Abstract
Exercise reduces inflammation, fatigue, and aids overall health. Additionally, physical fitness has been associated with desirable changes in the community composition of the athlete gut microbiome, with health-associated taxa being shown to be increased in active individuals. Here, using a combination of in silico and in vitro methods, we investigate the antimicrobial activity of the athlete gut microbiome. In vitro approaches resulted in the generation of 284 gut isolates with inhibitory activity against Clostridioides difficile and/or Fusobacterium nucleatum, and the most potent isolates were further characterized, and potential bacteriocins were predicted using both MALDI-TOF MS and whole-genome sequencing. Additionally, metagenomic reads from the faecal samples were used to recover 770 Metagenome Assembled Genomes (MAGs), of which 148 were assigned to be high-quality MAGs and screened for the presence of putative bacteriocin gene clusters using BAGEL4 software, with 339 gene clusters of interest being identified. Class I was the most abundant bacteriocin class predicted, accounting for 91.3% of predictions, Class III had a predicted abundance of 7.5%, and Class II was represented by just 1% of all predictions.
Keywords:
bacteriocins; microbiome; antimicrobial-peptides; athletes; in vitro; in silico; metagenomics 1. Introduction
Physical fitness has been associated with a better quality of life and, in general, fewer reported days of illness [1]. Exercise has also been shown to have beneficial effects concerning risk reduction of cardiovascular disease [2], anti-inflammatory potential [3], mental health including depression [4], and microbiome modulation [5,6,7,8,9].
The intestinal microbiome is a rich and diverse ecosystem collectively composed of 100 trillion cells, including bacterial, fungal, viral, and archaeal cells [10], which can cooperate/compete with each other and the host [11]. In the last decade, it has been well-documented that athletes have a more diverse microbiome when compared to non-athletes, often associated with differences in the relative abundance of certain bacterial taxa, including but not restricted to Akkermansia, Faecalibacterium, Prevotella, and Veillonella [5,8,9,12]. There is some evidence that these changes in the athlete microbiome arise as a result of a long-term adaptation, as opposed to a short-term exercise intervention. Indeed, Cronin and colleagues investigated the impact of an 8-week exercise regime and found that changes in the microbiome were subtle [6]. These recent findings have led us to hypothesise that the gut microbiome of elite athletes could be a possible source of antimicrobial peptide (AMP) producing bacteria and could potentially be exploited to harness bacteria with potential novel functions and probiotic traits (i.e., bacteriocin production).
Bacteriocins are small, heat-stable, ribosomally-synthesised antimicrobial peptides produced by one bacteria that are active against other bacteria, to which the producer strain is immune [12]. Antimicrobial peptides and especially bacteriocins have received increasing interest due to their potential applications in the treatment of bacterial infections owing to the growing threat of antimicrobial resistance (AMR). Bacteriocins and bacteriocin-producing bacteria are promising tools regarding preventing or treating target bacterial infections as many bacteriocins have a narrow spectrum of activity, ergo causing minimal disruption to the microbiome as a whole [13,14]. Indeed, bacteriocin production can be regarded as a desirable probiotic trait as it can aid in (a) inhibiting the growth of various pathogens in the gut [15], (b) support the colonization of desirable species in the gut [16], and (c) act as signaling peptides through quorum sensing systems [17]. Quorum sensing systems play a vital role in biofilm formation, which could prolong the resident time of probiotic bacteria in the gut [18] and therefore influence the host’s health. Past studies have shown that bacteriocins are attractive alternatives to antibiotic treatment [19,20,21], and such therapies should be further studied. Bacteriocin functionality is reliant on several genes working in tandem. At a minimum, a functional bacteriocin gene cluster needs a structural gene and an immunity gene (to protect the bacteriocin producer strain) [22,23].
The intestinal microbiota is one of the richest storehouses for bacteriocin-producing bacteria. Two previous data mining projects have investigated the prevalence of bacteriocins in the human gut. Both studies have identified bacteriocin gene clusters within the Firmicutes, Proteobacteria, Bacteroides, and Actinobacteria phyla, highlighting the abundance and diversity of bacteriocins in the gut [24,25]. Another study investigated bacteriocin diversity on several body sites, including the gut microbiome. They found that the gut had a high abundance of bacteriocin producers from the following species; Bacteroides fragilis, Bacteroides dorei, Eubacterium rectale, Escherichia coli, and Blautia hansenii [26]. Previous work in the literature has shown the applications of bacteriocins in controlling important pathogens. A murine study by Corr et al. found that a Lactobacillus salivarius UCC118 gut isolate produced an Abp118 bacteriocin, which protected the mice against a Listeria monocytogenes infection through direct antagonism [16]. Similarly, Bacteriocin 21, produced by a gut commensal Enterococcus faecalis, has been shown to inhibit the growth of vancomycin-resistant enterococci in a mouse model [27]. Other in vitro studies have also demonstrated the impact of bacteriocin-producing strains on known human pathogens, namely Clostridioides difficile [28], Salmonella spp. [16,29], Enterococcus faecium [30], Helicobacter pylori [31], Bacillus cereus [32] and Campylobacter spp. [33].
For the purpose of this study, we selected two important gut pathogens, Fusobacterium nucleatum and C. difficle, as our indicator strains to mine the athlete gut microbiome for producers of antimicrobials. C. difficle is a Gram-positive [34] enteric pathogen causing C. difficile-associated diarrhoea (CDAD) [35]. It has also been shown to have the ability to disrupt the gut microbiota of colonised persons [36]. It has become clear that novel treatment options should be sought, with one study suggesting the mortality rate of CDAD can reach as high as 25% in the elderly populations [37]. It has been shown to affect both the elderly and younger immunocompromised populations [38]. F. nucleatum is a Gram-negative bacterium [39] associated with several intestinal pathologies, including colorectal cancer (CRC) development and progression [40]. There is considerable merit in identifying antimicrobials that could contribute to novel treatment options to control these targets. This study aimed to screen the microbiome of an Irish athlete cohort for potential novel bacteriocins, using both in vitro and in silico based approaches. Our in silico approach looked at the reconstruction of Metagenome Assembled Genomes (MAGs) from the sequenced faecal samples, followed by predicting bacteriocin gene clusters present within the assemblies using the BAGEL4 software. We have then used athlete faecal samples associated with the recovered MAGs to screen the gut microbiome of an athlete cohort for novel bacteriocin producers in vitro (Figure 1).
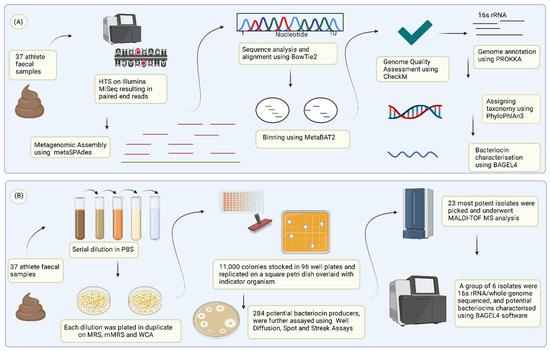
Figure 1.
In silico and in vitro based approaches used in this study to identify potential novel bacteriocins from the athlete’s gut. (A) Metagenomic data from 37 faecal samples in the form of paired-end reads were assembled, annotated, quality-checked, and binned to recover Metagenome-Assembled Genomes (MAGS) analysed using BAGEL4 for the presence of potential bacteriocin genes. (B) 37 faecal samples from elite Irish athletes were screened for novel bacteriocin-producing gut isolates. Potential bacteriocin producers were assayed further, and the spectrum of inhibition was assessed. Isolates exhibiting potential antimicrobial activity were brought forward for MALDI-TOF mass spectrophotometry, whole-genome sequencing (WGS), and bacteriocin biosynthetic gene clusters were predicted using BAGEL4 software. (Figure created with https://BioRender.com, accessed on 15 February 2022).
