Abstract
Human–wildlife coexistence may increase the potential risk of direct transmission of emergent or re-emergent zoonotic pathogens to humans. Intending to assess the occurrence of three important foodborne pathogens in wild animals of two wildlife conservation centers in Portugal, we investigated 132 fecal samples for the presence of Escherichia coli (Shiga toxin-producing E. coli (STEC) and non-STEC), Salmonella spp. and Campylobacter spp. A genotypic search for genes having virulence and antimicrobial resistance (AMR) was performed by means of PCR and Whole-Genome Sequencing (WGS) and phenotypic (serotyping and AMR profiles) characterization. Overall, 62 samples tested positive for at least one of these species: 27.3% for STEC, 11.4% for non-STEC, 3.0% for Salmonella spp. and 6.8% for Campylobacter spp. AMR was detected in four E. coli isolates and the only Campylobacter coli isolated in this study. WGS analysis revealed that 57.7% (30/52) of pathogenic E. coli integrated genetic clusters of highly closely related isolates (often involving different animal species), supporting the circulation and transmission of different pathogenic E. coli strains in the studied areas. These results support the idea that the health of humans, animals and ecosystems are interconnected, reinforcing the importance of a One Health approach to better monitor and control public health threats.
1. Introduction
Wildlife–livestock–human interfaces represent critical points for cross-species interaction and consequent transmission and emergence/re-emergence of pathogens into new host populations [1]. In fact, more than 1400 species of human pathogens have been recognized, of which more than half are of zoonotic origin. These pathogens can have a broad range of hosts, and it is a complex task to assess direct and indirect economic losses in sectors such as public health, animal health, and the environment [2,3,4]. As such, the implementation of integrative wildlife surveillance programs, following a One Health approach, is critical to learn about the interconnectedness of different zoonotic pathogens present in humans, animals and the environment and, consequently, to enhance the detection and control of public health threats [5,6].
According to the Zoonoses Directive 2003/99/EC, it is mandatory for European Union (EU) Member states (MS) to collect and report relevant data on zoonoses, zoonotic agents, antimicrobial resistance (AMR) and foodborne outbreaks to the European Food Safety Authority (EFSA). In 2020, zoonoses data from 36 European countries (27 MS and 9 non-MS) were reported. The analysis of the results showed that campylobacteriosis and salmonellosis were the most common reported zoonoses, with the total number of confirmed human cases being stable between 2015 and 2020. Shiga toxin-producing Escherichia coli (STEC) was the fourth most reported foodborne gastrointestinal infection in humans. Regarding STEC, after an increase of cases from 2015 to 2019, a reduction of infections was observed in 2020, probably due to the impact of the restrictions imposed by the COVID-19 pandemic [4].
Nowadays, there is evidence that wild animals are important reservoirs in the transmission of zoonotic agents, both to other animals and to humans. Wild boars (Sus scrofa) have a worldwide geographic distribution, with a significant increase in Europe in the last decades [7]. In Portugal, wild boars and deer (red deer, Cervus elaphus; fallow deer, Cervus dama) are mainly found in limited and controlled areas. In general, both species of these wild animals are considered to be potential vectors in the transmission of severe pathogens to humans, either by direct contact, for instance, in the context of hunting activities, or by indirect contamination of foodstuffs, water or environmental areas frequented by people or pets, via urine or feces. Commensal and pathogenic strains may also represent a serious hazard and an important risk for human health, regarding their antimicrobial resistance [8].
Although some studies which have focused on wild animals as reservoirs for presumptive zoonoses have been published in Europe, from a One Health perspective, they are scarce and have mainly focused on one pathogen [9,10,11,12,13,14,15,16,17,18,19,20].
The main goals of this study were to assess the occurrence of pathogenic E. coli, Salmonella spp., and Campylobacter spp. in wild animals (wild boar, red deer, fallow deer, hedgehogs (Erinaceus europaeus), and genets (Genetta genetta)) from two different populations and habitats, and to characterize the serotypes, the associated virulence markers, the antimicrobial resistance (AMR) profiles and the genetic diversity of the obtained isolates. Furthermore, as wild animals can act as efficient AMR reservoirs, as well as sources of epidemiological information and potential links between strains from human, livestock and natural environments, determination of the AMR profile of a fraction of the non-pathogenic E. coli was carried out. In addition, a comprehensive genomic study targeting pathogenic E. coli strains was performed.
To our best knowledge, this is the first study in Portugal, and one of the few in Europe, that simultaneously evaluates the presence of, and characterizes, E. coli (STEC and non-STEC), Salmonella spp. and Campylobacter spp. in wildlife, using both culture dependent methodologies and Whole-Genome Sequencing (WGS).
2. Materials and Methods
2.1. Study Areas
The selected sampling areas were two natural wildlife conservation centers, located in distinct geographical regions: Tapada Nacional de Mafra (TNM), in the Lisbon district, and The Centro de Recuperação de Fauna do Parque Biológico de Gaia (CRFPBG), in the Oporto district. TNM is a protected green area of about 1200 hectares, home to free-roaming wild boar and deer, among other animals. It is a national hunting zone (in a controlled and limited way) and used for rural tourism and leisure. A 17.5-km-long river, which originates in a nearby village, crosses through TNM to flow out into the Atlantic Ocean. CRFPBG is a 35-ha park located in an agroforestry area, and shelters hundreds of species in their natural habitats. The CRFPBG recuperates wild animals that have suffered any form of injuries or which were illegally held in captivity.
2.2. Study Population and Sample Collection
Animal fecal samples (N = 132) analyzed in this study were collected between July 2020 and June 2021, in a non-controlled sampling. The sampling was conducted with disposable sterile tools and the samples placed in sterile stool sample containers. Fresh feces of wild boar, red deer and fallow deer were collected at TNM (N = 113), under the supervision of a local biologist. Collection points were spaced apart to increase the likelihood of screening different individuals. Due to the COVID-19 pandemic restrictions, sampling was carried out in July, October, and November 2020, and later in May 2021. Feces of hedgehogs and genets were collected from animals rescued by CRFPBG (N = 19) between November 2020 and June 2021, excluding December 2020, and February and March 2021. All samples were kept refrigerated and processed immediately after arriving at the laboratory.
2.3. Isolation Methodology
For E. coli, a pre-enrichment in Buffered Peptone Water (1:10 dilution; BPW-Oxoid, Basingstoke, Hampshire, UK) was performed at 37 °C overnight and seeded in Tryptone Bile X-Glucuronide (TBX, Biokar Diagnostics, Pantin, France) and CHROMagar STEC (CHROMagar, Paris, France) agar plates for 20 h at 44 °C and 24 h at 37 °C, respectively. Five to ten colonies were selected and sub-cultured on Tryptone Soy Agar (TSA, Biokar Diagnostics) overnight at 37 °C. Suspicious colonies were confirmed by biochemical identification on the VITEK 2 compact system (bioMérieux, Marcy L’Etoile, France) or by amplification of E. coli 16S rRNA, according to Sabat et al. (2000) [21]. All positive isolates were stored at −80 °C in Tryptone Soy Broth (TSB, Biokar Diagnostics) with 20% glycerol.
