Abstract
Because of the ever-increasing multidrug resistance in microorganisms, it is crucial that we find and develop new antibiotics, especially molecules with different targets and mechanisms of action than those of the antibiotics in use today. Translation is a fundamental process that uses a large portion of the cell’s energy, and the ribosome is already the target of more than half of the antibiotics in clinical use. However, this process is highly regulated, and its quality control machinery is actively studied as a possible target for new inhibitors. In bacteria, ribosomal stalling is a frequent event that jeopardizes bacterial wellness, and the most severe form occurs when ribosomes stall at the 3′-end of mRNA molecules devoid of a stop codon. Trans-translation is the principal and most sophisticated quality control mechanism for solving this problem, which would otherwise result in inefficient or even toxic protein synthesis. It is based on the complex made by tmRNA and SmpB, and because trans-translation is absent in eukaryotes, but necessary for bacterial fitness or survival, it is an exciting and realistic target for new antibiotics. Here, we describe the current and future prospects for developing what we hope will be a novel generation of trans-translation inhibitors.
1. Introduction
Protein synthesis, or translation, is a fundamental biological process that occurs on ribonucleoprotein nanomachines named ribosomes. The bacterial ribosome is, therefore, a major antibiotic target, and many types of inhibitors can stop bacterial growth by binding its functional centers and interfering with the ribosome’s ability to synthesize proteins [1]. However, bacteria have evolved a wide set of mechanisms to resist the inhibitory effect of antibiotics, including those that target the ribosome. Indeed, resistance mechanisms have been identified for nearly every antibiotic currently in clinical use. Combined with the fact that pharmaceutical companies have not developed more than a few antibiotics recently, infections that are treatable now will probably, once again, become life threatening [2].
It is generally accepted that among the most important bacteria to target, those in the ESKAPE pathogens group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are of enormous interest when it comes to drug discovery [3,4]. They are the leading cause of nosocomial infections throughout the world, and most are multidrug-resistant isolates [5]. The World Health Organization (WHO) regularly issues global reports on antimicrobial resistance (AMR) surveillance [6], and the topic has ranked in the top 10 global health issues over the past few years [4,7].
To combat this crisis, we need new antibiotics, and, most importantly, we need new classes of antibiotics with novel mechanisms of action [8]. To do this, we must first identify new molecular processes that can be targeted. Ideally, these should be conserved among pathogenic bacteria; indispensable to the survival, or at least to the fitness, of the bacteria; sufficiently specific, so that they can distinguish between bacterial species and minimize microbiotal damage; not targeted by current antibiotics; absent in eukaryotes to limit toxicity. In fact, trans-translation, the primary quality control mechanism for rescuing stalled ribosomes in bacteria, appears to be a perfect candidate, allowing us to target this key cellular process in a totally new way. Here, we discuss the potential of targeting this pathway with novel antimicrobial compounds.
2. Ribosomal Stalling: From No-Go to Non-Stop
Several phenomena can cause the production of aberrant mRNA molecules that lead to the accumulation of stalled ribosomes in bacteria. The most frequently observed are spontaneous mutations in their corresponding genes, as well as transcription defects after the RNA polymerase prematurely terminates transcription, or does not correctly transcribe the stop codon [9]. Other phenomena include mRNA degradation, caused by either endo- or 3′–5′ exo-ribonucleases, or by environmental stresses that result in chemical and physical damage [10]. “Non-stop” situations (readthrough) can also occur when a canonical stop codon is translated in the presence of non-sense suppressor tRNA [11,12], aberrant frameshifts [13], or translational error-inducing drugs [14]. In bacteria, translation initiation mainly relies on the binding of the ribosomal binding site, the Shine–Dalgarno (SD) sequence to the 3′-end of 16S ribosomal RNA. This, therefore, means that translation can start before transcription is actually complete, and that non-stop events, such as degradation, can occur both before translation starts or while the ribosome advances along the mRNA [15]. It must be noted that another type of defective translation event can also appear during certain stressful conditions (e.g., starvation), which, during translation, slow or stop ribosomes upstream from the stop codon. Due to the presence of a stop codon, this situation is called “no-go” instead of “non-stop.” Even though this process could eventually be reversed, it is problematic if it occurs for too long, as endonucleases, such as RelE (the toxin component of the type II RelE–RelB toxin–antitoxin system), will cut the mRNA within the ribosomal A site to facilitate tmRNA-mediated rescue, and conserve the energy and nutrients being used to combat stress [16]. The “no-go” then becomes “non-stop,” and triggers the same quality control mechanisms for ribosomal release. In all of these cases, the rescue of non-stop ribosomes is essential in most or all bacteria [17], suggesting that interference with non-stop quality control mechanisms is surely a promising antibiotic development path.