2. Materials and Methods
2.1. Subject Recruitment and Sample Collection
An existing bank of elite athlete faecal samples was used in this study (O’Donovan et al. [7]). The recruitment criteria were as follows: Irish athletes/athletes representing Ireland, preparing for and/or participating in international competitions (including the Olympics). The ethical approval for the study was granted by the clinical research ethics committee (Project code: APC073). All subjects gave written informed consent before the study. Stool samples were collected from male (n = 23) and female (n = 14) athletes and stored anaerobically at −80 °C prior to culture-based analysis.
2.2. Recovery of Metagenome Assembled Genomes (MAGs) and Antimicrobial Peptide Production Analysis
Metagenomic data from 37 faecal samples in the form of paired-end reads were obtained from a previous study [7]. The raw data are available in the European Nucleotide Archive (ENA) under the accession number PRJEB32794. Human reads were removed with BMTagger [41], the resulting shotgun fastq files were converted to BAM files using Picard Tools (http://broadinstitute.github.io/picard/). The BAM files were then quality trimmed and duplicates removed using SAMTools v1.9 [42]. Metagenome assembly was performed de novo using MetaSPAdes assembler 3.13 [43], followed by sequence analysis and alignment using BowTie2 v.2.3.4 [44]. Genome binning was performed using MetaBAT2 [45]. The quality (completeness and contamination) of constructed MAGs was determined using CheckM [46]. MAGs with <90% Completeness and >5% Contamination were deemed Low-Quality and were dismissed from further analysis, while those with >90% Completeness and <5% Contamination were deemed High-Quality and were brought forward for further analysis [47]. High-Quality MAGs were annotated using PROKKA v.1.13 [48] and assigned taxonomy with PhyloPhlan3 v.3.0, SGB.Dec19 database, using the default 5% as the assignment threshold [49]. The presence of antimicrobial peptide gene clusters was assessed using BAGEL4 [50]. A gene set was considered a putative bacteriocin gene cluster if it contained a minimum of: transport/immunity gene, modification gene (for post-translationally modified peptides), leader-cleavage peptide, and a structural peptide [23].
2.3. Isolation of Bacterial Isolates Producing Antimicrobial Peptides
For this study, culturing conditions were used to target the isolation of bifidobacteria, lactobacilli, and culturable gut anaerobic species, whereby one gram of frozen faecal sample was suspended in Phosphate Buffered Saline (PBS) and serially diluted ten-fold. Each dilution was spread-plated onto various selective agars with a final agar concentration of 1.5% (w/v). Bifidobacteria were isolated anaerobically on De Man, Rogosa, and Sharpe agar (MRS; BDTM DifcoTM Trafalgar Scientific Ltd., Leicester, UK) supplemented with 0.05% (w/v) L-cysteine hydrochloride (Sigma, London, UK) (noted as mMRS agar). mMRS agar was further supplemented with Mupirocin (Sigma, London, UK) at 200 µg/mL of medium. Lactobacillus species were isolated on LBS agar (BDTM DifcoTM Trafalgar Scientific Ltd.,Leicester, UK) aerobically and anaerobically. Obligate anaerobic species were isolated on Wilkins-Chalgren Media (Sigma, London, UK) in an anaerobic chamber. Plates were incubated at 37 °C for 24–48 h. The isolates were sub-cultured in their respective liquid growth medium with 10% (v/v) glycerol and stored in 96 well plates at −80 °C.
2.4. Antimicrobial Activity Assays
Frozen bacterial stocks were replicated into 96 well plates containing the relevant liquid growth medium using a multi-pin stamper (Boekel Scientific, Feasterville-Trevose, PA, USA) and incubated at 37 °C for 24–48 h anaerobically. Liquid cultures were then sub-cultured by replication onto large petri dishes containing the corresponding growth medium solidified with 1.5% (w/v) agar and incubated for 24–48 h until isolated colonies were visible. Petri-dishes were overlaid with growth medium solidified with agar (0.8% w/v) seeded with 1% overnight inoculum of different indicator strains. The indicator strains and their respective growth conditions are summarised in Table 1. Colonies showing possible bacteriocin activity were selected for further characterisation.

Table 1.
Bacterial indicators used in this study and their respective growth conditions.
2.5. Characterisation of Antimicrobial Activity of Putative Bacteriocin-Producers
2.5.1. Well Diffusion Assay (WDA)
Pure cultures of potential bacteriocin-producers were obtained by inoculating 10 mL of a sterile liquid medium with frozen stock cultures and incubating for 24 h. Cell-free supernatant (CFS) was prepared from 2 mL of overnight culture by centrifugation for 3 min at 14,000 rpm. Wells were made in agar plates containing appropriate growth medium solidified with agar (0.8% w/v) and seeded with overnight cultures of indicator strains (200 µL inoculum/20 mL soft media). 50 µL of CFS of the putative bacteriocin-producing strain was pipetted into the well. Plates were left to dry and incubated overnight [51]. Zones of inhibition around the wells were assessed. Strains exhibiting antimicrobial activity were kept for further investigation and were genetically characterised using 16S rRNA sequencing and/or molecular masses of the active peptides were confirmed using MALDI-TOF MS. Peptide masses were compared with the Bactibase online database (http://bactibase.hammamilab.org/main.php, accessed on 2 June 2021) [52].
2.5.2. Identification of Putative Bacteriocin-Producing Strains Using 16S rDNA Analysis
Genomic DNA was extracted from 10 mL of liquid culture using a Qiagen PowerFaecal Pro DNA extraction kit (Qiagen, United Kingdom). For PCR reactions, Platinum Master Mix (Fisher Scientific, Ireland) was used with universal bacterial primers CO1 5′-AGTTTGATCCTGGCTCAG-3′ and CO2 5′-TACCTTGTTACGACT-3′ (PCR run conditions: 94 °C × 2 min (×1 cycle), 94 °C × 30 s, 52 °C × 30 s, 72 °C × 1 min (×30 cycles), 72 °C × 5 min (×1 cycle)). PCR reactions were purified using the Qiagen PCR Cleanup Kit (Qiagen, Manchester, UK). Sequencing of the amplicons was performed by GATC Biotech (Koln, Germany). Species designation was carried out using the 16S ribosomal RNA sequences database on the Basic Local Alignment Search Tool (BLAST), using >97% sequence identity.
2.5.3. Shotgun Whole-Genome Sequencing (WGS) and Analysis
Genomic DNA was extracted from 10 mL of overnight liquid cultures using GeneElute™ Bacterial Genome DNA Kit (Sigma-Aldrich, Arklow, Ireland). The concentration of extracted DNA was confirmed using a Qubit® 2.0 Fluorometer (ThermoFisher Scientific, Dublin, Ireland) according to standard protocols, and samples were then standardised to 0.2 ng/µL of DNA. Standardised DNA was then prepared for whole-genome sequencing using the Nextera XT DNA protocol (Illumina, San Diego, CA, USA), using their standard protocol guide and sequenced on Illumina NextSeq platform following standard Teagasc protocols. The paired-end reads underwent quality control using trim_galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and assembly into contigs using the SPAdes v.3.13 [53] software, using default settings, genes were predicted and annotated using PROKKA v.1.13 [48]. The assembled contigs were analysed using BAGEL4 [50] to assess antimicrobial activity, taxonomy assignment was performed using the atpA gene and confirmed using the GTDB-Tk software v.1.3 [54]. Raw sequence reads are available under the accession number PRJEB48530. Antimicrobial genes present were identified using the CARD Resistance Gene Identifier (RGI) database [55].