Isolation of Salmonella spp. was achieved according to ISO 6579-1:2017 [22]. After a pre-enrichment step, as previously described, a selective enrichment of 1 mL culture was performed in 10 mL of Muller–Kauffmann tetrathionate–novobiocin broth (MKTTn, Biokar Diagnostics) at 37 °C for 24 h. Simultaneously, 50 µL were added to the center of a Modified Semi-solid Rappaport–Vassiliadis agar plate (MSRV, Biokar Diagnostics) and incubated at 41.5 °C for 24 h. Finally, IRIS Salmonella agar (Biokar Diagnostics) was used for plating-out for 24 h at 37 °C, and colonies of presumptive Salmonella spp. were isolated on TSA at 37 °C for 24 h. The identity of suspicious isolates was confirmed by biochemical identification on the VITEK 2 compact system. All positive isolates were stored at −80 °C, as previously described.
Campylobacter spp. isolation was based on ISO 10272-1:2006-1 [23]. A test portion of about 1 g was added to 10 mL of Bolton broth enrichment medium (Bolton Broth with Bolton broth selective supplement, and 5% Horse Blood Lysed, Oxoid) and incubated in a microaerobic atmosphere at 37 °C for 4–6 h and, then, at 41.5 °C for 48 h. Plating-out was performed by applying five drops (≈100 µL) of the enrichment culture to a 0.65-µm pore-size filter (Nitrocellulose membrane filters, Whatman, Little Chalfont, Buckinghamshire, UK) placed over a plate of Columbia Agar + 5% Sheep blood (COS; bioMérieux). The filter was left in contact with the surface of the agar for 15 min at room temperature. Furthermore, about 100 µL of the enrichment culture was spread over a plate of modified Charcoal Cefoperazone Deoxycholate agar (mCCDA, Oxoid). Plates were incubated in a microaerobic atmosphere at 41.5 °C for 48 h. Three to five presumed Campylobacter colonies were sub-cultured on COS and their identification confirmed by oxidase activity and MALDI-TOF mass spectrometry (VITEK® MS, bioMérieux). All positive isolates were stored, as previously described.
2.4. Bacterial Typing and Antimicrobial Susceptibility Testing
Identification of potentially pathogenic E. coli was performed by screening for the presence of eae, aggR, elt, estp, and ipaH virulence genes (VG) by multiplex PCR (modified from Persson 2007, Boisen 2012, and Fujioka 2013) [24,25,26] and for the presence of Shiga toxins stx1 and stx2 [27]. Detection was initially performed in pools with a maximum of 10 colonies from each sample. Whenever a PCR-positive result was detected, all the pool isolates were analyzed individually. A single colony was boiled for 10 min, cooled for 1 min, centrifuged at 16.200× g for 10 min, and the supernatant was used as a DNA template. The amplification of VG was performed in a reaction volume of 25 µL and using the HotStar Taq master mix (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. For Shiga toxins detection, two separate reactions with a final volume of 20 µL and using the HotStar Taq master mix were prepared. Primers and PCR profile are presented in the Supplementary Material Table S1. Strains of External Quality Assessments (organized by the Statens Serum Institut, Copenhagen, Denmark) were used as positive controls. Amplicons were visualized on a 2.5% agarose gel in 0.5x TBE buffer at 100 V for 30–45 min. An E. coli isolate was classified as potentially pathogenic (STEC; EPEC, Enteropathogenic E. coli; EAEC, Enteroaggregative E. coli; ETEC, Enterotoxigenic E. coli; EIEC, Enteroinvasive E. coli) when at least one of the tested genes was detected.
Antimicrobial susceptibility testing (AST) was performed according to the Kirby–Bauer method in all pathogenic E. coli (N = 52) and in 42.3% (N = 33) of the non-pathogenic E. coli isolates (total of 85 isolates), following the European Committee on Antimicrobial Susceptibility Testing recommendations [28]. For the testing, the following panel of 18 antimicrobials was used: Ampicillin (AMP), Amoxicillin-Clavulanic Acid (AMC), Azithromycin (AZM), Cefepime (FEP), Cefotaxime (COX), Cefoxitin (FOX), Ceftazidime (CZD), Ceftriaxone (CRO), Chloramphenicol (CHL), Erythromycin (ERY), Gentamicin (GMN), Meropenem (MEM), Nalidixic Acid (NAL), Ciprofloxacin (CIP), Sulfamethoxazole (SMX), Tetracycline (TET), Tigecycline (TGC), and Trimethoprim (TMP). The results were interpreted according to the EUCAST epidemiological cut-off values (ECOFFs) [28]. An isolate was classified as multidrug-resistant (MDR) when it presented resistance to three or more antimicrobial classes.
Salmonella isolates were serotyped by the slide agglutination method for O and H antigens (SSI Diagnostica, Hillerod, Denmark; Sifin diagnostics, Berlin, Germany), according to the Kauffmann–White–Le Minor scheme [29]. AST was performed, as previously described, for E. coli to 17 antimicrobials, including Pefloxacin (PEF) instead of CIP, and excluding ERY.
For Campylobacter spp., AST was conducted as previously described for CIP, ERY, TET, GMN according to EUCAST 2021 [29], and for AMP and AMC according to Comité de l’antibiogramme de la Société Française de Microbiologie [30].
2.5. Whole-Genome Sequencing, in Silico Typing and Screening of Virulence/AMR Genes
Genomic DNA was extracted from fresh cultures of all the pathogenic isolates using the ISOLATE II Genomic DNA Kit (Bioline, London, England, UK), and quantified in the Qubit fluorometer (Invitrogen, Waltham, MA, USA) with the dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. Salmonella enterica enterica ser. Veneziana and Campylobacter hyointestinalis isolates were not sequenced, as they are currently not considered to be relevant pathogens in Portugal. DNA was then subjected to the NexteraXT library preparation protocol (Illumina, San Diego, CA, USA) prior to cluster generation and paired-end sequencing (2 × 250 bp or 2 × 150 bp) on either a MiSeq or a NextSeq 550 instrument (Illumina), according to the manufacturer’s instructions. FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 October 2022)) was used for quality control and Trimmomatic v0.38 [31] for trimming low-quality bases. For E. coli and Salmonella, sequencing reads were submitted to the Center for Genomic Epidemiology web server (https://cge.cbs.dtu.dk (accessed on 6 October 2022)) for identification of antimicrobial resistance genes (ResFinder 4.1), in silico Multilocus Sequence Typing (MLST) (MLST 2.0), E. coli virulence genes identification (VirulenceFinder 2.0), and in silico E. coli (SerotypeFinder 2.0) and Salmonella serotyping (SeqSero 1.2). For Campylobacter, in silico MLST was determined at the PubMLST platform (https://pubmlst.org/ (accessed on 6 October 2022)).
Sequencing reads were deposited in the European Nucleotide Archive (ENA) under the bioproject PRJEB54735 (E. coli), PRJEB32515 (Salmonella) and PRJEB46733 (C. coli). Accession numbers for each isolate are listed in Supplementary Material Table S2.
