Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov.
Abstract
1. Introduction
2. Materials & Methods
2.1. Isolates and Reference Strains
2.2. DNA Extraction, 16S rRNA, gyrB and lepA Genes Sequencing, Phylogenetic Analyses
2.3. Genome Sequencing and Calculating Overall Genome Relatedness Index
2.4. Bioinformatics Analyses and Whole-Genome Phylogeny
2.5. MALDI Analysis
2.6. Chemotaxonomic Characterization
2.7. Repetitive Sequence-Based PCR Fingerprinting (rep-PCR)
2.8. Phenotypic Characteristics, Electron Microscopy and Violacein Determination
3. Results & Discussion
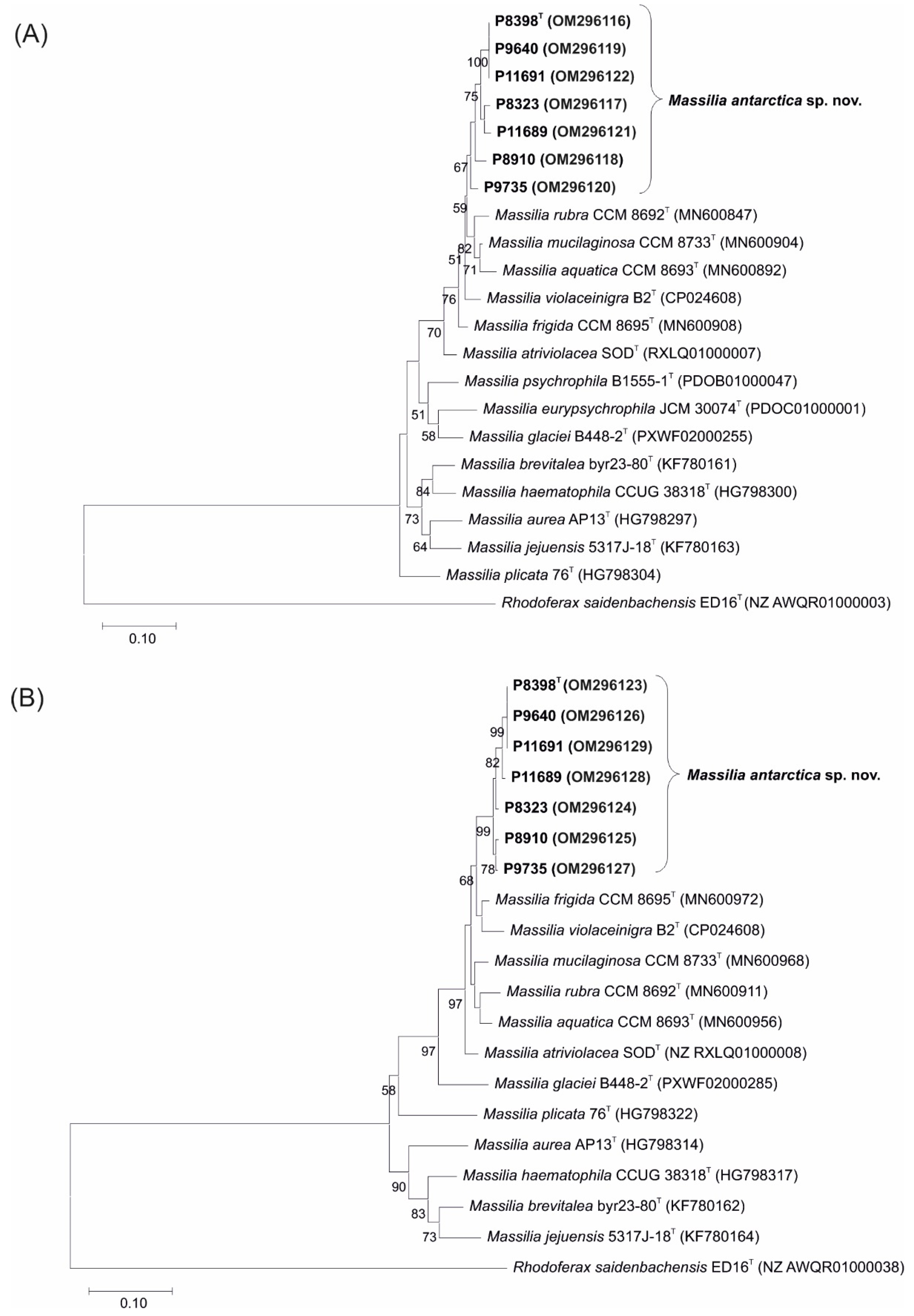
3.1. 16S rRNA, gyrB and lepA Gene Sequencing and Phylogenetic Relationship
3.2. Genome-Based Phylogeny and Basic Genome Characterization
3.3. MALDI-TOF MS
3.4. Chemotaxonomic Characterization
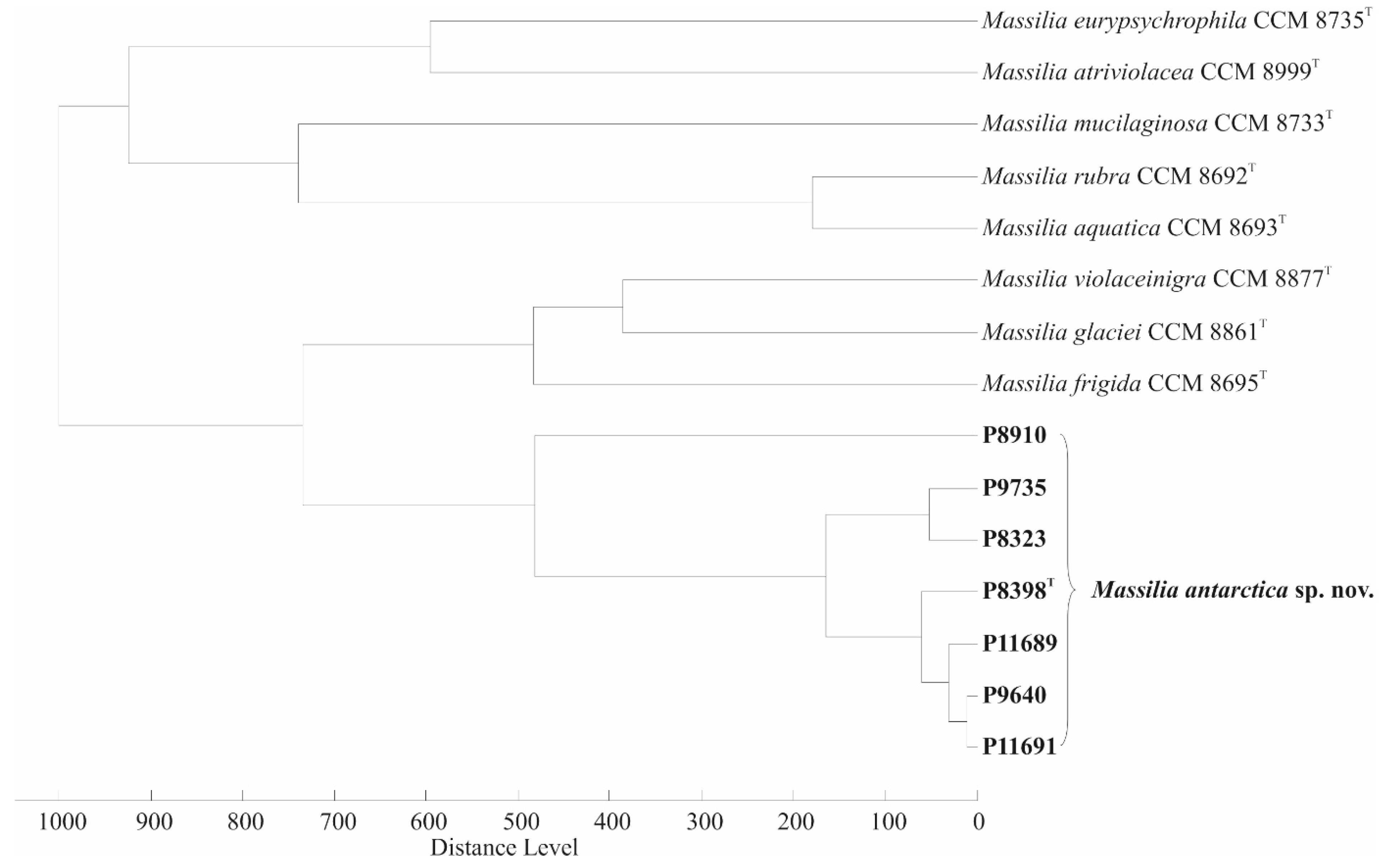
3.5. Repetitive PCR-Based Fingerprinting
3.6. Phenotypic Characteristics and Description of Novel Species
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnes, D.K.A. Polar marine ecosystems. In Life at Extremes: Environments, Organisms, and Strategies for Survival; Bell, E.M., Ed.; Cabi: London, UK, 2012; pp. 1–9. [Google Scholar]
- Peeters, K.; Verleyen, E.; Hodgson, D.A.; Convey, P.; Ertz, D.; Vyverman, W.; Willems, A. Heterotrophic bacterial diversity in aquatic microbial mat communities from Antarctica. Polar Biol. 2012, 35, 543–554. [Google Scholar] [CrossRef][Green Version]
- Sanyal, A.; Antony, R.; Samui, G.; Thamban, M. Microbial communities and their potential for degradation of dissolved organic carbon in cryoconite hole environments of Himalaya and Antarctica. Microbiol. Res. 2018, 208, 32–42. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Birtles, R.J.; Mallet, M.-N.; Raoult, D. Massilia timonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 1998, 36, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Family II. Oxalobacteraceae fam. nov. In Bergey’s Manual of Systematic Bacteriology, the Proteobacteria, Part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2005; Volume 2, p. 623. [Google Scholar]