3. Trans-Translation Components Are Major Targets for Interference
Despite the recent discovery of several back-up systems (see [18], for a complete review), trans-translation is the principal and most sophisticated quality control mechanism for avoiding inefficient protein synthesis on stalled non-stop bacterial ribosomes. It mainly relies on the complex between tmRNA and SmpB, the two main actors in the process.
3.1. Transfer-Messenger RNA (tmRNA)
Having both transfer and messenger RNA functions, tmRNAs are chimeric RNA molecules that are typically 260 to 420 nucleotides in length (363 nts in Escherichia coli). The ssrA gene, which encodes tmRNA, has been found in nearly all bacterial genomes [19]. tmRNA is always first transcribed as a precursor, and it is subsequently processed at its CCA 5′- and 3′-ends [20,21,22,23]. The number of tmRNA molecules per cell has been estimated to be 500–700, roughly 5% of the total number of ribosomes, as estimated from the ratio of tmRNA-to-5S ribosomal RNA [24,25]. As with classical tRNA, the T-loop undergoes some base modifications, with the TrmA enzyme catalyzing 5-methyluridine and TruB enabling pseudouridine production [26,27]. The classic mature tmRNA is composed of a tRNA-like domain (TLD), a messenger-like domain (MLD), and a large, halo-shaped pseudoknot (PK) ring (Figure 1).
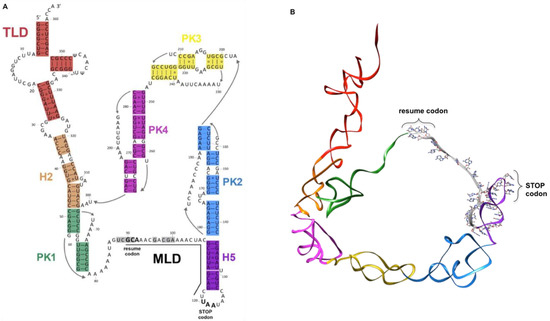
Figure 1.
tmRNA. (A) Organization of the secondary structure of Escherichia coli tmRNA. The internal open reading frame is underlined. (B) 3D structure of the E. coli tmRNA molecule. In both panels, the tRNA-like domain (TLD, red) is followed by helix H2 (orange). The pseudoknot ring is composed of PK1 (dark green), PK2 (steel blue), PK3 (yellow), PK4 (magenta), helix H5 (purple), and the mRNA-like domain (MLD, grey). The resume and stop codons are indicated.
The TLD portion plays the same role as in classical tRNA; the acceptor stem is always recognized and aminoacylated by alanine tRNA synthetase (AlaRS) after recognition of a G3:U base pair, a motif also present in canonical tRNAAla [28,29,30]. The domain displays a classical T-loop, and a small D-loop without a stem. It is also devoid of an anticodon loop, since no codon will need to be recognized within the vacant decoding site of a stalled ribosome [20,26,31]. In fact, this ostensible problem is overcome by SmpB; when interacting with the TLD region, it mimics codon–anticodon recognition and allows tmRNA to accommodate into the ribosomal A site (see below). The MLD is the RNA portion that contains the internal open reading frame (ORF) of tmRNA, which encodes the aminoacidic sequence A*ANDENYALAA in E. coli, with the first A* being carried by the TLD. This sequence is added to the stalled protein during trans-translation. The tag sequence displays strong phylogenetic conservation, with the consensus sequence A*AN----ALAA. The final three alanines (AxAA) are crucial, allowing for specific recognition of the tagged protein by proteases. The nature of the RNA sequence upstream from the resume codon allows for the correct placement of the codon into the decoding center. Accordingly, mutations in this region can lead to reading frameshifts or a loss of tmRNA function [32,33,34]. In fact, the structural elements that precede the resume codon, rather than the sequence itself, are important for the reinitiation of translation [35,36,37,38,39]. Once the ORF is completely translated, the tagged peptide is specifically degraded by several proteases. In addition to this classical single-chain conformation, tmRNA also exists (in alpha-proteobacteria, cyanobacteria, and some beta-proteobacteria lineages) as a two-piece molecule, a formation caused by a circular gene permutation that splits it into two molecules [40,41]. In this case, the TLD, MLD, and PK1 are similar to those of “one-piece” tmRNA, but the loop containing the tag reading frame is broken, and there are fewer pseudoknots [42].