2.6. Core-Genome MLST Clustering Analysis of Pathogenic E. coli
Core-genome MLST (cgMLST) analysis was performed for the 52 pathogenic E. coli isolates. To this end, we performed de novo genome assembly with the INNUca pipeline v4.2.2 (https://github.com/B-UMMI/INNUca (accessed on 6 October 2022)) [32] using default parameters. De novo genome assembly was performed with SPAdes v3.14 [33]. Reads were aligned with Bowtie v2.2.9 [34] and the assembly was polished with Pilon v1.23 [35]. Species confirmation/contamination screening was performed with Kraken2 v2.0.7 [36]. As the sample Ec-TM86 revealed traces of contamination with Morganella morganii, the E. coli-classified contigs were retrieved with an in-house script (https://github.com/vmixao/scripts/kraken_results2filter_assembly.py (accessed on 6 October 2022)), yielding a final assembly with the expected genome size for E. coli.
Allele-calling was performed with chewBBACA v2.8.5 [37] using the latest version of the 7601-loci wgMLST schema, curated and available at Chewie-NS website (https://chewbbaca.online/, last change date on 31 May 2021) [38]. First, a global clustering analysis was performed with ReporTree v1.0.0 (https://github.com/insapathogenomics/ReporTree (accessed on 6 October 2022)) [39] over the core-genome of the dataset (i.e., a sub-schema comprising loci called in all samples, n = 2567 loci), using GrapeTree (MSTreeV2 method) [40]. Clusters of closely related isolates were determined at a distance threshold equivalent to 0.34% of the total number of alleles of the applied cgMLST schema (i.e., 0.0034 × 2567 loci = ~9 allelic differences - ADs), as this threshold can provide a proxy to the identification of genetic clusters with potential epidemiological concordance (i.e., “outbreaks”) for E. coli [32]. Of note, the choice of a loci panel called in 100% of the isolates for this first clustering analysis, and resulted from preliminary analyses showing that a more flexible threshold for core-schema definition (including loci that were only called in 95% and 98% of samples) did not result in a substantial increase of the core-genome size, or, thus, in resolution. The schema size was also not impacted by the inclusion/exclusion of the curated Ec-TM86 assembly. Second, an in-depth clustering analysis for each ST was performed using a ST-specific core-genome schema (i.e., a sub-schema comprising the loci called in all the isolates of the same ST) and applying the same threshold for cluster definition. By maximizing the shared genome, this dynamic approach allowed increasing the resolution power and confidence concerning the initially detected clusters. Summary reports for the determined genetic clusters was automatically generated by ReporTree v1.0.0 [39], including cluster composition, timespan and distribution of host species or AMR phenotypes. In order to assess the genetic differences at SNP level between the isolates Ec-TM26 and Ec-TM69, which presented different AMR phenotypes but similar allelic profiles, an additional SNP analysis was conducted using Snippy v4.6.0 (https://github.com/tseemann/snippy (accessed on 6 October 2022)), setting “--mapqual 20 --mincov 10 --minfrac 0.51 --basequal 20” and using the Ec-TM26 assembly as reference.
3. Results
3.1. Detection and Characterization of Isolates
In the present study, 132 fecal samples from wild animals (51 from wild boar, 50 from fallow deer, 12 from red deer, 18 from hedgehogs, and 1 from a genet) were evaluated for the presence of pathogenic E. coli and Salmonella spp., and 118 samples for Campylobacter spp. (39 wild boar, 50 fallow deer, 12 red deer, 16 hedgehog, and 1 genet) (Table 1).

Table 1.
Isolation and characterization of STEC and non-STEC E. coli, Salmonella spp. and Campylobacter spp. in 132 tested samples.
Overall, 62 of the 132 tested samples (47.0%) tested positive for at least one of the evaluated pathogenic bacterial species. In total, pathogenic E. coli (STEC and non-STEC), Salmonella spp. and Campylobacter spp. were detected in 37.9% (50/132), 3.0% (4/132) and 6.8% (8/118) of the samples, respectively (Table 1). There were no samples presenting two different pathogenic bacterial species simultaneously.
Pathogenic E. coli was recovered in 17.6% (9/51) of the wild boar samples, 68.0% (34/50) of fallow deer samples, 50% (6/12) of red deer samples, and 5.5% (1/18) of the hedgehog samples (Table 1). STEC was the most frequently detected pathotype (27.3%; 36/132), being identified in wild boar, fallow deer and red deer samples, followed by EPEC (9.1%; 12/132), identified in wild boar, fallow deer, red deer and hedgehog samples, and ETEC (2.3%; 3/132), found only in wild boar samples (Table 1). Two fallow deer samples contained two different pathogenic E. coli isolates: one sample contained one EPEC and one STEC isolate, and the other contained two different STEC isolates (Ec-TM29 and EcTM30, respectively; Table S2).
Although Salmonella was rarely found in the studied animals, it is noteworthy that there was the identification of three distinct serovars: two cases of S. Schleissheim (1.5%; 2/132), recovered from wild boar and red deer samples, one S. Enteritidis (0.8%; 1/132) recovered from a wild boar sample, and one S. Veneziana (0.8%; 1/132) isolated from a hedgehog sample (Table 1).
Regarding Campylobacter, C. hyointestinalis was detected in seven wild boar samples (5.9%; 7/118) and Campylobacter coli in one genet’s sample (0.8%; 1/118) (Table 1).
3.2. Antimicrobial Susceptibility Testing and in Silico Genotyping
The S. Schleissheim isolates belonged to ST53 and S. Enteritidis to ST11. All Salmonella isolates were phenotypically susceptible to the 17 tested antimicrobials. WGS of the S. Schleissheim and S. Enteritidis, revealed the presence of the aac(6′)-Iaa gene.
The Campylobacter coli isolate was identified as belonging to ST1595. The isolate was resistant to CIP, TET and AMP, and harbored the corresponding resistance determinants, the gyrA (Thr86Ile) mutation, and the tetO and blaOXA-61 genes, respectively.
Among the 52 E. coli pathogenic isolates, 15 O antigens and 10 H antigens were identified in silico. The most prevalent serotypes were O146:H21 and O75:H8 (10 STEC isolates each), followed by serotypes O146:H28 (8 STEC isolates) and O27:H30 (5 STEC isolates) (Table 2).

Table 2.
Distribution of serotypes, sequence types, pathotypes and virulence determinants among the 52 pathogenic E. coli isolates, determined by PCR and WGS.
E. coli MLST analysis identified 16 ST, with ST13 being the most common (11 isolates) followed by ST442 (10 isolates), ST738 (8 isolates) and ST753 (5 isolates) (Table 2).
For E. coli, the antimicrobial resistance level was low, being phenotypically detected in 4.7% of the tested isolates (4/85; 5.8% in pathogenic and 3.0% in non-pathogenic E. coli isolates), of which one was classified as MDR. In silico genotyping confirmed the found pathogenic E. coli AMR phenotypes (Table S2). One wild boar ETEC isolate (Ec-TM17) was resistant to TET and SMX (tet(B) and sul2 genes); one red deer EPEC isolate (Ec-TM100) was resistant to AMP, CIP and NAL (blaTEM-1D, qnrB36, qnrB19, qnrB82, gyrA), one MDR wild boar EPEC isolate (Ec-TM26) was resistant to AMP, CHL and SMX (blaTEM-1B, floR, sul2), and one non-pathogenic E. coli isolate (Ec-PBG26), recovered from the genet faecal sample, was resistant to SMX.