- Kämpfer, P.; Lodders, N.; Martin, K.; Falsen, E. Revision of the genus Massilia La Scola et al. 2000, with an emended description of the genus and inclusion of all species of the genus Naxibacter as new combinations, and proposal of Massilia consociata sp. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Du, J.; Won, K.; Yang, J.-E.; Yin, C.; Kook, M.; Yi, T.-H. Massilia arvi sp. nov., isolated from fallow-land soil previously cultivated with Brassica oleracea, and emended description of the genus Massilia. Int. J. Syst. Evol. Microbiol. 2015, 65, 3690–3696. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, R.; Zhang, G.; Zhang, D.; Zhang, W.; Chen, T.; Liu, G. Massilia arenae sp. nov., isolated from sand soil in the Qinghai-Tibetan Plateau. Int. J. Syst. Evol. Microbiol. 2020, 70, 2435–2439. [Google Scholar] [CrossRef]
- Yang, E.; Zhao, M.; Li, S.; Wang, Y.; Sun, L.; Liu, J.; Wang, W. Massilia atriviolacea sp. nov., a dark purple-pigmented bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 2135–2141. [Google Scholar] [CrossRef]
- Ren, M.; Li, X.; Zhang, Y.; Jin, Y.; Li, S.; Huang, H. Massilia armeniaca sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 2319–2324. [Google Scholar] [CrossRef]
- Sun, L.-N.; Yang, E.-D.; Cui, D.-X.; Ni, Y.-W.; Wang, Y.-B.; Sun, D.-D.; Wang, W.-Y. Massilia buxea sp. nov., isolated from a rock surface. Int. J. Syst. Evol. Microbiol. 2017, 67, 4390–4396. [Google Scholar] [CrossRef]
- Feng, G.-D.; Yang, S.-Z.; Li, H.-P.; Zhu, H. Massilia putida sp. nov., a dimethyl disulfide-producing bacterium isolated from wolfram mine tailing. Int. J. Syst. Evol. Microbiol. 2016, 66, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liu, Y.; Gu, Z.; Xu, B.; Wang, N.; Jiao, N.; Liu, H.; Zhou, Y. Massilia eurypsychrophila sp. nov. a facultatively psychrophilic bacteria isolated from ice core. Int. J. Syst. Evol. Microbiol. 2015, 65, 2124–2129. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, B.; Liu, Y.; Gu, Z.; Shen, L.; Liu, K.; Wang, N.; Xing, T.; Liu, H.; Zhou, Y.; Li, J. Massilia psychrophila sp. nov., isolated from an ice core. Int. J. Syst. Evol. Microbiol. 2016, 66, 4088–4093. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Liu, Y.; Xu, B.; Wang, N.; Jiao, N.; Shen, L.; Liu, H.; Zhou, Y.; Liu, X.; Li, J.; et al. Massilia glaciei sp. nov., isolated from the Muztagh Glacier. Int. J. Syst. Evol. Microbiol. 2017, 67, 4075–4079. [Google Scholar] [CrossRef]
- Gallego, V.; Sanchez-Porro, C.; Garcia, M.T.; Ventosa, A. Massilia aurea sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 2006, 56, 2449–2453. [Google Scholar] [CrossRef]