3.2. SmpB
SmpB is a small, basic protein of ~160 amino acids, encoded by the smpB gene. In the E. coli genome, it is located just upstream from the ssrA gene that codes for tmRNA [43]. SmpB binds to tmRNA with high affinity, and is its most important partner during ribosome rescue. In fact, in its absence, tmRNA can no longer accommodate its TLD portion into the vacant A site [44,45,46,47]. A comparison between the SmpB proteins in the various ESKAPE bacteria reveals that the six proteins conserve the same fold, but the sequence and length of the C-terminal tail differs, especially in S. aureus, A. baumannii, and E. cloacae (Figure 2).
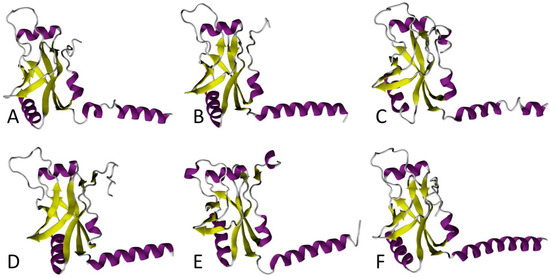
Figure 2.
Comparison between 3D models of SmpB in ESKAPE bacteria. In each conformation, the tmRNA TLD contact region is on the left side, and the helix-shaped C-terminal end is shown to the right. The α-helices and β-strands are purple and yellow, respectively. These models of Enterococcus faecium (A), Staphylococcus aureus (B), Klebsiella pneumoniae (C), Acinetobacter baumannii (D), Pseudomonas aeruginosa (E), and Enterobacter cloacae (F) were all computed with the I-TASSER program using E. coli SmpB as the structural template (PDB 7ACJ).
The SmpB body is arranged in an oligonucleotide/oligosaccharide binding (OB) domain that folds in a classical fashion into a β-barrel made up of six antiparallel β-strands, which is also typical of other RNA-binding proteins, such as IF1 or bS1 [48,49,50]. By interacting with the tmRNA TLD, SmpB mimics the missing D-loop and anticodon stem–loop present in classical tRNA [51,52]. The interesting shape assumed by the tmRNA–SmpB complex is important for its entry into the ribosome, as it simulates the codon–anticodon pairing, which then promotes the reactivity of a cognate tRNA. Of the ~160 amino acids, the last 30 C-terminal residues form a tail, which is unstructured in solution, but folds into an α-helix during trans-translation. This C-terminal tail is rich in positively charged side-chain residues, essential for contacts with the tmRNA helix H5, as well as for interactions with the negatively charged nucleotides within the decoding site of the 30S ribosomal subunit [53,54,55,56,57]. Indeed, by inserting into the mRNA entry channel, the C-terminal tail is instrumental in selecting the stalled ribosomes with empty mRNA entry channels. The recent cryo-electron microscopy (cryo-EM) structures of E. coli tmRNA–SmpB bound to a stalled ribosome [56,57], and the previous crystallographic study of trans-translation in Thermus thermophilus [58], both show that, just as in canonical translation, the presence of the protein in the decoding center induces reorientation of nucleotides A1492 and A1493 in helix 44. Besides its main RNA-binding site on the TLD, SmpB also has a secondary RNA-binding site, which later binds the MLD to ensure that the resume codon is correctly positioned in the ribosomal A site [56]. These results confirm the long-predicted importance of SmpB in the trans-translation partnership [53,55,59].
4. The Molecular Process of Trans-Translation
During trans-translation (Figure 3), the tmRNA–SmpB complex is first brought to the ribosome with EF-Tu•GTP. Stalled ribosomes are selected by SmpB, whose C-terminal tail probes the mRNA entrance channel [58]. In this pre-accommodation state, GTP hydrolysis in EF-Tu is favored, as are conformational changes in the ribosomal subunits, and this induces the accommodation of tmRNA–SmpB into the vacant ribosomal A site. After transpeptidation occurs between the stalled incomplete peptide and the tmRNA alanine, a swap between the tmRNA MLD and the non-stop mRNA allows translation to resume. The C-terminal tail of SmpB, which was involved in ribosomal vacant A site recognition, then rotates by 60° to allow the MLD to move into the mRNA channel, as well as to allow ejection of the problematic mRNA. The ribosome translates the MLD until it reaches the stop codon, after which the stalled ribosomes are recycled (see [60], for the structural details of the process). The incomplete peptides are tagged with a signal sequence that results in quick proteolysis. Two AAA+ proteolytic enzymes (ATPases associated with various cellular activities), ClpXP and ClpAP, are able to degrade the tagged proteins by converting ATP hydrolysis energy into mechanical work [61]. FtsH, a hexameric protease anchored to the internal side of the cytoplasmic membrane, is also involved in degrading a small subset of tagged proteins present in the inner membrane [43]. On the other hand, the energy-independent protease Tsp takes over the tmRNA-tagged substrates in the periplasm [62]. The problematic mRNAs are degraded by RNase R [63]. This enzyme, of 92 kDa, belongs to the RNase II superfamily, a group of exoribonucleases able to degrade the RNA molecules in the 3′ → 5′ direction [64], as well as digest various RNA substrates [65,66]. However, the details of how the ribonuclease works with the complex to promptly recognize and handle problematic mRNA is still unclear.