All 52 pathogenic E. coli isolates contained virulence genes encoding toxins (Table 2), as well as the genes used for the primarily pathotype classification, e.g., by PCR. The most frequently detected genes were ehxa (76.9%), stx2b (71.2%), subA (59.6%), mchF (51.9%), and ast A (50.0%). Other genes are mentioned in Table 2 and Table S2. Among STEC isolates, 70.3% (26/37) presented only the stx2 gene (stx2b) and 29.7% (11/37) the stx1 and stx2 genes (stx1c and stx2b), and all of them were eae-negative (Table 2, Table S2).
The three isolates encoding the major number of virulence factors were all EPEC isolates: Ec-TM26 (ST137; O145:HND; MDT), the Ec-TM57 (ST29; O70:H11), and the Ec-TM65 (ST29; O26:H11) (Table 2).
3.3. Core Genome MLST Clustering Analysis of Pathogenic E. coli Isolates
A key step in One Health surveillance is the assessment of the genetic diversity of the circulating pathogens, including the identification of strains circulating among different host species and/or across long periods of time. To assess whether different E. coli pathogenic isolates (collected at different time points and/or sources) cluster together at high resolution level (thus, potentially representing the same circulating strain), a first cgMLST analysis comprising the 52 isolates was performed (Figure 1).
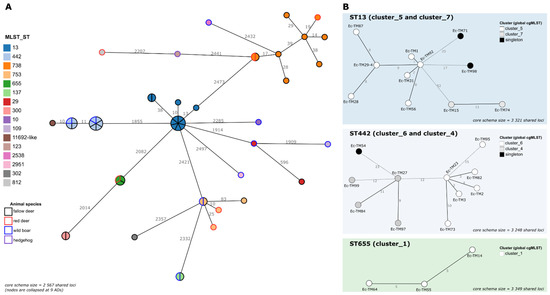
Figure 1.
The cgMLST analysis of pathogenic E. coli. (A) The global Minimum Spanning Tree (MST) was reconstructed, based on the core-genome of the 52 isolates (i.e., a sub-schema of the original 7601-loci wgMLST schema comprising loci called in all samples). Each circle (node) represents a single isolate or a cluster of isolates linked by ≤9 ADs (corresponding to the 0.34% threshold applied for cluster investigation). Each division in a node corresponds to a single isolate. Node colors reflect the ST (inner color) and the animal species (outer line color) of each isolate. Lines connect nodes with ADs above the applied threshold, with the numbers representing the ADs between nodes. (B) For each ST of interest (details in Table 3), an MST was constructed, based on a dynamic cgMLST analysis, enrolling a maximized ST-specific core-genome schema (i.e., a sub-schema comprising the loci called in all the isolates of the same ST). Only the MSTs corresponding to STs harboring clusters with >2 isolates are presented (see details for the others in Table 3). Each circle (node) represents a unique allelic profile, with colors reflecting the genetic cluster to which the isolate belonged in the global cgMLST analysis. Straight and dotted lines reflect nodes linked with the ADs below/equal, and above, the 0.34% threshold applied for cluster investigation, respectively. The numbers in the connecting lines represent the ADs between isolates. All MSTs were constructed using the GrapeTree MSTreeV2 algorithm implemented in the ReporTree pipeline (https://github.com/insapathogenomics/ReporTree (accessed on 6 October 2022)), using the chewieNS schema (https://chewbbaca.online/ (accessed on 6 October 2022)). Data visualization was adapted from the GrapeTree dashboard.
A threshold of 0.34% ADs (9 ADs in 2567 shared loci) was applied to the generated MST as a proxy to identify clusters of very closely related isolates [32]. Despite the low number of samples present in our dataset, 57.7% (30/52) of the isolates integrated clusters, in a total of 9 genetic clusters (2 to 7 isolates per cluster) across seven STs and two pathotypes (Table 3). Six out of the nine genetic clusters comprised isolates collected from different animal species, but the largest cluster (cluster_5; STEC) was exclusively isolated from fallow deer (Figure 1, Table 3, Supplementary Material Table S2). Of note, the time span of most of the clusters extended almost up to four months.

Table 3.
Genetic clusters identified among pathogenic E. coli through cgMLST analyses.
To increase the clustering resolution, an additional cgMLST analysis per ST, maximizing the number of shared loci under evaluation, was performed. This in-depth assessment not only confirmed the detected clusters but also provided further clues about extra isolate/cluster linkages (Figure 1 and Table 3). Of particular note was a well-confirmed genetic cluster (cluster_9) comprising two EPEC isolates, collected from different animal species, with different antibiotic-resistance phenotypes (Table 3), one of them being susceptible (Ec-TM69) to the tested antimicrobials and the other (Ec-TM26) being resistant to AMP, CHL and SMX (Table 3, Supplementary Material Table S2). Finely comparative cgMLST and SNP analyses of these isolates confirmed their high genetic relatedness (≤5 ADs/SNPs), while suggesting horizontal gene transfer as the source of the Ec-TM26 resistance phenotypes. Indeed, AMP- and SMX-resistance could be linked to the presence of blaTEM-1B and sul2 genes, respectively, both co-localizing in the same putative mobile genetic element, which also harbored the aph(6)-Id and aph(3′’)-Ib genes, linked to streptomycin resistance (phenotype not tested). The CHL resistance, associated with the presence of the floR gene, again fell into a putative mobile genetic element. Despite the near isogenicity of Ec-TM26 and Ec-TM69 in the extended core genome, these elements were confirmed to be absent in the susceptible Ec-TM69.
4. Discussion
The occurrence of pathogenic E. coli in fecal samples from wild ungulates in Europe is extremely variable. The frequency value found in this study was in accordance with the ones reported in other studies done recently in Italy [10] and Poland [16], but higher than studies found in Spain [11] and Portugal [15]. Indeed, sometimes, even studies within the same country report different outcomes [18,41] and, in some cases, researchers have not found pathogenic E. coli in analyzed samples [42,43,44]. This heterogeneity may be related to numerous factors, such as the proximity of urban areas, and the number of animals per ha, or may even be related to the season in which sample collection was conducted. The high frequency of pathogenic E. coli found in TNM ungulates may be justified by the fact that the studied population is from a geographically limited area that is in close contact with humans (through visits and hunting activities) and with a high concentration of animals per ha.
For Salmonella and Campylobacter, the comparison of the results found in this study with other European studies performed in ungulate fecal samples shows highly variable outcomes. Regarding Salmonella, several studies have reported the absence of this pathogen in tested samples [18,42,43]. However, there are several other research works reporting frequency values between 1.1 and 10.8% [12,17,45,46,47,48], and at least one reporting a higher value (17.5%) in wild boars, particularly in populations co-habiting with cattle, where the rate increases to 35.7% [49]. In the case of Campylobacter, there are several studies reporting frequency values below 5% [17,42,43,50]. However, there is at least one study, in Spain, which referred to a rate of 15.2% in wild artiodactyl species [51] and another one, carried out in Italy, which reported a 91.66% frequency value of C. coli in wild boars [19].