- Lu, H.; Deng, T.; Liu, F.; Wang, Y.; Yang, X.; Xu, M. Duganella lactea sp. nov., Duganella guangzhouensis sp. nov., Duganella flavida sp. nov. and Massilia rivuli sp. nov., isolated from a subtropical stream in PR China and proposal to reclassify Duganella ginsengisoli as Massilia ginsengisoli comb. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 4822–4830. [Google Scholar] [CrossRef] [PubMed]
- Orthová, I.; Kampfer, P.; Glaeser, S.P.; Kaden, R.; Busse, H.-J. Massilia norwichensis sp. nov., isolated from an air sample. Int. J. Syst. Evol. Microbiol. 2015, 65, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Lodders, N.; Martin, K.; Falsen, E. Massilia oculi sp. nov., isolated from a human clinical specimen. Int. J. Syst. Evol. Microbiol. 2012, 62, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Wery, N.; Gerike, U.; Sharman, A.; Chaudhuri, J.B.; Hough, D.W.; Danson, M.J. Use of a packed-column bioreactor for isolation of diverse protease-producing bacteria from Antarctic soil. Appl. Environ. Microbiol. 2003, 69, 1457–1464. [Google Scholar] [CrossRef]
- Cong, B.; Yin, X.; Deng, A.; Shen, J.; Tian, Y.; Wang, S.; Yang, H. Diversity of cultivable microbes from soil of the Fildes Peninsula, Antarctica, and their potential application. Front. Microbiol. 2020, 11, 570836. [Google Scholar] [CrossRef]
- Holochová, P.; Mašlaňová, I.; Sedláček, I.; Švec, P.; Králová, S.; Kovařovic, V.; Busse, H.-J.; Staňková, E.; Barták, M.; Pantůček, R. Description of Massilia rubra sp. nov., Massilia aquatica sp. nov., Massilia mucilaginosa sp. nov., Massilia frigida sp. nov., and one Massilia genomospecies isolated from Antarctic streams, lakes and regoliths. Syst. Appl. Microbiol. 2020, 43, 126112. [Google Scholar] [CrossRef]
- Sajjad, W.; Din, G.; Rafiq, M.; Iqbal, A.; Khan, S.; Zada, S.; Ali, B.; Kang, S. Pigment production by cold-adapted bacteria and fungi: Colorful tale of cryosphere with wide range applications. Extremophiles 2020, 24, 447–473. [Google Scholar] [CrossRef] [PubMed]
- Dieser, M.; Greenwood, M.; Foreman, C. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Zhang, Y.-Q.; Li, W.-J.; Zhang, K.-Y.; Tian, X.-P.; Jiang, Y.; Xu, L.-H.; Jiang, C.-L.; Lai, R. Massilia dura sp. nov., Massilia albidiflava sp. nov., Massilia plicata sp. nov. and Massilia lutea sp. nov., isolated from soils in China. Int. J. Syst. Evol. Microbiol. 2006, 56, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, J.; Pang, H.; Zhang, Y.; Li, Y.; Fan, J. Massilia flava sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 580–585. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Wang, S.; Zhao, B.; Lou, K.; Xing, X.-H. Massilia violaceinigra sp. nov., a novel purple-pigmented bacterium isolated from glacier permafrost. Int. J. Syst. Evol. Microbiol. 2018, 68, 2271–2278. [Google Scholar] [CrossRef]
