as a first step towards oil spills zooremediation. Sci. Total Environ. 2021, 761, 143209. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Li, S.; Li, B. Removal of Toxic Organic Pollutants from Coke Plant Wastewater by UV-Fenton. J. Residuals Sci. Technol. 2016, 13, S143–S148. [Google Scholar] [CrossRef][Green Version]
- Rameshraja, D.; Srivastava, V.C.; Kushwaha, J.P.; Mall, I.D. Quinoline adsorption onto granular activated carbon and bagasse fly ash. Chem. Eng. J. 2012, 181–182, 343–351. [Google Scholar] [CrossRef]
- Zhu, H.; Ma, W.; Han, H.; Xu, C.; Han, Y.; Ma, W. Degradation characteristics of two typical N-heterocycles in ozone process: Efficacy, kinetics, pathways, toxicity and its application to real biologically pretreated coal gasification wastewater. Chemosphere 2018, 209, 319–327. [Google Scholar] [CrossRef]
- Singh, L.; Rekha, P.; Chand, S. Comparative evaluation of synthesis routes of Cu/zeolite Y catalysts for catalytic wet peroxide oxidation of quinoline in fixed-bed reactor. J. Environ. Manag. 2018, 215, 1–12. [Google Scholar] [CrossRef]
- Bai, Q.; Yang, L.; Li, R.; Chen, B.; Zhang, L.; Zhang, Y.; Rittmann, B.E. Accelerating Quinoline Biodegradation and Oxidation with Endogenous Electron Donors. Environ. Sci. Technol. 2015, 49, 11536–11542. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Yu, S.; Wang, J. Degradation of pyridine and quinoline in aqueous solution by gamma radiation. Radiat. Phys. Chem. 2018, 144, 322–328. [Google Scholar] [CrossRef]
- Yi, R.; Yi, C.; Du, D.; Zhang, Q.; Yu, H.; Yang, L. Research on quinoline degradation in drinking water by a large volume strong ionization dielectric barrier discharge reaction system. Plasma Sci. Technol. 2021, 23, 085505. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, Y. State of the art of biological processes for coal gasification wastewater treatment. Biotechnol. Adv. 2016, 34, 1442. [Google Scholar] [CrossRef]
- Oberoi, A.S.; Philip, L.; Bhallamudi, S.M. Biodegradation of Various Aromatic Compounds by Enriched Bacterial Cultures: Part B-Nitrogen-, Sulfur-, and Oxygen-Containing Heterocyclic Aromatic Compounds. Appl. Biochem. Biot. 2015, 176, 1746–1769. [Google Scholar] [CrossRef]
- Zhuang, H.; Han, H.; Xu, P.; Hou, B.; Jia, S.; Wang, D.; Li, K. Biodegradation of quinoline by Streptomyces sp N01 immobilized on bamboo carbon supported Fe3O4 nanoparticles. Biochem. Eng. J. 2015, 99, 44–47. [Google Scholar] [CrossRef]
- Wu, B.; Wang, J.; Hu, Z.; Yuan, S.; Wang, W. Anaerobic biotransformation and potential impact of quinoline in an anaerobic methanogenic reactor treating synthetic coal gasification wastewater and response of microbial community. J. Hazard. Mater. 2020, 384, 121404. [Google Scholar] [CrossRef]
- Wang, J.; Han, L.; Shi, H.; Qian, Y. Biodegradation of quinoline by gel immobilized Burkholderia sp. Chemosphere 2001, 44, 1041–1046. [Google Scholar] [CrossRef]
- Wang, J.; Quan, X.; Han, L.; Qian, Y.; Hegemann, W. Microbial degradation of quinoline by immobilized cells of Burkholderia pickettii. Water Res. 2002, 36, 2288–2296. [Google Scholar] [CrossRef]
- Tuo, B.; Yan, J.; Fan, B.; Yang, Z.; Liu, J. Biodegradation characteristics and bioaugmentation potential of a novel quinoline-degrading strain of Bacillus sp. isolated from petroleum-contaminated soil. Bioresour. Technol. 2012, 107, 55–60. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, M.; Cheng, F.; Geng, Q. Biodegradation characterization and immobilized strains’ potential for quinoline degradation by Brevundimonas sp. K4 isolated from activated sludge of coking wastewater. Biosci. Biotechnol. Biochem. 2015, 79, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Dinamarca, M.A.; Eyzaguirre, J.; Baeza, P.; Aballay, P.; Canales, C.; Ojeda, J. A new functional biofilm biocatalyst for the simultaneous removal of dibenzothiophene and quinoline using Rhodococcus rhodochrous and curli amyloid overproducer mutants derived from Cobetia sp. strain MM1IDA2H-1. Biotechnol. Rep. 2018, 20, e286. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Chen, F.; Fu, J.; Sheng, G.; Sun, G. Microbial metabolism of quinoline by Comamonas sp. World J. Microb. Biot. 2004, 20, 539–543. [Google Scholar] [CrossRef]
- Sugaya, K.; Nakayama, O.; Hinata, N.; Kamekura, K.; Ito, A.; Yamagiwa, K.; Ohkawa, A. Biodegradation of quinoline in crude oil. J. Chem. Technol. Biot. 2001, 76, 603–611. [Google Scholar] [CrossRef]
- Bai, Y.; Sun, Q.; Zhao, C.; Wen, D.; Tang, X. Quinoline biodegradation and its nitrogen transformation pathway by a Pseudomonas sp strain. Biodegradation 2010, 21, 335–344. [Google Scholar] [CrossRef]
- Qiao, L.; Wang, J. Biodegradation characteristics of quinoline by Pseudomonas putida. Bioresour. Technol. 2010, 101, 7683–7686. [Google Scholar] [CrossRef]
- Qiu, J.; Zhang, J.; Zhang, Y.; Wang, Y.; Tong, L.; Hong, Q.; He, J. Biodegradation of Picolinic Acid by a Newly Isolated Bacterium Alcaligenes faecalis Strain JQ135. Curr. Microbiol. 2017, 74, 508–514. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Bao, Y.; Zhang, K.; Qiu, J.; He, Q.; Zhu, J.; He, J. Enhanced degradation of dicamba by an anaerobic sludge acclimated from river sediment. Sci. Total Environ. 2021, 777, 145931. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, D.; Fan, L.; Ni, J. Degradation of quinoline by Rhodococcus sp. QL2 isolated from activated sludge. J. Hazard. Mater. 2008, 160, 289–294. [Google Scholar] [CrossRef]
- Oren, A. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 2011, 13, 1908–1923. [Google Scholar] [CrossRef]
- Luo, Y.; Yue, X.; Wei, P.; Zhou, A.; Kong, X.; Alimzhanova, S. A state-of-the-art review of quinoline degradation and technical bottlenecks. Sci. Total Environ. 2020, 747, 141136. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J.; Feng, Y.C.; Bollag, J. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 1996, 60, 483. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Bauder, R.; Speer, M.; Rommel, T.O.; Lingens, F. Microbial-metabolism of quinoline and related-compounds II. degradation of quinoline by Pseudomonas fluorescens 3, Pseudomonas-putida 86 and Rhodococcus spec b1. Biol. Chem. Hoppe-Seyler 1989, 370, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Wu, M.; Tang, Q.; Song, Z.; Zhou, H.; Bao, Y.; Liu, L.; Qu, Y. Comparative characterization and functional genomic analysis of two Comamonas sp. strains for biodegradation of quinoline. J. Chem. Technol. Biot. 2020, 95, 2017–2026. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).