As AMR poses a major worldwide threat to human health [52], the determination of the AMR profile of all pathogenic isolates, as well as of a part of the commensal E. coli, was carried out. In compliance with this study, the presence of several antimicrobial resistance genes had already been detected in other E. coli, Salmonella and Campylobacter isolates from fecal samples of wild ungulates in Europe [10,17,19,43,45,47,49,51]. In 2018 and 2019, in accordance with Commission Implementing Decision 2013/652/EU, phenotypic AMR was monitored in E. coli isolates obtained from fecal samples of the most relevant food-producing animals at slaughter (fattening pigs, calves under 1 year of age, broilers and fattening turkeys). The goal was to provide information on the reservoirs of resistant bacteria that could potentially be transferred between animal populations and from animals to humans [8]. According to the European Union Summary Report on Antimicrobial Resistance in zoonotic bacteria and indicator bacteria from humans, regarding animals and food in 2018/2019 [8], all antimicrobial resistance traits, identified in this study (sulfamethoxazole, tetracycline, chloramphenicol, ampicillin, ciprofloxacin and nalidixic acid), were already highly disseminated among food-producing animals, reinforcing the importance of a One Health approach to better monitor and control public health threats. It is also important to highlight that the three AMR Escherichia coli isolates belonged to serotypes already found in confirmed cases of E. coli human infections [53,54,55] and that the MDR isolate belonged to serogroup O145, which was in the “top five” most frequent serogroups reported in confirmed cases of human STEC infections in EU/EEA in 2020 [56].
Regarding serotyping and genotypic characterization of the isolated Salmonella, and its potential relation with human infection, it is important to notice that Salmonella Enteritidis, one of the identified serotypes, is among the most prevalent foodborne pathogen worldwide [57]. ST11, the one found in this study, is the major Salmonella Enteritidis ST and has been associated with recent multi-country outbreaks in Europe [58,59]. Although, according to the diversity in Enterobase (as of August 2022), Salmonella Schleissheim has been detected in humans, cattle, poultry, wild animals, foodstuffs and environmental samples, the S. Schleissheim isolated from wild animals (wild boar and red deer), in the present study, belonged to ST53, which, thus far, has only been reported in foodstuffs (https://enterobase.warwick.ac.uk/species/index/senterica (accessed on 6 October 2022)). Salmonella Veneziana serotype, isolated here from a hedgehog, has been considered a non-significant serotype in human disease. However, according to the diversity in Enterobase (as of August 2022), it has already been isolated from humans (https://enterobase.warwick.ac.uk/species/index/senterica (accessed on 6 October 2022)) and has already been potentially associated with a case of acute terminal Ileitis [60].
Concerning the isolated Campylobacter species, C. coli, identified in one genet sample, is the second most frequently reported Campylobacter species in human infections [61]. The identified sequence type, ST1595, is documented (based on the PubMLST collection as of August 2022) to have already been found in isolates from humans with campylobacteriosis, pigs, poultry (broilers and turkeys), waters, and other environmental samples (https://pubmlst.org/organisms/campylobacter-jejunicoli (accessed on 6 October 2022)). Campylobacter hyointestinalis serovar, isolated here from 7 wild boars, has already been isolated from several different animal species and is a member of the “emerging Campylobacter spp.” group that can also cause disease in humans [62].
With reference to E. coli typing results, it is relevant to mention that three of the identified O antigens belong to the top 20 most frequent serogroups reported in confirmed cases of human STEC infections in EU/EEA, 2015–2017 (O145, O146, O121) [63]. Moreover, serotype O26:H11, which has emerged as one of the most common non-O157 STEC strains causing human diseases in many countries [64], was detected in a fallow deer within this study. Contrarily to the isolates found in humans, the isolate collected in this study lacked Stx toxin genes. The absence of Stx-harboring phages in E. coli O26:H11 has already been found in isolates from healthy cattle and sheep at slaughter in Switzerland [65]. This genome alteration may promote E. coli resilience outside the host and enable adaptation to stress conditions in the gastrointestinal tract [66]. As in this study, serotypes O146:H28 and O27:H30 were already found in E. coli isolates in fecal samples from Iberian Peninsula ungulates [11,15]. In the scope of our work, as previously stated by Dias et al. [15], O27:H30 serotype was recovered from fecal samples of both deer and wild boar, confirming that the previously described association of this serotype with deer [11] no longer exists.
Looking at the toxin genes identified on the 52 pathogenic E. coli isolates, in agreement with other European studies performed on wild ungulates [15,16,41,67], our results showed that stx2 had a higher prevalence (71.2%) than stx1 (21.2%) among pathogenic E. coli isolates. Only 11 of the 37 STEC isolates contained both stx1 and stx2 genes, and none of them contained only stx1 gene. Although clinical epidemiological studies suggest that stx2 is more often associated with severe disease and development of hemolytic–uremic syndrome (HUS) than stx1, pointing out for a high pathogenicity of the isolated STECs, the stx2 subtype found in this study was always stx2b, a subtype with a potency similar to that of stx1 [68].
Analysis of STEC in Europe showed that stx2b, alone, or together with stx1c, is common in STEC from deer dropping and wildlife populations [9] but does not appear to cause severe human illness. Nevertheless, some studies have reported that 10 to 15% of human clinical samples from diarrheal illnesses are positive for stx1c and/or stx2b [69,70].
Heat-stable enterotoxin b gene (stb), which is known to be mainly found in association with porcine ETEC [71], was also only detected in the wild boar ETEC isolates (Table 2), one of them also showing the presence of heat-stable enterotoxin a (sta1) gene.
Two of the most common E. coli STs detected (442 and 738), were previously associated with cases of human disease. ST442 has been significantly associated with hemolytic uremic syndrome [72] and ST738 was one of the STs isolated during 2010–2014 from human cases of infection in Switzerland [70].
Regarding cgMLST analysis of the 52 E. coli pathogenic isolates, it is noteworthy that nine genetic clusters were detected from which six enrolled isolates collected from different animal species, evidencing direct or indirect transmission of the E. coli isolates between animals cohabiting in the studied natural conservation center. This was expected, since the TNM ungulates live in a geographically limited area with high animal population density. Moreover, there are several drinking fountains shared by these animals, and, in the summer, there is a food supplementation program that brings the animals that inhabit TNM closer together, thus, likely increasing the contact rate.
Another important observation regarded a genetic cluster (cluster_9) that comprised two nearly isogenic E. coli isolates with different antibiotic-resistance phenotypes (susceptible vs. MDR) (Table 3), a feature possibly acquired by horizontal gene transfer. One cannot discard the hypothesis that the circulation and transmission of AMR determinants might be linked to the proximity between humans and the studied population during visits and hunting activities. The river that crosses TNM may also be a wildlife–livestock–human interface to take in consideration. In fact, during this study, three samples from TNM river water were also analyzed and it is important to highlight the presence, in one of the samples, of an O157:H7, ST11 E. coli strain, which is known to be the most relevant pathogenic E. coli in humans. This isolate was susceptible to all tested antibiotics. All the other samples tested negative for pathogenic E. coli.