- Myeong, N.R.; Seong, H.J.; Kim, H.-J.; Sul, W.J. Complete genome sequence of antibiotic and anticancer agent violacein producing Massilia sp. strain NR 4-1. J. Biotech. 2016, 223, 36–37. [Google Scholar] [CrossRef]
- Sedláček, I.; Holochová, P.; Sobotka, R.; Busse, H.-J.; Švec, P.; Králová, S.; Šedo, O.; Pilný, J.; Staňková, E.; Koublová, V.; et al. Classification of violacein-producing psychrophilic group of isolates associated with freshwater in Antarctica and description of Rugamonas violacea sp. nov. Microbiol. Spectr. 2021, 9, e00452-21. [Google Scholar] [CrossRef] [PubMed]
- Kýrová, K.; Sedláček, I.; Pantůček, R.; Králová, S.; Holochová, P.; Mašlaňová, I.; Staňková, E.; Kleinhagauer, T.; Gelbíčová, T.; Sobotka, R.; et al. Rufibacter ruber sp. nov., isolated from fragmentary rock. Int. J. Syst. Evol. Microbiol. 2016, 66, 4401–4405. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ma, J. The Global Catalogue of Microorganisms (GCM) 10K type strain sequencing project: Providing services to taxonomists for standard genome sequencing and annotation. Int. J. Syst. Evol. Microbiol. 2019, 69, 895–898. [Google Scholar] [CrossRef]
- Shi, W.; Sun, Q.; Fan, G.; Hideaki, S.; Moriya, O.; Itoh, T.; Zhou, Y.; Cai, M.; Kim, S.-G.; Lee, J.-S.; et al. gcType: A high-quality type strain genome database for microbial phylogenetic and functional research. Nucleic Acids Res. 2021, 49, D694–D705. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Staals, R.; Morales, S.; Fineran, P.; Brown, C.M. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef]
- Busse, H.-J.; Auling, G. Polyamine pattern as a chemotaxonomic marker within the Proteobacteria. Syst. Appl. Microbiol. 1988, 11, 1–8. [Google Scholar] [CrossRef]
- Busse, H.-J.; Bunka, S.; Hensel, A.; Lubitz, W. Discrimination of members of the family Pasteurellaceae based on polyamine patterns. Int. J. Syst. Evol. Microbiol. 1997, 47, 698–708. [Google Scholar] [CrossRef]
- Tindall, B.J. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 1990, 66, 199–202. [Google Scholar] [CrossRef]
- Tindall, B. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 1990, 13, 128–130. [Google Scholar] [CrossRef]
- Altenburger, P.; Kämpfer, P.; Makristathis, A.; Lubitz, W.; Busse, H.-J. Classification of bacteria isolated from a medieval wall painting. J. Biotechnol. 1996, 47, 39–52. [Google Scholar] [CrossRef]