2.5.4. MALDI-TOF Mass Spectrometry
Single colonies of each strain were inoculated into 5 mL volumes of MRS broth and incubated at 37 °C overnight. 250 µL of each inoculum was used to inoculate 25 mL volumes of MRS, which were in turn incubated at 37 °C overnight. 50 µL aliquots of each cell-free culture supernatant (CFS) were plated on Lactobacillus bulgaricus and Listeria innocua indicator plates. The 25 mL inocula were used to inoculate 600 mL volumes of XAD MRS (MRS passed through a column containing XAD) and MRS and incubated as described. Individual cultures were centrifuged at 11,000× g for 20 min, and cells were separated from supernatant. Cells were mixed with 150 mL 70% propan-2-ol and stirred at room temperature for 3–4 h. The cell extract was centrifuged again, and the supernatant was retained for purification attempts. MALDI-TOF analysis was undertaken on strains of interest.
2.6. Targeted Assembly of Metagenome Assembled Genomes
Five Enterococcus species (E. faecalis, E. faecium, Enterococcus durans, E. mundtii, and Enterococcus hirae) commonly found in the human gut [56] were selected, and representative genomes were downloaded from RefSeq. We downloaded 1866 E. faecalis, 2374 E. faecium, 124 E. durans, 48 E. mundtii, and 386 E. hirae genomes.
Five separate reference genome databases were created for each bacterial species mentioned above. Assembled contigs were blasted against each reference database, and output was filtered to ascertain the contigs that aligned with the database, with >95% identity and >50% coverage. The faSomeRecords.py script (downloaded from: https://github.com/santiagosnchez/faSomeRecords, accessed on 9 November 2021) was used to extract the aligned contigs and subsequently convert them into a single multifasta file to represent a single MAG. The MAGs were then genome quality assessed using the CheckM software [46], where a threshold of Completeness and Contamination was set to >90% and <5%, respectively.
2.7. Statistical Analysis
Multidimensional Scaling analysis of Bray-Curtis distance was performed on metagenomic data (paired-end reads) from 37 Irish athletes [7] and 21 Low BMI controls previously used in a study by Barton et al. [57]. Raw Reads from Barton et al. are available under the accession number PRJEB15388. Statistical analysis and figure visualization were performed in RStudio 3.0.1, using the following packages “ggplot2”, “vegan”, “reshape”, “harry potter”, and “dplyr”.
3. Results
3.1. Assessment and Recovery of Metagenome Assembled Genomes (MAGs) for Bacteriocinogenic Potential
Metagenomic sequencing data from 37 faecal samples (obtained from [7]) yielded 770 MAGs in total. For this study, 148 High-Quality MAGs (>90% Completeness and <5% Contamination) were used and assigned taxonomy using PhyloPhlAn3. A majority of the MAGs were unclassified at the genus level after taxonomic assignment. The most abundant genera recovered were Lachnospiraceae_unclassified, Bacteroides, Ruminococcus, and Coprococcus (Figure 2). The bacteriocinogenic potential of the MAGs recovered from the athlete faecal metagenomic data was then assessed using BAGEL4. The data shows that 91.3% of predicted bacteriocins corresponded to Class I bacteriocins, followed by representatives of Class III (antimicrobial proteins >10 kDa in mass), and 7.5% and 1% to Class II (Figure 3A). The sactipeptide sub-class was particularly abundant, representing 76% of the predicted Class I bacteriocins; Lasso peptides were the second most abundant sub-class (12%) with the remaining sub-classes predicted at >1% abundance (Auto-Inducing Peptides, LanM, Thiopeptide, Lanthipeptides, Linardin, Cyanobactin, and LAP bacteriocins). Class III was represented by Zoocin A-like clusters (90% of predicted Class III bacteriocins) and Closticin_574. Lastly, the Class II group of predicted bacteriocins consisted of Class IIa, Class IId and Class II unclassified (Figure 3B).
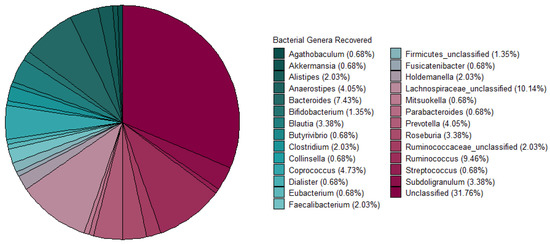
Figure 2.
Bacterial genera recovered from Metagenome Assembled Genomes. The majority of MAGs were unclassified, followed by a high abundance of; Lachnospiraceae_unclassified, Bacteroides, Ruminococcus, and Coprococcus, using PhyloPHhlAn3 to assign taxonomy.
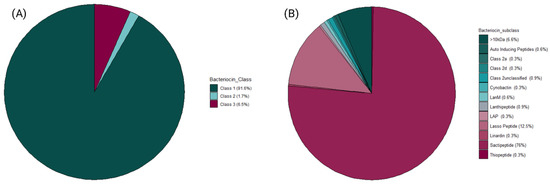
Figure 3.
Frequency of bacteriocin classes and their subsequent t subclasses predicted by BAGEL4 from the recovered MAGs. Class I, II, and III bacteriocins were predicted, with Class_1 (including Sactipeptide sub-group) found to be the most abundant predicted bacteriocin class within the athlete gut sampled in this study. Sactipeptides and Lasso peptides were amongst the most predicted bacteriocin sub-classes, followed by >10 kDa. (A) Contains all bacteriocin classes predicted by BAGEL4 software (B) Contains all bacteriocin sub-classes predicted by BAGEL4 software.
3.2. Detection of Bacteriocin-Producing Bacterial Isolates from Athlete Faecal Samples
Due to the high abundance of anaerobic species recovered in our in silico analysis, we decided to culture these using WCA agar (Figure 2). We also observed bifidobacteria genus MAGs in our samples and therefore decided to use mMRS agar for isolation. Under this approach, approximately 11,000 colonies of different morphologies were isolated from 37 athlete faecal samples. The samples used in this study were from elite athletes who were preparing to partake in international competitions (including the Rio Olympics [7]). mMRS agar with added mupirocin was used as it was previously shown to successfully isolate Bifidobacterium spp. from faecal samples [58,59], and approximately 3500 presumptive bifidobacteria were isolated. BD LBS Agar was employed to isolate 1500 presumptive Lactobacillus isolates, and finally, obligate anaerobic species (6000 isolates) were recovered on WCA, which is commonly used for the isolation and enumeration of anaerobic species [60,61]. Gut isolates stocked in 96-well plates were replicated onto agar plates, and the resulting colonies were screened for antimicrobial activity using a soft agar overlay seeded with indicator strains. In several instances, putative antimicrobial activity was displayed by a majority of isolates, thus precluding identification of individual producer strains. To circumvent this, isolates that showed the most potent antimicrobial activity were selected, and well assays were performed. This resulted in a total of 284 potential bacteriocin-producing gut isolates being identified that were active against at least one of the four indicator strains used. Initially, isolates recovered on MRS and mMRS were screened for activity against L. innocua and L. bulgaricus. However, the screening detected just one colony with antagonistic activity against L. bulgaricus, representing a very low isolation frequency of 0.02%. Similarly, a very low isolation frequency (0.04%) was noted for isolates with antimicrobial activity against L. innocua. Subsequent use of WCA medium to recover gut isolates exhibiting antimicrobial activity against C. difficile and F. nucleatum yielded higher isolation frequencies of 2.4% (C. difficile, 145 isolates) and 2.26% (F. nucleatum, 136 isolates) (summarised in Table 2).

Table 2.
Isolation frequency of intestinal gut isolates using different indicator organisms.