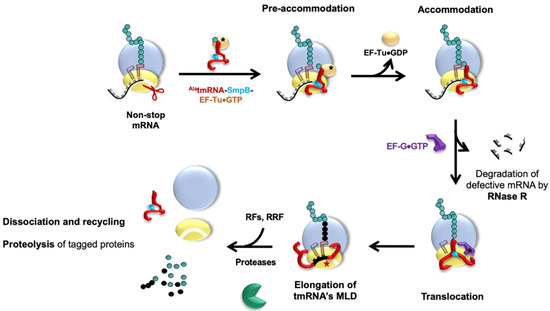
Figure 3.
The complete trans-translation quality control cycle. Pre-accommodation state: tmRNA associates with its partner SmpB to form a complex. Elongation factor EF-Tu•GTP binds to AlatmRNA–SmpB, thereby forming the quaternary complex needed to rescue the ribosome stalled on a non-stop mRNA. To recognize these ribosomes, this quaternary complex enters the vacant ribosomal A site. There, SmpB mimics a codon–anticodon pairing while its C-terminal tail inserts into the mRNA channel. The EF-Tu•GDP is then released after GTP hydrolysis. Accommodation: The AlatmRNA–SmpB complex is accommodated into the A site, triggering the peptidyl transfer reaction. Translocation: Thanks to GTP hydrolysis, EF-G•GTP helps shift the tmRNA–SmpB into the P site. EF-G•GDP is released, and the non-stop mRNA is ejected then degraded by RNase R. Elongation: The tmRNA open reading frame is placed into the A site, and new tRNAs arrive at the ribosome to resume translation. Termination: The tmRNA–SmpB complex moves towards the E site, and the TLD and SmpB are promptly ejected. Translation of the MLD continues until translation of the tmRNA-encoded tag is terminated at the stop codon with the help of the release factors (RFs). The ribosomal subunits are then dissociated by the ribosome recycling factor (RRF), and the nascent peptide is degraded by ClpXP/ClpAP/FtsH/Tsp proteases. All of the components are now recycled and ready for a new round.
5. Trans-Translation as a Target for New Antimicrobial Compounds
Considering that trans-translation is absent in eukaryotes, tmRNA–SmpB is an especially promising target for novel antibiotics. Obviously, when it is essential to the survival of pathogenic bacteria, the trans-translation machinery is an excellent specific target for use in developing molecules to kill bacteria directly [35,67,68]. When non-lethal, because alternative rescue factors can take over the rescue process, deletion of tmRNA and/or SmpB induces various phenotypes, including loss of virulence or loss of antibiotic tolerance [69,70,71,72]. These hypersensitive mutants are not viable in the presence of low doses of some protein synthesis inhibitors (chloramphenicol, lincomycin, spiramycin, tylosin, erythromycin, and spectinomycin) that do not otherwise significantly affect the growth of wild-type cells [69,70,73]. Strikingly, mutants deleted for tmRNA are also more sensitive to antibiotics that do not target translation than wild-type cells, such as inhibitors of cell wall synthesis. This is probably because these drugs stress the bacteria, and this is handled more efficiently when trans-translation is active [74]. In all of these cases, it is possible that trans-translation inhibitors could be used in combination with already commercialized antibiotics, in order to diminish their minimal inhibitory concentration (MIC) in pathogens, or even to reenable the use of antibiotics no longer used because of resistance. Finally, trans-translation is also important for persister survival, as well as tolerance to a variety of antibiotics and stresses [75]. Despite the enormous potential and extensive research into how it works and how this pathway can be targeted for treatments against bacterial infection, there are currently no drugs on the market that use this mechanism. Since this review focuses on trans-translation, we will only discuss the possible strategies for specifically impairing that process via targeting tmRNA, SmpB, and/or the ribosome itself. However, we must mention that a global strategy should not overlook the possibility of altering the activity of supporting actors, such as the highly conserved aminoacyl-tRNA synthetase (AlaRS) enzyme, serine protease ClpP, or ribonuclease RNase R.