- Stolz, A.; Busse, H.-J.; Kampfer, P. Pseudomonas knackmussii sp. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note 101; Microbial ID, Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Švec, P.; Pantůček, R.; Petráš, P.; Sedláček, I.; Nováková, D. Identification of Staphylococcus spp. using (GTG)5-PCR fingerprinting. Syst. Appl. Microbiol. 2010, 33, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Carlone, G.M.; Valadez, M.J.; Pickett, M.J. Methods for distinguishing gram-positive from gram-negative bacteria. J. Clin. Microbiol. 1982, 16, 1157–1159. [Google Scholar] [CrossRef]
- Sedláček, I.; Králová, S.; Kýrová, K.; Mašlaňová, I.; Busse, H.-J.; Staňková, E.; Vrbovská, V.; Němec, M.; Barták, M.; Holochová, P.; et al. Red-pink pigmented Hymenobacter coccineus sp. nov., Hymenobacter lapidarius sp. nov. and Hymenobacter glacialis sp. nov., isolated from rocks in Antarctica. Int. J. Syst. Evol. Microbiol. 2017, 67, 1975–1983. [Google Scholar] [CrossRef]
- Da, X.; Jiang, F.; Chang, X.; Ren, L.; Qiu, X.; Kan, W.; Zhang, Y.; Deng, S.; Fang, C.; Peng, F. Pedobacter ardleyensis sp. nov., isolated from soil in Antarctica. Int. J. Syst. Evol. Microbiol. 2015, 65, 3841–3846. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; ASM Press: Washington, DC, USA, 2010. [Google Scholar]
- Barrow, G.I.; Feltham, R.K.A. Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd ed.; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Kosina, M.; Barták, M.; Mašlaňová, I.; Pascutti, A.V.; Šedo, O.; Lexa, M.; Sedláček, I. Pseudomonas prosekii sp. nov., a novel psychrotrophic bacterium from Antarctica. Curr. Microbiol. 2013, 67, 637–646. [Google Scholar] [CrossRef]
- Margesin, R.; Gander, S.; Zacke, G.; Gounot, A.M.; Schinner, F. Hydrocarbon degradation and enzyme activities of cold-adapted bacteria and yeasts. Extremophiles 2003, 7, 451–458. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement (M100-S25); Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 35, No. 3. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of Mics and Zone Diameters, Version 12.0; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022. [Google Scholar]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Vasu, K.; Nagaraja, V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Białkowska, A.; Majewska, E.; Olczak, A.; Twarda-Clapa, A. Ice binding proteins: Diverse biological roles and applications in different types of industry. Biomolecules 2020, 10, 274. [Google Scholar] [CrossRef]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ´omic´ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta Biomembr. 