3.3. Identification of Putative Bacteriocin-Producing Strains Isolated from Faecal Samples
Gut isolates exhibiting putative bacteriocin activity were narrowed down further based on the size of the zone produced, and the six most promising gut isolates, recovered from either mMRS or WCA, were brought forward for further characterisation (summarized in Table 3). We recovered three isolates from mMRS showing putative antimicrobial activity against L. innocua and/or L. bulgaricus. We also brought forward the three most potent isolates recovered from WCA with antagonistic activity against F. nucleatum and/or C. difficile (see Table 3).

Table 3.
Most promising gut isolates showing antagonistic activity against indicators of choice.
The three mMRS-recovered gut isolates were identified using shown to be Enterococcus species based on 16S rRNA Sanger sequencing (see Table 3). We also endeavoured to determine the mass of the putative bacteriocins produced by each isolate using MALDI-TOF MS, compared them with the Bactibase online database, and all three masses aligned with well-characterised bacteriocins (see Supplementary Figure S1). The bacteriocin produced by isolate LW003 showed antagonistic activity against L. bulgaricus and had a molecular mass of 5207 Da and 5218 Da, which corresponds to the individual components of the two-peptide bacteriocin, enterocin 62-6 of the class IIc bacteriocin classification group [49]. The other two enterococci isolates, LW001 and LW002, exhibited activity against L. innocua and were found to produce molecules with masses of 3979 Da and 3977 Da, respectively, which correlate to the previously characterised enterocin Q, a leaderless class IIc bacteriocin [50]. The three aforementioned gut isolates were not brought forward for whole-genome sequencing due to the high occurrence of antagonistic activity against C. difficile and/or F. nucleatum, which held a greater interest for the context of the project.
As noted above, we selected the three most potent WCA isolates for further analysis and subjected these were subjected to whole-genome sequencing. The two anti-Fusobacterium nucleatum gut isolates (DPC7280 and DPC7281) were identified as E. faecalis, and the anti-Clostridioides difficile isolate (DPC7282) was assigned as an E. mundtii (see Table 3). The two E. faecalis gut isolates active against F. nucleatum possessed a putative gene cluster corresponding to the class III bacteriocin enterolysin A [62]. In addition to enterolysin A, strain DPC7280 harbours bacteriocin gene clusters predicted to encode enterocin Nkr-5-3b and a potentially novel functional sactipeptide operon (see Figure 4). The predicted sactipeptide operon carries an ABC transporter permease, a protein often associated with bacteriocin transportation across the membrane [52], ABC transporter binding protein possibly associated with self-immunity [53], and a SPASM domain-containing protein, which could be involved in peptide modification [54], however, a structural gene was not be identified (see Figure 4). Finally, the E. mundtii isolate (DPC7282) harbours a gene cluster corresponding to that which encodes enterocin CRL35 bacteriocin, belonging to a class IIa bacteriocin with demonstrated activity against Listeria species [55]. The molecular masses corresponding to bacteriocins encoded by these clusters were not detected through colony mass spectrometry analysis (see Supplementary Figure S1).
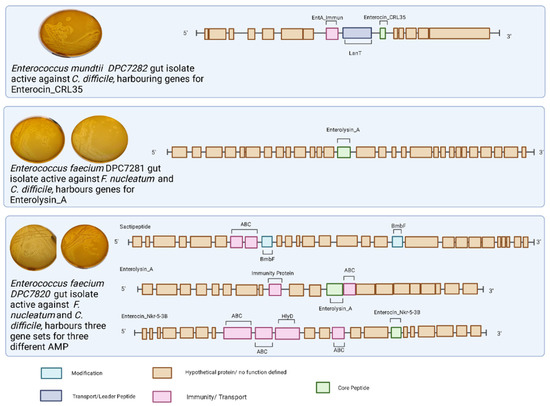
Figure 4.
BAGEL4 outputs for gut isolates active against F. nucleatum and/or C. difficile. Three bacterial isolates recovered from WCA underwent whole-genome sequencing and taxonomy assignment using GTDB-Tk software. Putative bacteriocin gene clusters were annotated and predicted using the BAGEL4 software. Figure created with (https://BioRender.com, accessed on 15 February 2022).
3.4. Assessment of Potential Bacteriocin-Producing Strains for Antimicrobial Resistance Genes (ARGs)
The three isolates active against C. difficile and/or F. nucleatum were assessed for the presence of ARGs using the CARD RGI database (Table 3). Both E. faecalis isolates DPC7280and DPC7281 were found to harbour genes for dfrE and efrA. Additionally, isolate DPC7281 was found to carry six additional ARG genes; tetM, ErmB, E. faecalis chloramphenicol acetyltransferase, aad(6), SAT-4, and APH(3′)-IIIa. Isolate E. mundtii (DPC7282) did not contain any ARGs.
3.5. Targeted Assembly of Metagenome Assembled Genomes (MAGs)
Due to the overwhelming recovery of Enterococcus spp. isolates in the in vitro screen, we subsequently specified our MAG assemblies to target the Enterococcus genus. In this targeted bioinformatics approach, contigs obtained from our metagenomic assembly were blasted against our five different reference databases, which represent the five species of interest (E. faecalis, E. faecium, E. durans, E. mundtii, and E. hirae). We have chosen these Enterococcus species of interest in consideration of the most frequent and abundant Enterococcus spp. associated with the human gut microbiome [43]. Results are presented in the form of BLAST hits (see Table 4/see Supplementary Figure S2). Metagenomic contigs aligning to the E. faecium reference database numbered 670,970,682 BLAST hits, followed by E. faecalis with 73,372,776 hits recovered, E. mundtii with 2,732,290 hits recovered, E. hirae with 13,166,269, and finally, E. durans with 9,623,616 hits. We recovered 40 bins/MAGs for each species of interest, resulting in 200 bins/MAGs recovered. A set of two E. faecalis MAGs were recovered from the low-quality MAG category (<30% Completeness, <10% Contamination); the remaining 198 MAG bins possessed <13% Completeness and therefore could not be assigned to any quality group (see Supplementary Excel S1).

Table 4.
The total amount of BLAST that aligned to the Enterococcus spp. pangenomes, recovered from targeted binning.
4. Discussion
This study is one of the first to target the athlete gut microbiome, a high diversity niche for a potential source of novel antimicrobial agents. In this study, we aimed to identify AMP-producing strains in the athlete’s gut. Elite athletes and their associated microbiomes could be viewed as potentially different and more diverse than the general population [5,8,12]. We have also verified this using healthy controls and beta diversity measures. (see Supplementary Figure S3).
The combined use of in vitro and in silico approaches allowed for a broader investigation of the bacteriocinogenic potential of the athlete gut. Our in silico analysis recovered a large abundance of anaerobic gut species, often associated with the athlete gut microbiome (i.e., Bacteroides spp. [63], Collinsella spp. [64,65], Coprococcus spp. [66,67], Eubacterium spp. [8], Prevotella spp. [8] and Ruminococcus spp [8,62,65,66]. (Figure 2)). For the purpose of re-isolation of the aforementioned species, we have decided to use WCA agar, widely used in isolation of strict anaerobic gut species.
We have also predicted the presence of a myriad of bacteriocin classes and sub-classes embedded in the metagenome-assembled genomes; Class I bacteriocins were particularly abundant at 91%, out of which 76% of all predictions belonged to sactipeptides. This agrees with a previous study by Walsh et al. [25], where they found a high abundance of sactipeptides within the human gut microbiome. Class III bacteriocins were abundant at 7.5% of all bacteriocins, and Class II bacteriocins comprised just 1% of all predictions.