6. Antibiotics Targeting Trans-Translation: Are We There Yet?
6.1. Oxadiazole Compounds
In 2013, based on a luciferase assay, Keiler’s group performed a high-throughput screening assay on a library of 663,000 candidate compounds. This led to the identification of 1,3,4-oxadiazole and tetrazol-based compounds as broad-spectrum antibiotics that specifically inhibited the pathway [67]. The most promising compound was the oxadiazole KKL-35 (Figure 4), which displays an antibiotic effect against very distantly related bacteria, suggesting that it may have antibiotic activity against a broad spectrum of species, thus paving the way for the development of the first class of small molecules inhibiting trans-translation. How KKL-35 targets trans-translation could not be easily identified. KKL-35 binds poorly to tmRNA and SmpB, suggesting that the compound probably affects a later step in the quality control process. Indeed, later biochemical experiments, using Mycobacterium smegmatis and Staphylococcus aureus cells, highlighted KKL-2098, an analog of KKL-35 that incorporates a photoreactive azide group and a terminal alkyne moiety. KKL-2098 targets helix 89 of 23S rRNA, but in a region not targeted by conventional antibiotics. It binds to a pocket adjacent to the peptidyl transfer center (PTC), without inhibiting canonical translation [68,76]. More recently, this result was confirmed by cryo-EM (EMDB with the accession code EMD-20121). Despite a rather low occupancy, KKL-2098, cross-linked to a non-stop ribosome, binds near the PTC and significantly alters the conformation of the ribosomal protein bL27. This suggests that 1,3,4-oxadiazoles may, at least in part, inhibit trans-translation by preventing tmRNA–SmpB binding at the A site, or by interfering with the translocation of the complex from the A to the P site [77]. In another oxadiazole example, a Bacillus subtilis proteomic response library was used to show that KKL-35 and other oxadiazole derivatives induce responses that are similar to those of ionophores, which disturb metal homeostasis, and to other agents, causing oxidative stress responses. This activity could be linked to the importance of trans-translation in cells undergoing oxidative stress [78].
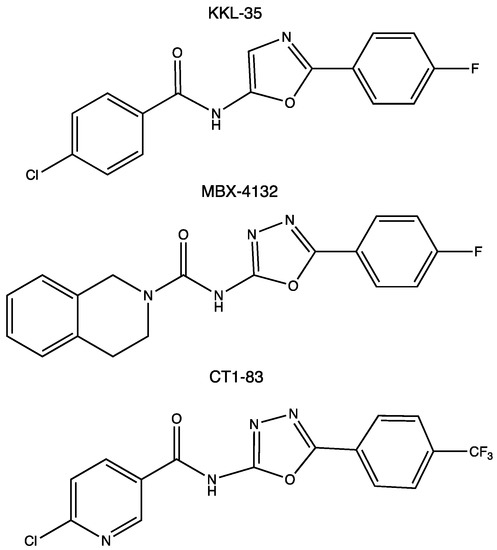
Figure 4.
Chemical structures of the experimental oxadiazole compounds KKL-35, MBX-4132, and CT1-83.
In 2017, our group developed a new double-fluorescence reporter system for the simultaneous and specific quantification of bacterial trans-translation, as well as proteolysis, in E. coli [79]. However, when we tested KKL-35, we did not observe any significant changes in fluorescence levels, despite its strong antibiotic activity, suggesting that trans-translation is not its only target, or that the molecule is rapidly metabolized (certainly due to amide bond fragility, see below) and the resulting products of degradation act on another target in E. coli. These data were supported by the fact that the inhibitory activity of KKL-35 is similar in both a ΔarfA strain (in which trans-translation is essential) and in a ΔssrA strain deprived of trans-translation. Furthermore, in the human pathogen Legionella pneumophila (which causes Legionnaires’ disease), the antibiotic activity of KKL-35 is not related to the specific inhibition of trans-translation, as it remained active against L. pneumophila mutants expressing an alternate ribosome-rescue system and lacking tmRNA [80].
Because the characterization of a new antibiotic target in living cells can be slow, difficult, and treacherous (as shown with KKL-35), we recently constructed a system to detect trans-translation in vitro [81]. It is based on an engineered tmRNA variant that reassembles green fluorescent protein (GFP) when trans-translation is active. This system is, thus, adapted for the high-throughput screening of chemical compounds by fluorescence, and the limited number of reaction components allows for the direct detection of the relevant targets of trans-translation, which are as follows: tmRNA, SmpB, and the ribosome itself. Based on this simple system, we demonstrated that several 1,3,4-oxadiazole compounds do, indeed, inhibit trans-translation in vitro, though only moderately [81,82]. In KKL-35, replacing the benzene of the chloro-aryl moiety with a pyridine group (compound CT1-83, see Figure 4) results in much stronger inhibition of trans-translation.