2004, 1666, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Obruča, S.; Dvořák, P.; Sedláček, P.; Koller, M.; Sedlář, K.; Pernicová, I.; Šafránek, D. Polyhydroxyalkanoates synthesis by halophiles and thermophiles: Towards sustainable production of microbial bioplastics. Biotechnol. Adv. 2022, 107906. [Google Scholar] [CrossRef] [PubMed]
| Strain Number | Year of Isolation | Locality (GPS) |
|---|---|---|
| P8323 (CCM 8944) | 2017 | Seals stream, nearby Monolith lake, James Ross Island (63°53′10″ S, 57°56′50″ W) |
| P8398T (CCM 8941T) | 2017 | Glacial stream, Komárek valley, James Ross Island (63°49′51″ S, 57°49′48″ W) |
| P8910 (CCM 9189) | 2017 | Small stream nearby Whiskey glacier, James Ross Island (63°55′03″ S, 57°56′25″ W) |
| P9640 | 2018 | Lakelet nearby sea coast, Eagle Island (63°37′29″ S, 57°25′52″ W) |
| P9735 | 2018 | Lakelet below moraine Triangular, James Ross Island (63°51′42″ S, 57°51′07″ W) |
| P11689 | 2019 | Small lake, approx. 200 m SW from Rožmberk lake, James Ross Island (63°49′17″ S, 57°50′28″ W) |
| P11691 | 2019 | Small lake, approx. 200 m SW from Rožmberk lake, James Ross Island (63°49′17″ S, 57°50′28″ W) |
| Species | Strain | Gene Sequence Similarities of P8398T | ANI and dDDH Values of P8398T | Gene Sequence Similarities of P8910 | ANI and dDDH Values of P8910 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | gyrB | lepA | ANI | dDDH | 16S rRNA | gyrB | lepA | ANI | dDDH | ||
| M. antarctica sp. nov. | P8398T | 100 | 100 | 100 | 100 | 100 | 99.9 | 97.0 | 97.8 | 97.0 | 73.9 |
| M. antarctica sp. nov. | P8910 | 99.9 | 97.0 | 97.8 | 97.0 | 73.9 | 100 | 100 | 100 | 100 | 100 |
| M. aquatica | CCM 8693T | 99.7 | 95.8 | 95.0 | 87.6 | 33.7 | 99.8 | 96.0 | 95.0 | 87.7 | 33.7 |
| M. atriviolacea | CCM 8999T | 99.5 | 94.4 | 95.1 | 87.9 | 34.0 | 99.5 | 94.5 | 94.9 | 87.7 | 33.8 |
| M. eurypsychrophila | CCM 8735T | 98.8 | 89.1 | 88.2 | 79.3 | 23.1 | 98.7 | 89.1 | 87.4 | 79.2 | 23.0 |
| M. frigida | CCM 8695T | 100 | 96.0 | 96.4 | 92.1 | 46.4 | 99.9 | 96.4 | 96.4 | 92.2 | 46.3 |
| M. glaciei | CCM 8861T | 98.9 | 90.9 | 88.9 | 79.0 | 23.4 | 99.0 | 90.7 | 88.5 | 79.0 | 23.1 |
| M. mucilaginosa | CCM 8733T | 99.7 | 96.5 | 95.5 | 88.0 | 34.7 | 99.6 | 96.6 | 95.1 | 88.0 | 34.6 |