It is interesting to note that all the bacteriocin-producing gut isolates brought forward for further analysis in this study were found to be Enterococcus spp. Enterococcus-selective media was not used in the present study; there is no evidence in the literature to support the increased abundance of Enterococcus spp. in athlete cohorts [5,8,9,12], quite on the contrary—one study found Enterococcus spp. to be decreased within the exercise group of a murine model [68]. It is possible that the rich composition of Wilkins-Chalgren medium (WCA) combined with the generally non-fastidious requirements of members of the Enterococcus genus allowed for their overgrowth and subsequent overrepresentation of the Enterococcus spp. in the library of culturable isolates.
The frequency of isolation of strains with activity against the indicators L. innocua and L. bulgaricus is comparable to that for previous studies [69,70], however, the frequency of isolation observed for C. difficile appears to be higher than current observations in the literature [71]. Screening of gut isolates against F. nucleatum has not yet been addressed, and therefore no data exists for direct comparison.
As noted, it is possible that Enterococcus spp. overgrew their commensal counterparts during culturing and are subsequently overrepresented within the biobank community of isolates. This could be attributed to the fact that obligate anaerobic species were isolated on WCA, an agar used to isolate a wide variety of anaerobic species. This combined with the fact that Enterococcus spp. are generally less fastidious than other anaerobic microbiome commensals, and could potentially attribute to the subsequently higher isolation of Enterococcus spp. isolation. We also suspect a high incidence of repeated isolation of the Enterococcus species with activity against C. difficile and/or F. nucleatum, which could explain the higher isolation frequency observed with the aforementioned enteric pathogen indicators.
In contrast, Bifidobacterium spp. and Lactobacillus spp. were isolated on genus-specific culture media, leaving little room for Enterococcus spp. to dominate the culturing environment. Nevertheless, our observations support the possibility that the Enterococcus spp. isolated in this study tended to exhibit strong antagonistic activity against C. difficile and/or F. nucleatum.
The Enterococcus spp. isolates with activity against L. innocua (LW001 and LW002) and/or L. bulgarcius (LW003) had molecular masses that corresponded to those of enterocin Q and enterocin 62-6, respectively, which is not surprising as enterocin Q is known to inhibit species of the Listeria genus [72]. Similarly, enterocin 62-6 has been demonstrated to inhibit Gram-positive bacterial species [73].
This is, to our knowledge, the first report of an E. mundtii gut isolate harbouring the gene cluster for enterocin CRL35 showing activity against C. difficile. Enterocin CRL35 displayed strong antagonism against C. difficile and little activity against F. nucleatum, implying it has a narrow spectrum. This aligns with previous studies that illustrated the narrow spectrum usually observed in Class IIa bacteriocins (reviewed by [74]). A literature search presented limited information regarding the functionality of the bacteriocin; however, its ability to inhibit the gut pathogen Listeria monocytogenes has been well documented [75]. However, the ability of Enterococcus spp. to inhibit the growth of C. difficile has been well-documented. Bacteriocin biosynthetic gene clusters corresponding to those for duracin 61A, enterocin AS-48, enterocin A/B/P, and Q amongst others (as reviewed by [76] have all been noted to possess antimicrobial activity against C. difficile.
Additionally, we also show for the first time that an E. faecium gut isolate harbouring genes encoding enterolysin A inhibits F. nucleatum, a gut pathogen associated with colorectal cancer [77]. This would appear to support the findings of a recent human trial investigating the administration of a multi-strain probiotic cocktail including E. faecalis in colorectal cancer patients, where a 5-fold decrease in F. nucleatum was observed in the probiotic-supplementation group of the study [78]. Enterolysin A is known to inhibit the growth of several enterococci, pedicocci, lactococci, and lactobacilli [62,79], as well as Listeria, Bacillus, and Staphylococcus species [62]. Similarly, a second E. faecium isolate displayed antagonistic activity against both F. nucleatum and C. difficile. BAGEL4 predicted the presence of three biosynthetic gene clusters, a putative sactipeptide, enterolysin A, and enterocin NKR-5-3B. Sactipeptides have previously been shown to inhibit the growth of C. difficile and may contribute to this isolates activity [80]. Enterocin NKR-5-3B is a circular bacteriocin that displays a broad spectrum of activity, inhibiting a wide range of Gram-positive species [81]. The presence of the enterocin NKR-5-3B gene cluster in the genome of the enterococcal gut isolate DPC7280 may account for its inhibitory activity against C. difficile. Enterococci spp. bacteriocins are well known for their inhibitory activity against Listeria spp. Previous studies demonstrated the ability of helveticin [82], hiraecin S [83], enterocin 1146 [84], and bacteriocins RZS C5 and RZS C13 produced by E. faecium [85] to all have antimicrobial activity against Listeria spp. strains.
We have also assessed the AMR profile of the sequenced genomes active against F. nucleatum and/or C. difficile. In recent years enterococci have become resistant to many commonly used antibiotics, i.e., erythromycin and tetracycline [86], and therefore, interest in its antimicrobial resistance profile is of growing importance. Interestingly, E. mundtii DPC7282 strain did not contain any ARGs. E. mundtii usually carries a less significant ARG profile when compared to other members of the enterococci genus [87], with some studies reporting no ARGs present within the E. mundtii genomes [88,89]. Isolates DPC7281 and DPC7280 were resistant to diaminopyrimidine (dfrE) and possessed a multidrug efflux pump (efrA). Additionally, isolate DPC7281 was found to carry resistance genes to six additional antibiotics; tetracycline (tetM), macrolides (ErmB), chloramphenicol (E. faecalis chloramphenicol acetyltransferase), aminoglycosides (aad(6) and APH(3′)-IIIa), and streptothricin (SAT-4). The presence of the aforementioned genes has been previously noted in the literature as common resistance mechanisms in enterococci genomes [90,91,92,93,94,95,96,97,98].
Initial MAG recovery did not yield any genomes assigned to the Enterococcus genus. We then specified our genome assembly using a targeted binning approach. Even though we have not recovered any bins/MAGs that matched our threshold (Completeness > 90%, Contamination < 5%), we have recovered millions of alignments/hits corresponding to the targeted Enterococcus spp. of interest, which are commonly found in the human gut. These results confirm that reads corresponding to Enterococcus spp. are present in the microbiome of elite athletes. However, due to a suspected low abundance of enterococci in the athlete gut, we did not recover any high-quality MAGs corresponding to the genus [68]. Our findings are substantiated by recent studies concluding that Enterococcus spp. genomes are difficult to assemble and recover from metagenomic samples due to high gene divergence and high genome plasticity [99]. E. faecalis in particular has been shown to have high levels of genome plasticity, insertions/deletions and repetitive regions, which can hinder successful assembly [99,100].
This study highlights the merits and disadvantages of both in silico and in vitro based approaches. In silico screening allowed for a broader representation of the taxonomical and functional composition of the niche at hand, without the inherent bias introduced by culturing microorganisms present in the samples. Conversely, in vitro approaches allowed for the isolation of bacterial species and relatively rapid assessment of their clinical relevance using established assays for antagonistic activity against a selection of important enteric pathogens. Ultimately, both approaches contain distinct inherent biases. In silico analysis, in this instance the BAGEL4 tool, is reliant on homology and can predict spurious matches on that basis. Another important drawback is that in silico evaluation of metagenomic datasets, particularly the presence of potential antimicrobial gene clusters as in the present study, is dependent on the degree of success of assembly of the metagenome, which can vary according to the genomic composition of constituent species. Similarly, the isolation and in vitro screening process is influenced by the choice of indicator species for antagonistic assays, antimicrobial expression conditions of certain microbial taxa, including environmental and other microbial and host factors, and the degree to which a species exhibits obligate or facultative growth in vitro. These dynamic factors are not apparent in in silico analysis yet may contribute to false negatives in the isolation pipeline as well as the overrepresentation of certain species in the pool of isolates. It is also vital to recognize that identified isolates with potential antimicrobial activity require verification of the mode of action, for instance, through cloning and functional expression of putative bacteriocin genes or purification/direct chemical synthesis.