5. Conclusions
In conclusion, we can say that a wide range of factors, related to the transmission and ecology of diseases (namely increasing pressure of humans on natural ecosystems and rising interactions between the different species), has reached a plateau without precedent and this is a major concern for the control of wildlife diseases. Animal health surveillance is recognized as a key element in preventing public health risks related to emerging zoonotic diseases. From a public health perspective, the present research highlights and confirms that wild animals constitute important reservoirs of zoonotic pathogens like Escherichia coli, Salmonella spp. and Campylobacter spp., including resistant and MDR strains. These results reinforce the importance of a One Health approach, showing that a better understanding of community ecology is essential for a better understanding of the epidemiological links between all actors in the wildlife–livestock–human continuum. To our best knowledge, this is the first study in Portugal, and one of the few in Europe, that simultaneously performs the phenotypic and genotypic characterizations of three of the most common foodborne bacteria (Campylobacter spp., Salmonella spp., and pathogenic E. coli) isolated in fecal samples from wild animals.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10112132/s1, Table S1: Primers and PCR profile for detection of E. coli virulence genes and Shiga toxins; Table S2: Whole-genome sequencing data of STEC and non-STEC E. coli, Salmonella spp. and Campylobacter spp. isolates.
Author Contributions
Conceptualization, A.P., L.S., M.O. and R.B.; methodology, A.P., L.S., S.R., M.F., R.C., A.C., R.F., T.L., C.M., A.S., V.S., M.O. and R.B.; validation, A.P., L.S., C.B.C., M.S., M.O. and R.B; formal analysis, A.P., L.S., V.M., V.B., J.P.G., M.O. and R.B.; writing—original draft preparation, A.P. and R.B.; writing—review and editing, A.P., L.S., S.R., M.F., R.C., A.C., R.F., T.L., C.M., V.M., V.B., A.S., V.S., C.B.C., J.P.G., M.S., M.O. and R.B.; project administration, A.P. and M.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No 773830: One Health European Joint Programme, as part of the DiSCoVeR project (Discovering the sources of Salmonella, Campylobacter, VTEC and Antimicrobial Resistance). S.R., R.C. and V.M. were beneficiaries of fellowships from the same Programme on behalf of ADONIS (S.R.), FedAMR (R.C.) and BeOne (V.M) projects.
Institutional Review Board Statement
Ethical review and approval were waived for this study, since the sampling approach was not invasive or harmful for the animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All supporting data and protocols have been provided within the article or through supplementary data files. Supplementary Material is available with the online version of this article.
Acknowledgments
The authors express their gratitude to the Technology and Innovation Unit of the National Institute of Health Doutor Ricardo Jorge for performing the Next Generation Sequencing runs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hassel, J.M.; Begon, M.; Ward, M.J. Urbanization and Disease Emergence: Dynamics at the Wildlife-Livestock-Human Interface. Trends Ecol. Evol. 2017, 32, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.J.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.B.; Häsler, B.; Rushton, J. Economic Aspects of Zoonoses: Impact of Zoonoses on the Food Industry. In Zoonoses-Infections Affecting Humans and Animals; Springer: Dordrecht, The Netherlands, 2014; pp. 1107–1126. [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, 6971. [Google Scholar]
- Aguirre, A.A.; Dabritz, H.; Hill, D.; Klein, P.N.; Lepczyk, C.; Lilly, E.L.; McLeod, R.; Milcarsky, J.; Murphy, C.E.; Su, C.; et al. The One Health Approach to Toxoplasmosis: Epidemiology, Control, and Prevention Strategies. Ecohealth 2019, 16, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Latif, A.A.; Mukaratirwa, S. Zoonotic origins and animal hosts of coronaviruses causing human disease pandemics: A review. Onderstepoort J. Vet. Res. 2020, 87, 1895. [Google Scholar] [CrossRef]
- Massei, G.; Kindberg, J.; Licoppe, A.; Gačić, D.; Šprem, N.; Kamler, J.; Baubet, E.; Hohmann, U.; Monaco, A.; Ozolinš, J.; et al. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag. Sci. 2015, 71, 492–500. [Google Scholar] [CrossRef] [PubMed]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 6490. [Google Scholar]
- Mora, A.; López, C.; Dhabi, G.; López-Beceiro, A.M.; Fidalgo, L.E.; Diaz, E.A.; Martínez-Carrasco, C.; Mamani, R.; Herrera, A.; Blanco, J.E.; et al. Seropathotypes, Phylogroups, Stx subtypes, and intimin types of wildlife-carried, shiga toxin-producing Escherichia coli strains with the same characteristics as human-pathogenic isolates. Appl. Environ. Microbiol. 2012, 78, 2578–2585. [Google Scholar] [CrossRef]
- Bertelloni, F.; Cilia, G.; Bogi, S.; Ebani, V.V.; Turini, L.; Nuvoloni, R.; Cerri, D.; Fratini, F.; Turchi, B. Pathotypes and Antimicrobial Susceptibility of Escherichia Coli Isolated from Wild Boar (Sus scrofa) in Tuscany. Animals 2020, 10, 744. [Google Scholar] [CrossRef]
- Alonso, C.A.; Mora, A.; Díaz, D.; Blanco, M.; González-Barrio, D.; Ruiz-Fons, F.; Simón, C.; Blanco, J.; Torres, C. Occurrence;and characterization of stx and/or eae-positive Escherichia coli isolated from wildlife, including a typical EPEC strain from a wild boar. Vet. Microbiol. 2017, 207, 69–73. [Google Scholar] [CrossRef]
- Sannö, A.; Aspán, A.; Hestvik, G.; Jacobson, M. Presence of Salmonella spp., Yersinia enterocolitica, Yersinia pseudotuberculosis and Escherichia coli O157:H7 in wild boars. Epidemiol. Infect. 2014, 142, 2542–2547. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, S.; Martinez, R.; García, A.; Vidal, D.; Blanco, J.; Blanco, M.; Blanco, J.E.; Mora, A.; Herrera-León, S.; Echeita, A.; et al. Detection and characterisation of O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in wild boars. Vet. Microbiol. 2010, 143, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Porrero, M.C.; Mentaberre, G.; Serrano, E.; Mateos, A.; Cabal, A.; Domínguez, L.; Lavín, S. Escherichia coli O157:H7 in wild boars (Sus scrofa) and Iberian ibex (Capra pyrenaica) sharing pastures with free-ranging livestock in a natural environment in Spain. Vet. Q 2015, 35, 102–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dias, D.; Caetano, T.; Torres, R.T.; Fonseca, C.; Mendo, S. Shiga toxin-producing Escherichia coli in wild ungulates. Sci. Total Environ. 2019, 651 Pt 1, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Szczerba-Turek, A.; Socha, P.; Bancerz-Kisiel, A.; Platt-Samoraj, A.; Lipczynska-Ilczuk, K.; Siemionek, J.; Kończyk, K.; Terech-Majewska, E.; Szweda, W. Pathogenic potential to humans of Shiga toxin-producing Escherichia coli isolated from wild boars in Poland. Int. J. Food Microbiol. 2019, 300, 8–13. [Google Scholar] [CrossRef]
- Navarro-Gonzalez, N.; Casas-Díaz, E.; Porrero, C.M.; Mateos, A.; Domínguez, L.; Lavín, S.; Serrano, E. Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 2013, 167, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Wacheck, S.; Fredriksson-Ahomaa, M.; König, M.; Stolle, A.; Stephan, R. Wild Boars as an Important Reservoir for Foodborne Pathogens. Foodborne Pathog Dis 2010, 7, 307–312. [Google Scholar] [CrossRef]
- Marotta, F.; Di Marcantonio, L.; Janowicz, A.; Pedonese, F.; Di Donato, G.; Ardelean, A.; Nuvoloni, R.; Di Giannatale, E.; Garofolo, G. Genotyping and Antibiotic Resistance Traits in Campylobacter jejuni and coli from Pigs and Wild Boars in Italy. Front. Cell Infect. Microbiol. 2020, 10, 592512. [Google Scholar] [CrossRef]
- 20 Mokracka, J.; Krzymińska, S.; Ałtunin, D.; Wasyl, D.; Koczura, R.; Dudek, K.; Dudek, M.; Chyleńska, Z.A.; Ekner-Grzyb, A. In vitro virulence characteristics of rare serovars of Salmonella enterica isolated from sand lizards (Lacerta agilis L.). Antonie Van Leeuwenhoek 2018, 111, 1863–1870. [Google Scholar] [CrossRef]
- Sabat, G.; Rose, P.; Hickey, W.J.; Harkin, J.M. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 2000, 66, 844–849. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain-Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017.