However, because of the rapid hydrolysis of the amide bond of KKL-35 in liver microsomes, it cannot be used in animals. A recent structure–activity relationship (SAR) program thus led to the development of a new uriedo-oxadiazole derivate, MBX-4132 (Figure 4). This compound is much more stable and not significantly less potent, able to inhibit trans-translation both in vitro and in vivo, and clears multidrug-resistant Neisseria gonorrhoeae in infected mice [77]. While the oxadiazole strategy has been deeply studied, its cellular targets and mode of action remain uncertain, which justifies further investigation, as well as the continued search for other molecules.
6.2. Pyrazinamide
In 2011, it was proposed that pyrazinamide (PZA), a mainstay of anti-tuberculosis combination therapy [83], inhibits trans-translation [84]. Using proteomic studies, pyrazinoic acid (POA), the hydrolyzed and active form of PZA, was shown to bind to the ribosomal protein S1, encoded by the rpsA gene [84]. Interestingly, POA only inhibits trans-translation and not canonical translation, and this inhibition depends strictly on wild-type M. tuberculosis S1. Crystal structures of the S1–POA complex revealed that the residues Lys303, Phe307, Phe310, and Arg357 in the S1 domain directly interact with POA, and that mutations on these locations blocked the interaction with the drug, and diminished the binding between S1 and tmRNA [85]. However, the action of PZA on S1 and trans-translation in M. tuberculosis was called into question, and experiments suggest that this drug directly targets a critical player in the metabolism of coenzyme A instead [86]. A recent study seems to confirm this hypothesis, since no measurable binding between POA and S1 could be recovered, despite the use of a wide panel of biophysical methods, including nuclear magnetic resonance (NMR) spectroscopy, isothermal titration calorimetry (ITC), microscale thermophoresis (MST), and electrophoretic mobility shift assays (EMSA) [87].
6.3. Peptides and Oligonucleotides
Peptide aptamers (PA) are combinatorial proteins that consist of a stable scaffold protein and random amino acids designed to bind to specific targets, in order to disrupt their activity [88]. In a recent study into the ways to vaccinate and protect zebrafish against infection, PAs were developed to target SmpB in Aeromonas veronii [89]. These opportunistic bacteria depend on trans-translation for virulence, and they are commonly found in aquaculture, where they cause wound infection, diarrhea, and septicemia [90]. The aptamers directed against SmpB were selected from a PA library, and the leading aptamer PA-1 (sequence: GGVTFLVNTYPNGVQSRAGG) was shown to specifically target SmpB, and to knockdown its functioning. When PA-1 was introduced into A. veronii, the engineered strain was much less virulent and could be used as a potential attenuated live vaccine, thereby providing a novel strategy for preventing A. veronii infection [89]. A second aptamer PA-2 (sequence: IGQEWGLGVRGPLSAK) was demonstrated to interact not only with SmpB, but also with the alternative rescue factor ArfA, resulting in the dysfunction of both rescue factors [91]. Considering the expected conservation of the fold in SmpB (see Figure 2), PA-1 and PA-2 could theoretically target a wide range of different bacteria.
Another peptide strategy involves using a peptide that mimics the SmpB C-terminal tail to compete with endogenous SmpB for binding in the vacant ribosomal A site, thus preventing tmRNA recruitment and, in turn, inhibiting trans-translation. We showed that the peptide that corresponds to the C-terminal extremity of E. coli SmpB (sequence: GKKQHDKRSDIKEREWQVDKARIMKNAHR) acts as a potent trans-translation inhibitor in vivo [79].
Finally, the most obvious strategy is to use antisense oligonucleotides directed towards the genes encoding SmpB or tmRNA (ssrA gene), or towards the mature tmRNA itself. This approach is already in use in vitro by various laboratories, often as an internal control, with an antisense oligonucleotide targeting the tmRNA MLD and, thereby, very efficiently inhibiting trans-translation (for an example, see [92]).