5. Conclusions
In conclusion, the gut microbiome of the elite athletes in this study appeared to be a rich source of AMPs with potential applications in human health.
In silico approaches can be used to provide a broad overview of the bacterial taxa present and their potential metabolites, which can inform the design of the in vitro screen. Similarly, in vitro results can also validate in silico results, as shown in the present study through an Enterococcus-specific approach.
Our in silico analysis identified a broad range of potential bacteriocin classes present in the athlete gut, suggesting the athlete gut could be used to harness novel natural bacteriocin-producers for potential development as alternatives to existing antibiotics.
Putative bacteriocin-producing gut isolates identified in this study through in vitro analysis could be harnessed as an alternative treatment against relevant gut/enteric pathogens (F. nucleatum and C. difficile), especially E. mundtii isolate, which was shown to harbour no ARGs.
Therefore, we suggest a tandem deployment of in silico and in vitro approaches to broadly interrogate the niche at hand.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10040701/s1, Excel S1. MAG bins recovered with Targeted Approach, Figure S1. MALDI-TOF MS analysis of potential bacteriocin producing gut isolates, Figure S2. Bar chart representing the BLAST hits per sample recovered, for each species of interest, Figure S3. Multidimensional scalling analysis of Bray Curtis distance, at species level between elite Irish athletes and Low BMI non-athlete controls.
Author Contributions
Conceptualization, O.O., P.D.C. and C.M.G.; methodology, O.O., C.J.W., C.M.G., E.M.L., P.M.O. and L.W.; software, O.O., C.J.W. and L.W.; validation, O.O., C.M.G. and P.D.C.; formal analysis, L.W., O.O. and C.M.G.; data curation, L.W.; writing—original draft preparation, L.W.; writing—review and editing, O.O., C.M.G. and P.D.C.; visualization, L.W.; supervision, O.O., C.M.G. and P.D.C.; project administration, C.M.G. and O.O.; funding acquisition, O.O., C.M.G. and P.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Teagasc Walsh Scholar Scheme. This research was conducted with the financial support of Science Foundation Ireland (SFI) under Grant Numbers SFI/12/RC/2273, 13/SIRG/2160, and 11/PI/1137.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Raw reads and assembled contigs are available under the accession number PRJEB48530.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brown, D.W.; Heath, G.W.; Balluz, L.; Giles, W.H.; Ford, E.S.; Mokdad, A.H. Associations between Physical Activity Dose and Health-Related Quality of Life. Med. Sci. Sports Exerc. 2004, 36, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Siscovick, D.S.; Fried, L.; Mittelmark, M.; Rutan, G.; Bild, D.; O’Leary, D.H.; Cardiovascular Health Study Research Group. Exercise Intensity and Subclinical Cardiovascular Disease in the Elderly: The Cardiovascular Health Study. Am. J. Epidemiology 1997, 145, 977–986. [Google Scholar] [CrossRef]
- Ostrowski, K.; Rohde, T.; Asp, S.; Schjerling, P.; Pedersen, B.K. Pro- and anti-inflammatory cytokine balance in strenuous exercise in humans. J. Physiol. 1999, 515, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Craft, L.L.; Landers, D.M. The Effect of Exercise on Clinical Depression and Depression Resulting from Mental Illness: A Meta-Analysis. J. Sport Exerc. Psychol. 1998, 20, 339–357. [Google Scholar] [CrossRef]
- Clarke, S.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Cronin, O.; Barton, W.; Skuse, P.; Penney, N.C.; Garcia-Perez, I.; Murphy, E.F.; Woods, T.; Nugent, H.; Fanning, A.; Melgar, S.; et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. mSystems 2018, 3, e00044-18. [Google Scholar] [CrossRef]
- O’Donovan, C.M.; Madigan, S.M.; Garcia-Perez, I.; Rankin, A.; Sullivan, O.O.; Cotter, P. Distinct microbiome composition and metabolome exists across subgroups of elite Irish athletes. J. Sci. Med. Sport 2019, 23, 63–68. [Google Scholar] [CrossRef]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 2017, 5, 98. [Google Scholar] [CrossRef]
- Scheiman, J.; Luber, J.M.; Chavkin, T.; Macdonald, T.; Tung, A.; Pham, L.-D.; Wibowo, M.C.; Wurth, R.C.; Punthambaker, S.; Tierney, B.; et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 2019, 25, 1104–1109. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Rainey, P.; Rainey, K. Evolution of cooperation and conflict in experimental bacterial populations. Nature 2003, 425, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.; Hill, C.; Ross, R. Bacteriocins: Developing innate immunity for food. Nat. Rev. Genet. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Hatakka, K.; Saxelin, M. Probiotics in Intestinal and Non-Intestinal Infectious Diseases—Clinical Evidence. Curr. Pharm. Des. 2008, 14, 1351–1367. [Google Scholar] [PubMed]
- Rea, M.C.; Dobson, A.; O’Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Ross, R.P. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108, 4639–4644. [Google Scholar] [PubMed]
- Walsh, M.C.; Gardiner, G.E.; Hart, O.; Lawlor, P.G.; Daly, M.; Lynch, B.; Richert, B.T.; Radcliffe, S.; Giblin, L.; Hill, C.; et al. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiol. Ecol. 2008, 64, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Corr, S.C.; Li, Y.; Riedel, C.U.; O’Toole, P.W.; Hill, C.; Gahan, C.G.M. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 2007, 104, 7617–7621. [Google Scholar] [CrossRef] [PubMed]
- Van Hemert, S.; Meijerink, M.; Molenaar, D.; Bron, P.A.; De Vos, P.; Kleerebezem, M.; Marco, M.L. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 2010, 10, 293. [Google Scholar]
- Deng, Z.; Luo, X.M.; Liu, J.; Wang, H. Quorum Sensing, Biofilm, and Intestinal Mucosal Barrier: Involvement the Role of Probiotic. Front. Cell. Infect. Microbiol. 2020, 10, 504. [Google Scholar] [CrossRef]
- Khalaf, H.; Nakka, S.S.; Sandén, C.; Svärd, A.; Hultenby, K.; Scherbak, N.; Aili, D.; Bengtsson, T. Antibacterial effects of Lactobacillus and bacteriocin PLNC8 αβ on the periodontal pathogen Porphyromonas gingivalis. BMC Microbiol. 2016, 16, 188. [Google Scholar] [CrossRef]
- Ovchinnikov, K.V.; Kranjec, C.; Thorstensen, T.; Carlsen, H.; Diep, D.B. Successful Development of Bacteriocins into Therapeutic Formulation for Treatment of MRSA Skin Infection in a Murine Model. Antimicrob. Agents Chemother. 2020, 64, e00829-20. [Google Scholar] [CrossRef]
- Nascimento, J.S.; Ceotto, H.; Nascimento, S.B.; Giambiagi-Demarval, M.; Santos, K.R.N.; Bastos, M.C.F. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett. Appl. Microbiol. 2006, 42, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Mathiesen, G.; Huehne, K.; Kroeckel, L.; Axelsson, L.; Eijsink, V.G. Characterization of a New Bacteriocin Operon in Sakacin P-Producing Lactobacillus sakei, Showing Strong Translational Coupling between the Bacteriocin and Immunity Genes. Appl. Environ. Microbiol. 2005, 71, 3565–3574. [Google Scholar] [PubMed]