- ISO 10272-1:2006; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for Detection and Enumeration of Campylobacter spp.—Part 1: Detection Method. ISO: Geneva, Switzerland, 2006.
- Persson, S.; Olsen, K.E.; Scheutz, F.; Krogfelt, K.A.; Gerner-Smidt, P. A method for fast and simple detection of major diarrhoeagenic Escherichia coli in the routine diagnostic laboratory. Clin. Microbiol. Infect. 2007, 13, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Boisen, N.; Scheutz, F.; Rasko, D.A.; Redman, J.C.; Persson, S.; Simon, J.; Kotloff, K.L.; Levine, M.M.; Sow, S.; Tamboura, B.; et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J. Infect. Dis. 2012, 205, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Otomo, Y.; Ahsan, C.R. A novel single-step multiplex polymerase chain reaction assay for the detection of diarrheagenic Escherichia coli. J. Microbiol. Methods 2013, 92, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- EUCAST-The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0. Available online: http://www.eucast.org (accessed on 4 October 2022).
- Grimont, P.A.D.; Weill, F.X. Antigenic formulae of the Salmonella serovars. In WHO Collaborating Centre for Reference and Research on Salmonella, 9th ed.; WHO Collaborating Centre for reference and research on Salmonella, Institute Pasteur: Paris, France, 2007; pp. 1–166. [Google Scholar]
- EUCAST-The European Committee on Antimicrobial Susceptibility Testing. Comité de l’antibiogramme de la Société Française de Microbiologie. Recommandations V.1.1 Avril. Available online: sfm-microbiologie.org (accessed on 4 October 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Llarena, A.-K.; Ribeiro-Gonçalves, B.F.; Silva, D.N.; Halkilahti, J.; Machado, M.P.; Da Silva, M.S.; Jaakkonen, A.; Isidro, J.; Hämäläinen, C.; Joenperä, J.; et al. INNUENDO: A Cross-sectoral Platform for the Integration of Genomics in the Surveillance of Food-borne Pathogens. EFSA Support. Publ. 2018, 15, 1498E. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Langmead, B. Aligning Short Sequencing Reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.4. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A Complete Suite for Gene-by-Gene Schema Creation and Strain Identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef] [PubMed]
- Mamede, R.; Vila-Cerqueira, P.; Silva, M.; Carriço, J.A.; Ramirez, M. Chewie Nomenclature Server (chewie-NS): A Deployable Nomenclature Server for Easy Sharing of Core and Whole Genome MLST Schemas. Nucleic Acids Res. 2020, 49, D660–D666. [Google Scholar] [CrossRef] [PubMed]
- Mixão, V.; Pinto, M.; Sobral, D.; Di Pasquale, A.; Gomes, J.P.; Borges, V. ReporTree: A Surveillance-Oriented Tool to Strengthen the Linkage between Pathogen Genetic Clusters and Epidemiological Data. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Hofer, E.; Cernela, N.; Stephan, R. Shiga toxin subtypes associated with Shiga toxin-producing Escherichia coli strains isolated from red deer, roe deer, chamois, and ibex. Foodborne Pathog. Dis. 2012, 9, 792–795. [Google Scholar] [CrossRef] [PubMed]
- Kemper, N.; Aschfalk, A.; Höller, C. Campylobacter spp., Enterococcus spp., Escherichia coli, Salmonella spp., Yersinia spp., and Cryptosporidium oocysts in semi-domesticated reindeer (Rangifer tarandus tarandus) in Northern Finland and Norway. Acta Vet. Scand. 2006, 48, 7. [Google Scholar] [CrossRef][Green Version]
- Lillehaug, A.; Bergsjø, B.; Schau, J.; Bruheim, T.; Vikøren, T.; Handeland, K. Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet. Scand. 2005, 46, 23–32. [Google Scholar] [CrossRef]
- Schierack, P.; Römer, A.; Jores, J.; Kaspar, H.; Guenther, S.; Filter, M.; Eichberg, J.; Wieler, L.H. Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl. Environ. Microbiol. 2009, 75, 695–702. [Google Scholar] [CrossRef]
- Cilia, G.; Turchi, B.; Fratini, F.; Bilei, S.; Bossù, T.; De Marchis, M.L.; Cerri, D.; Pacini, M.I.; Bertelloni, F. Prevalence, Virulence and Antimicrobial Susceptibility of Salmonella spp., Yersinia enterocolitica and Listeria monocytogenes in European Wild Boar (Sus scrofa) Hunted in Tuscany (Central Italy). Pathogens 2021, 10, 93. [Google Scholar] [CrossRef]
- Sannö, A.; Rosendal, T.; Aspán, A.; Backhans, A.; Jacobson, M. Distribution of enteropathogenic Yersinia spp. and Salmonella spp. in the Swedish wild boar population, and assessment of risk factors that may affect their prevalence. Acta Vet. Scand. 2018, 60, 40. [Google Scholar]
- Zottola, T.; Montagnaro, S.; Magnapera, C.; Sasso, S.; De Martino, L.; Bragagnolo, A.; D’Amici, L.; Condoleo, R.; Pisanelli, G.; Iovane, G.; et al. Prevalence and antimicrobial susceptibility of Salmonella in European wild boar (Sus scrofa); Latium Region-Italy. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Molino, M.G.; Sánchez, A.G.; Pérez, D.R.; Blanco, P.G.; Molina, A.Q.; Pérez, J.R.; Cano, F.E.M.; Horrillo, R.C.; Salcedo, J.H.-M.; Llario, P.F. Prevalence of Salmonella spp. in tonsils, mandibular lymph nodes and faeces of wild boar from Spain and genetic relationship between isolates. Transbound. Emerg. Dis. 2019, 66, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Mentaberre, G.; Porrero, C.M.; Serrano, E.; Mateos, A.; López-Martín, J.M.; Lavín, S.; Domínguez, L. Effect of cattle on Salmonella carriage, diversity and antimicrobial resistance in free-ranging wild boar (Sus scrofa) in northeastern Spain. PLoS ONE 2012, 7, e51614. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Ugarte-Ruiz, M.; Porrero, M.C.; Zamora, L.; Mentaberre, G.; Serrano, E.; Mateos, A.; Lavín, S.; Domínguez, L. Campylobacter shared between free-ranging cattle and sympatric wild ungulates in a natural environment (NE Spain). Ecohealth 2014, 11, 333–342. [Google Scholar] [CrossRef]