7. Conclusions
Although trans-translation was discovered more than 25 years ago, and has been studied carefully ever since, with several attempts made to develop molecules to target it, the only chemical family that has displayed potential activity derives from 1,3,4-oxadiazole compounds. The recent development of sensitive and selective high-throughput screening assays that target ESKAPE pathogenic bacteria will undoubtedly help us to find new scaffolds that specifically target ribosome rescue [92]. Current studies work from scratch, by screening pharmacologically active small molecules from large chemical or natural product libraries [93], or are based on rational drug design, attempting to target the interactions between tmRNA, SmpB, and the ribosome (Figure 5), as recently described in cryo-EM structural studies [56,57]. Among the interactions discussed, the most promising targets may be the TLD–SmpB interface (in order to inhibit the tmRNA–SmpB interaction before the complex enters the ribosome); the mechanism of stalled ribosome recognition (to block or compete with SmpB C-terminal tail insertion into the empty mRNA channel); the SmpB–MLD binding site, which allows resume codon registration during translocation (to impede protein tagging). The possible specific integration of such molecules within pathogenic bacteria would be an extraordinary tool in the fight against multiresistance. There is no doubt that the groundwork already laid will soon respond to this increasingly urgent antibiotic resistance emergency.
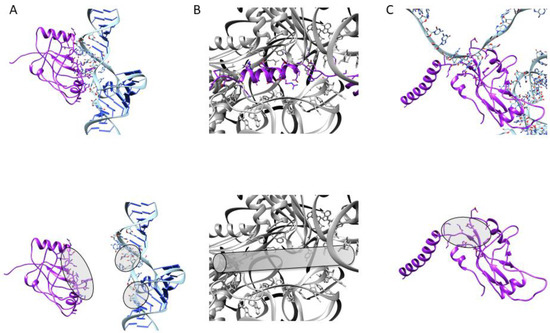
Figure 5.
Potential anti-trans-translation targets. To interfere with trans-translation, one can (A) inhibit the tmRNA–SmpB interaction by targeting the binding sites of either partner; (B) compete with the SmpB C-terminal tail for stalled ribosome recognition; (C) alter the tagging process by targeting the binding site between SmpB and the MLD.
Author Contributions
R.C.-S., G.D. and R.G. wrote the first draft of the manuscript. R.C.-S., G.D., O.D., E.G., A.J.M. and R.G. participated in manuscript revision and approved its final version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Centre National de la Recherche Scientifique (UMR CNRS 6290), the University of Rennes 1, the Agence Nationale pour la Recherche as part of the RIBOTARGET 18-JAM2-0005-03 project under the JPI AMR framework. Funding was also received from the CAPES-COFECUB program (RCD), the European Union’s ERASMUS+ program (GU), and the Britanny Region (GU). The APC was funded by CNRS.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Juliana Berland for insightful comments on the manuscript.
Conflicts of Interest
Reynald Gillet is a co-inventor of the screening system described here (patent application #EP/2018/063780).
References
- Vázquez-Laslop, N.; Mankin, A.S. Context-specific action of ribosomal antibiotics. Annu. Rev. Microbiol. 2018, 72, 185–207. [Google Scholar] [CrossRef]
- Ero, R.; Yan, X.F.; Gao, Y.G. Ribosome protection proteins—“New” players in the global arms race with antibiotic-resistant pathogens. Int. J. Mol. Sci. 2021, 22, 5356. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Kakkar, A.K.; Shafiq, N.; Singh, G.; Ray, P.; Gautam, V.; Agarwal, R.; Muralidharan, J.; Arora, P. Antimicrobial stewardship programs in resource constrained environments: Understanding and addressing the need of the systems. Front. Public Health 2020, 8, 140. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Janssen, B.D.; Diner, E.J.; Hayes, C.S. Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Bact. Regul. RNA 2012, 905, 291–309. [Google Scholar] [CrossRef][Green Version]
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Collier, J.; Binet, E.; Bouloc, P. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 2002, 45, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Yamamoto, Y.; Ogawa, K.; Abo, T.; Inokuchi, H.; Aiba, H. Bacterial SsrA system plays a role in coping with unwanted translational readthrough caused by suppressor tRNAs. Genes Cells 2002, 7, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.S.; Sauer, R.T. Cleavage of the a site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 2003, 12, 903–911. [Google Scholar] [CrossRef]
- Abo, T.; Ueda, K.; Sunohara, T.; Ogawa, K.; Aiba, H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 2002, 7, 629–638. [Google Scholar] [CrossRef]