| M. rubra | CCM 8692T | 99.9 | 95.4 | 94.9 | 88.0 | 34.4 | 99.9 | 95.5 | 94.5 | 88.1 | 34.4 |
| M. violaceinigra | CCM 8877T | 99.9 | 95.4 | 95.5 | 91.0 | 42.4 | 99.9 | 95.7 | 95.3 | 91.0 | 42.1 |
| Test | 1 * | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Violet pigment | + | - | - | - | - | + | + | - | - |
| Pink-red pigment | - | + | + | + | + | - | - | - | - |
| Oxidase | - | - | - | - | - | - | - | + | + |
| Growth at 5 °C on R2A | + | + | + | + | + | - | + | + | + |
| Growth in 0.5% NaCl | - | - | + | + | - | - | w | - | - |
| Acid from xylose | + | + | w | - | - | - | + | - | - |
| API ZYM: Lipase (C14) | - | - | - | - | - | + | w | - | + |
| Cystine arylamidase | - | - | - | - | - | + | - | - | + |
| BIOLOG GEN III: Arabitol | - | - | - | - | - | - | + | - | - |
| Pectin | - | - | - | - | - | - | + | - | - |
| Glucuronamide | - | + | + | w | + | + | - | - | + |
| Chloramphenicol (30 µg) | R | S | R | S | S | S | R | S | S |
| Hydrolysis of: Gelatine | + | + | + | + | + | + | + | + | - |
| Starch | + | - | + | - | + | + | + | + | - |
| ONPG | + | + | - | - | + | + | - | + | - |
| Tween 80 | + | + | + | + | - | + | + | - | + |
| Tyrosine | + | + | + | + | + | + | + | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedláček, I.; Holochová, P.; Busse, H.-J.; Koublová, V.; Králová, S.; Švec, P.; Sobotka, R.; Staňková, E.; Pilný, J.; Šedo, O.; et al. Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov. Microorganisms 2022, 10, 704. https://doi.org/10.3390/microorganisms10040704
Sedláček I, Holochová P, Busse H-J, Koublová V, Králová S, Švec P, Sobotka R, Staňková E, Pilný J, Šedo O, et al. Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov. Microorganisms. 2022; 10(4):704. https://doi.org/10.3390/microorganisms10040704
Chicago/Turabian StyleSedláček, Ivo, Pavla Holochová, Hans-Jürgen Busse, Vendula Koublová, Stanislava Králová, Pavel Švec, Roman Sobotka, Eva Staňková, Jan Pilný, Ondrej Šedo, and et al. 2022. "Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov." Microorganisms 10, no. 4: 704. https://doi.org/10.3390/microorganisms10040704
APA StyleSedláček, I., Holochová, P., Busse, H.-J., Koublová, V., Králová, S., Švec, P., Sobotka, R., Staňková, E., Pilný, J., Šedo, O., Smolíková, J., & Sedlář, K. (2022). Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov. Microorganisms, 10(4), 704. https://doi.org/10.3390/microorganisms10040704