- Carbonero, A.; Paniagua, J.; Torralbo, A.; Arenas-Montes, A.; Borge, C.; García-Bocanegra, I. Campylobacter infection in wild artiodactyl species from southern Spain: Occurrence, risk factors and antimicrobial susceptibility. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 115–121. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Collis, R.M.; Biggs, P.J.; Midwinter, A.C.; Browne, A.S.; Wilkinson, D.A.; Irshad, H.; French, N.P.; Brightwell, G.; Cookson, A.L. Genomic epidemiology and carbon metabolism of Escherichia coli serogroup O145 reflect contrasting phylogenies. PLoS ONE 2020, 15, e0235066. [Google Scholar] [CrossRef]
- Beutin, L.; Gleier, K.; Kontny, I.; Echeverria, P.; Scheutz, F. Origin and characteristics of enteroinvasive strains of Escherichia coli (EIEC) isolated in Germany. Epidemiol. Infect. 1997, 118, 199–205. [Google Scholar] [CrossRef]
- Foster, M.A.; Iqbal, J.; Zhang, C.; McHenry, R.; Cleveland, B.E.; Romero-Herazo, Y.; Fonnesbeck, C.; Payne, D.C.; Chappell, J.D.; Halasa, N.; et al. Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County, Tennessee. Diagn. Microbiol. Infect. Dis. 2015, 83, 319–324. [Google Scholar] [CrossRef]
- STEC Infection: Annual Epidemiological Report for 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/STEC-infection-AER-2020-JD-FINAL.pdf (accessed on 4 October 2022).
- Guard-Petter, J. The chicken, the egg and Salmonella Enteritidis. Environ. Microbiol. 2001, 3, 421–430. [Google Scholar] [CrossRef]
- ECDC and EFSA (European Centre for Disease Prevention and Control, European Food Safety Authority). Multi-Country Outbreak of Salmonella Enteritidis Sequence Type (ST)11 Infections Linked to Poultry Products in the EU/EEA and the United Kingdom–25 February Stockholm: ECDC/EFSA. Available online: https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/sp.efsa.2021.EN-6486 (accessed on 4 October 2022).
- ECDC and EFSA (European Centre for Disease Prevention and Control, European Food Safety Authority). Multi-Country Outbreak of Salmonella Enteritidis Sequence Type (ST)11 Infections Linked to Eggs and Egg Products –8 February. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/ROA_Salmonella-Enteritidis-ST11_2022_final.pdf (accessed on 4 October 2022).
- Dionisio, D.; Esperti, F.; Vivarelli, A.; Fabbri, C.; Apicella, P.; Meola, N.; Lencioni, P.; Vannucci, R. Acute terminal ileitis mimicking Crohn’s disease caused by Salmonella veneziana. Int. J. Infect. Dis. 2001, 5, 225–227. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; O’Donnell, A.J.; Akhter, R.N.; Fayaz, A.; Mack, H.J.; Rogers, L.E.; Biggs, P.J.; French, N.P.; Midwinter, A.C. Updating the genomic taxonomy and epidemiology of Campylobacter hyointestinalis. Sci. Rep. 2018, 8, 2393. [Google Scholar] [CrossRef] [PubMed]
- The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2017. Available online: https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/j.efsa.2018.5500 (accessed on 4 October 2022).
- Bielaszewska, M.; Mellmann, A.; Bletz, S.; Zhang, W.; Köck, R.; Kossow, A.; Prager, R.; Fruth, A.; Orth-Höller, D.; Marejková, M.; et al. Enterohemorrhagic Escherichia coli O26:H11/H: A new virulent clone emerges in Europe. Clin. Infect. Dis. 2013, 56, 1373–1381. [Google Scholar] [CrossRef]
- Zweifel, C.; Cernela, N.; Stephan, R. Detection of the emerging Shiga toxin-producing Escherichia coli O26:H11/H- sequence type 29 (ST29) clone in human patients and healthy cattle in Switzerland. Appl. Environ. Microbiol. 2013, 79, 5411–5413. [Google Scholar] [CrossRef]
- Mellmann, A.; Bielaszewska, M.; Karch, H. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 2009, 136, 1925–1938. [Google Scholar] [CrossRef]
- Szczerba-Turek, A.; Siemionek, J.; Socha, P.; Bancerz-Kisiel, A.; Platt-Samoraj, A.; Lipczynska-Ilczuk, K.; Szweda, W. Shiga toxin-producing Escherichia coli isolates from red deer (Cervus elaphus), roe deer (Capreolus capreolus) and fallow deer (Dama dama) in Poland. Food Microbiol. 2020, 86, 103352. [Google Scholar] [CrossRef]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef]
- de Boer, R.F.; Ferdous, M.; Ott, A.; Scheper, H.R.; Wisselink, G.J.; Heck, M.E.; Rossen, J.W.; Kooistra-Smid, A.M.D. Assessing the public health risk of Shiga toxin-producing Escherichia coli by use of a rapid diagnostic screening algorithm. J. Clin. Microbiol. 2015, 53, 1588–1598. [Google Scholar] [CrossRef]
- Fierz, L.; Cernela, N.; Hauser, E.; Nüesch-Inderbinen, M.; Stephan, R. Characteristics of Shigatoxin-Producing Escherichia coli Strains Isolated during 2010–2014 from Human Infections in Switzerland. Front. Microbiol. 2017, 8, 1471. [Google Scholar] [CrossRef]
- Duan, Q.; Yao, F.; Zhu, G. Major virulence factors of enterotoxigenic Escherichia coli in pigs. Ann. Microbiol. 2012, 62, 7–14. [Google Scholar] [CrossRef]
- Mellmann, A.; Fruth, A.; Friedrich, A.W.; Wieler, L.H.; Harmsen, D.; Werber, D.; Middendorf, B.; Bielaszewska, M.; Karch, H. Phylogeny and disease association of Shiga toxin-producing Escherichia coli O91. Emerg. Infect. Dis. 2009, 15, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).