- Nes, I.F.; Diep, D.B.; Brurberg, M.B.; Eijsink, V.; Holo, H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 1996, 70, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Drissi, F.; Buffet, S.; Raoult, D.; Merhej, V. Common occurrence of antibacterial agents in human intestinal microbiota. Front. Microbiol. 2015, 6, 441. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.J.; Guinane, C.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project’s reference genome database. BMC Microbiol. 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Zheng, J.; Gänzle, M.; Lin, X.B.; Ruan, L.; Sun, M. Diversity and dynamics of bacteriocins from human microbiome. Environ. Microbiol. 2014, 17, 2133–2143. [Google Scholar] [CrossRef]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.M.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef]
- Rea, M.; Clayton, E.; O’Connor, P.M.; Shanahan, F.; Kiely, B.; Ross, R.; Hill, C. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med Microbiol. 2007, 56, 940–946. [Google Scholar] [CrossRef]
- Casey, P.; Casey, G.; Gardiner, G.; Tangney, M.; Stanton, C.; Ross, R.; Hill, C.; Fitzgerald, G. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett. Appl. Microbiol. 2004, 39, 431–438. [Google Scholar] [CrossRef]
- Reenen, V.; Dicks, L.M.; Chikindas, M.L. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J. Appl. Microbiol. 1998, 84, 1131–1137. [Google Scholar] [CrossRef]
- Kim, T.-S.; Hur, J.-W.; Yu, M.-A.; Cheigh, C.-I.; Kim, K.-N.; Hwang, J.-K.; Pyun, Y.-R. Antagonism of Helicobacter pylori by Bacteriocins of Lactic Acid Bacteria. J. Food Prot. 2003, 66, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Soria, M.C.; Audisio, M.C. Inhibition of Bacillus cereus Strains by Antimicrobial Metabolites from Lactobacillus johnsonii CRL1647 and Enterococcus faecium SM21. Probiotics Antimicrob. Proteins 2014, 6, 208–216. [Google Scholar] [PubMed]
- Chaveerach, P.; Lipman, L.; van Knapen, F. Antagonistic activities of several bacteria on in vitro growth of 10 strains of Campylobacter jejuni/coli. Int. J. Food Microbiol. 2003, 90, 43–50. [Google Scholar] [CrossRef]
- Vedantam, G.; Clark, A.; Chu, M.; McQuade, R.; Mallozzi, M.; Viswanathan, V.K. Clostridium difficile infection: Toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 2012, 3, 121–134. [Google Scholar]
- George, W.L.; Goldstein, E.; Sutter, V.; Ludwig, S.; Finegold, S. ÆTIOLOGY OF ANTIMICROBIAL-AGENT-ASSOCIATED COLITIS. Lancet 1978, 311, 802–803. [Google Scholar] [CrossRef]
- Lim, S.; Knight, D.; Riley, T. Clostridium difficile and One Health. Clin. Microbiol. Infect. 2019, 26, 857–863. [Google Scholar] [CrossRef]
- Pépin, J.; Valiquette, L.; Alary, M.-E.; Villemure, P.; Pelletier, A.; Forget, K.; Pépin, K.; Chouinard, D. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: A changing pattern of disease severity. Can. Med. Assoc. J. 2004, 171, 466–472. [Google Scholar] [CrossRef]
- Starr, J. Clostridium difficile associated diarrhoea: Diagnosis and treatment. BMJ 2005, 331, 498–501. [Google Scholar] [CrossRef][Green Version]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef]
- Li, X.; Huang, J.; Yu, T.; Fang, X.; Lou, L.; Xin, S.; Ji, L.; Jiang, F.; Lou, Y. Fusobacterium nucleatum Promotes the Progression of Colorectal Cancer Through Cdk5-Activated Wnt/β-Catenin Signaling. Front. Microbiol. 2021, 11, 3231. [Google Scholar] [CrossRef]
- Rotmistrovsky, K.; Agarwala, R. BMTagger: Best Match Tagger for Removing Human Reads from Metagenomics Datasets. 2011. Available online: ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/ (accessed on 2 June 2021).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar]
- Ryan, M.P.; Rea, M.C.; Hill, C.; Ross, R.P. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 1996, 62, 612–619. [Google Scholar]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal gut bacteria: Distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 2011, 62, 449–459. [Google Scholar] [CrossRef]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [PubMed]
- Rosberg-Cody, E.; Ross, R.; Hussey, S.; Ryan, C.A.; Murphy, B.P.; Fitzgerald, G.F.; Devery, R.; Stanton, C. Mining the Microbiota of the Neonatal Gastrointestinal Tract for Conjugated Linoleic Acid-Producing Bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 4635–4641. [Google Scholar] [CrossRef]
- Wall, R.; Hussey, S.G.; Ryan, C.A.; O’Neill, M.; Fitzgerald, G.; Stanton, C.; Ross, R.P. Presence of two Lactobacillus and Bifidobacterium probiotic strains in the neonatal ileum. ISME J. 2007, 2, 83–91. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 2012, 7, e44595. [Google Scholar]
- Brazier, J.S.; Goldstein, E.J.; Citron, D.M.; Ostovari, M.I. Fastidious anaerobe agar compared with Wilkins-Chalgren agar, brain heart infusion agar, and brucella agar for susceptibility testing of Fusobacterium species. Antimicrob. Agents Chemother. 1990, 34, 2280–2282. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar]
- Jang, L.-G.; Choi, G.; Kim, S.-W.; Kim, B.-Y.; Lee, S.; Park, H. The combination of sport and sport-specific diet is associated with characteristics of gut microbiota: An observational study. J. Int. Soc. Sports Nutr. 2019, 16, 21. [Google Scholar] [CrossRef]
- Jaago, M.; Timmusk, U.S.; Timmusk, T.; Palm, K. Drastic Effects on the Microbiome of a Young Rower Engaged in High-Endurance Exercise After a Month Usage of a Dietary Fiber Supplement. Front. Nutr. 2021, 8, 654008. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.B.; Marques, C.; Abreu, R.; Figueiredo, P.; Calhau, C.; Brito, J.; Sousa, M. Gut microbiota of elite female football players is not altered during an official international tournament. Scand. J. Med. Sci. Sports 2021. published ahead of print. [Google Scholar] [CrossRef]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Z.; Hu, B.; Huang, W.; Yuan, C.; Zou, L. Response of Gut Microbiota to Metabolite Changes Induced by Endurance Exercise. Front. Microbiol. 2018, 9, 765. [Google Scholar] [CrossRef]
- Queipo-Ortuño, M.I.; Seoane, L.M.; Murri, M.; Pardo, M.; Gomez-Zumaquero, J.M.; Cardona, F.; Casanueva, F.; Tinahones, F.J. Gut Microbiota Composition in Male Rat Models under Different Nutritional Status and Physical Activity and Its Association with Serum Leptin and Ghrelin Levels. PLoS ONE 2013, 8, e65465. [Google Scholar] [CrossRef]
- Lakshminarayanan, B.; Guinane, C.; O’Connor, P.; Coakley, M.; Hill, C.; Stanton, C.; O’Toole, P.; Ross, R. Isolation and characterization of bacteriocin-producing bacteria from the intestinal microbiota of elderly Irish subjects. J. Appl. Microbiol. 2012, 114, 886–898. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2018, 95, fiy241. [Google Scholar] [CrossRef]
- Khattab, R.A.; Ahmed, N.A.; Ragab, Y.M.; Rasmy, S.A. Bacteria producing antimicrobials against Clostridium difficile isolated from human stool. Anaerobe 2020, 63, 102206. [Google Scholar] [CrossRef]
- Basanta, A.; Sánchez, J.; Gómez-Sala, B.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 2008, 125, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Dezwaan, D.C.; Mequio, M.J.; Littell, J.S.; Allen, J.P.; Rossbach, S.; Pybus, V. Purification and characterization of enterocin 62-6, a two-peptide bacteriocin produced by a vaginal strain of Enterococcus faecium: Potential significance in bacterial vaginosis. Microb. Ecol. Health Dis. 2007, 19, 241–250. [Google Scholar] [PubMed]
- Umu, Ö.C.O.; Bäuerl, C.; Oostindjer, M.; Pope, P.B.; Hernández, P.E.; Pérez-Martínez, G.; Diep, D.B. The Potential of Class II Bacteriocins to Modify Gut Microbiota to Improve Host Health. PLoS ONE 2016, 11, e0164036. [Google Scholar] [CrossRef]
- Minahk, C.; Farãas, M.E.; Sesma, F.; Morero, R.D. Effect of Enterocin CRL35 on Listeria monocytogenes cell membrane. FEMS Microbiol. Lett. 2000, 192, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Kang, H.-J.; Ivanova, I.V.; Holzapfel, W.H. Bacteriocins From LAB and Other Alternative Approaches for the Control of Clostridium and Clostridiodes Related Gastrointestinal Colitis. Front. Bioeng. Biotechnol. 2020, 8, 1088. [Google Scholar] [CrossRef]
- Lawrence, G.W.; Begley, M.; Cotter, P.D.; Guinane, C.M. Potential Use of Biotherapeutic Bacteria to Target Colorectal Cancer-Associated Taxa. Int. J. Mol. Sci. 2020, 21, 924. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Wu, W.; Qin, H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol. Med. Rep. 2015, 12, 6119–6127. [Google Scholar] [CrossRef]
- Hickey, R.M.; Twomey, D.P.; Ross, R.P.; Hill, C. Production of enterolysin A by a raw milk enterococcal isolate exhibiting multiple virulence factors. Microbiology 2003, 149, 655–664. [Google Scholar] [CrossRef][Green Version]
- Rea, M.C.; Sit, C.S.; Clayton, E.; O’Connor, P.M.; Whittal, R.M.; Zheng, J.; Vederas, J.C.; Ross, R.P.; Hill, C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. USA 2010, 107, 9352–9357. [Google Scholar] [CrossRef]
- Himeno, K.; Rosengren, K.J.; Inoue, T.; Perez, R.H.; Colgrave, M.L.; Lee, H.S.; Chan, L.Y.; Henriques, S.T.; Fujita, K.; Ishibashi, N.; et al. Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium. Biochemistry 2015, 54, 4863–4876. [Google Scholar] [CrossRef]
- McKay, A.M. Antimicrobial activity of Enterococcus faecium against Listeria spp. Lett. Appl. Microbiol. 1990, 11, 15–17. [Google Scholar] [CrossRef]
- Siragusa, G.R. Production of bacteriocin inhibitory to Listeria species by Enterococcus hirae. Appl. Environ. Microbiol. 1992, 58, 3508–3513. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Hill, C. Inhibition of Listeria in Buffer, Broth, and Milk by Enterocin 1146, a Bacteriocin Produced by Enterococcus faecium. J. Food Prot. 1992, 55, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Vlaemynck, G.; Herman, L.; Coudijzer, K. Isolation and characterization of two bacteriocins produced by Enterococcus faecium strains inhibitory to Listeria monocytogenes. Int. J. Food Microbiol. 1994, 24, 211–225. [Google Scholar] [CrossRef]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- Abriouel, H.; Ben Omar, N.; Molinos, A.C.; López, R.L.; Grande, M.J.; Martínez-Viedma, P.; Ortega, E.; Cañamero, M.M.M.; Galvez, A. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 2008, 123, 38–49. [Google Scholar] [CrossRef]
- Nawaz, F.; Khan, M.N.; Javed, A.; Ahmed, I.; Ali, N.; Ali, M.I.; Bakhtiar, S.M.; Imran, M. Genomic and Functional Characterization of Enterococcus mundtii QAUEM2808, Isolated From Artisanal Fermented Milk Product Dahi. Front. Microbiol. 2019, 10, 434. [Google Scholar] [CrossRef]
- Foka, F.E.T.; Ateba, C.N. Detection of Virulence Genes in Multidrug Resistant Enterococci Isolated from Feedlots Dairy and Beef Cattle: Implications for Human Health and Food Safety. BioMed Res. Int. 2019, 2019, 5921840. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, H.; Wang, Z. Distribution of acquired antibiotic resistance genes among Enterococcus spp. isolated from a hospital in Baotou, China. BMC Res. Notes 2019, 12, 27. [Google Scholar] [CrossRef]
- Portillo, A.; Larrea, F.R.; Zarazaga, M.; Alonso, A.; Martinez, J.L.; Torres, C. Macrolide Resistance Genes in Enterococcus spp. Antimicrob. Agents Chemother. 2000, 44, 967–971. [Google Scholar] [CrossRef]
- Jensen, L.B.; Frimodt-Møller, N.; Aarestrup, F. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 1999, 170, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Trieu-Cuot, P.; de Cespédès, G.; Bentorcha, F.; Delbos, F.; Gaspar, E.; Horaud, T. Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: Characterization of a new CAT determinant. Antimicrob. Agents Chemother. 1993, 37, 2593–2598. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Werner, G.; Hildebrandt, B.; Witte, W. Aminoglycoside-Streptothricin Resistance Gene Cluster aadE–sat4–aphA-3 Disseminated among Multiresistant Isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3267–3269. [Google Scholar] [CrossRef] [PubMed]
- Diarra, M.S.; Rempel, H.; Champagne, J.; Masson, L.; Pritchard, J.; Topp, E. Distribution of Antimicrobial Resistance and Virulence Genes in Enterococcus spp. and Characterization of Isolates from Broiler Chickens. Appl. Environ. Microbiol. 2010, 76, 8033–8043. [Google Scholar] [PubMed]
- Woegerbauer, M.; Zeinzinger, J.; Springer, B.; Hufnagl, P.; Indra, A.; Korschineck, I.; Allerberger, F. Prevalence of the aminoglycoside phosphotransferase genes aph(3′)-IIIa and aph(3′)-IIa in Escherichia coli, Enterococcus faecalis, Enterococcus faecium, Pseudomonas aeruginosa, Salmonella enterica subsp. enterica and Staphylococcus aureus isolates in Austria. J. Med. Microbiol. 2014, 63, 210–217. [Google Scholar]
- Coque, T.M.; Singh, K.V.; Weinstock, G.; Murray, B.E. Characterization of Dihydrofolate Reductase Genes from Trimethoprim-Susceptible and Trimethoprim-Resistant Strains of Enterococcus faecalis. Antimicrob. Agents Chemother. 1999, 43, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, H.; Ortega-Polo, R.; Zaheer, R.; Goji, N.; Amoako, K.K.; Brown, R.S.; Majury, A.; Liss, S.N.; McAllister, T.A. Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. 2020, 20, 20. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Pan, B.; Hao, P.; Li, Y.; Shao, Z.; Xu, X.; Li, X. Optimizing hybrid assembly of next-generation sequence data from Enterococcus faecium: A microbe with highly divergent genome. BMC Syst. Biol. 2012, 6, S21. [Google Scholar] [CrossRef]
- Zhong, Z.; Kwok, L.-Y.; Hou, Q.; Sun, Y.; Li, W.; Zhang, H.; Sun, Z. Comparative genomic analysis revealed great plasticity and environmental adaptation of the genomes of Enterococcus faecium. BMC Genom. 2019, 20, 602. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).