- Laursen, B.S.; Sørensen, H.P.; Mortensen, K.K.; Sperling-Petersen, H.U. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 101–123. [Google Scholar] [CrossRef]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Keiler, K.C.; Feaga, H.A. Resolving nonstop translation complexes is a matter of life or death. J. Bacteriol. 2014, 196, 2123–2130. [Google Scholar] [CrossRef]
- Müller, C.; Crowe-McAuliffe, C.; Wilson, D.N. Ribosome rescue pathways in bacteria. Front. Microbiol. 2021, 12, 652980. [Google Scholar] [CrossRef]
- Gueneau de Novoa, P.; Williams, K.P. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004, 32, D104–D108. [Google Scholar] [CrossRef]
- Komine, Y.; Kitabatake, M.; Yokogawa, T.; Nishikawa, K.; Inokuchi, H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 9223–9227. [Google Scholar] [CrossRef]
- Li, Z.; Pandit, S.; Deutscher, M.P. 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 2856–2861. [Google Scholar] [CrossRef] [PubMed]
- Lin-Chao, S.; Wei, C.L.; Lin, Y.T. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 1999, 96, 12406–12411. [Google Scholar] [CrossRef] [PubMed]
- Gimple, O.; Schön, A. In vitro and in vivo processing of Cyanelle tmRNA by RNase P. Biol. Chem. 2001, 382, 1421–1429. [Google Scholar] [CrossRef]
- Lee, S.Y.; Bailey, S.C.; Apirion, D. Small stable RNAs from Escherichia coli: Evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J. Bacteriol. 1978, 133, 1015–1023. [Google Scholar] [CrossRef]
- Moore, S.D.; Sauer, R.T. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 2005, 58, 456–466. [Google Scholar] [CrossRef]
- Felden, B.; Hanawa, K.; Atkins, J.F.; Himeno, H.; Muto, A.; Gesteland, R.F.; McCloskey, J.A.; Crain, P.F. Presence and location of modified nucleotides in Escherichia coli tmRNA: Structural mimicry with tRNA acceptor branches. EMBO J. 1998, 17, 3188–3196. [Google Scholar] [CrossRef]
- Ranaei-Siadat, E.; Fabret, C.; Seijo, B.; Dardel, F.; Grosjean, H.; Nonin-Lecomte, S. RNA-methyltransferase TrmA is a dual-specific enzyme responsible for C5-methylation of uridine in both tmRNA and tRNA. RNA Biol. 2013, 10, 572–578. [Google Scholar] [CrossRef]
- Hou, Y.M.; Schimmel, P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nat. Cell Biol. 1988, 333, 140–145. [Google Scholar] [CrossRef]
- Schimmel, P.; Ribas de Pouplana, L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem. Sci. 2000, 25, 207–209. [Google Scholar] [CrossRef]
- Ribas de Pouplana, L.; Schimmel, P. Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem. Sci. 2001, 26, 591–596. [Google Scholar] [CrossRef]
- Nameki, N.; Tadaki, T.; Himeno, H.; Muto, A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000, 470, 345–349. [Google Scholar] [CrossRef]
- Williams, K.P.; Martindale, K.A.; Bartel, D.P. Resuming translation on tmRNA: A unique mode of determining a reading frame. EMBO J. 1999, 18, 5423–5433. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ishii, M.; Tadaki, T.; Muto, A.; Himeno, H. Determinants on tmRNA for initiating efficient and precise trans-translation: Some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA 2001, 7, 999–1012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miller, M.R.; Healey, D.W.; Robison, S.G.; Dewey, J.D.; Buskirk, A.R. The role of upstream sequences in selecting the reading frame on tmRNA. BMC Biol. 2008, 6, 29, Published 30 June 2008. [Google Scholar] [CrossRef]
- Thibonnier, M.; Thiberge, J.M.; De Reuse, H. Trans-translation in Helicobacter pylori: Essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE 2008, 3, e3810. [Google Scholar] [CrossRef] [PubMed]
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283. [Google Scholar] [CrossRef]
- Yang, C.; Glover, J.R. The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS ONE 2009, 4, e4459. [Google Scholar] [CrossRef] [PubMed]
- Barends, S.; Zehl, M.; Bialek, S.; De Waal, E.; A Traag, B.; Willemse, J.; Jensen, O.N.; Vijgenboom, E.; Van Wezel, G.P. Transfer–messenger RNA controls the translation of cell-cycle and stress proteins in Streptomyces. EMBO Rep. 2010, 11, 119–125. [Google Scholar] [CrossRef]
- Katz, A.; Elgamal, S.; Rajkovic, A.; Ibba, M. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol. Microbiol. 2016, 101, 545–558. [Google Scholar] [CrossRef]
- Keiler, K.C.; Shapiro, L.; Williams, K.P. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. USA 2000, 97, 7778–7783. [Google Scholar] [CrossRef] [PubMed]
- Sharkady, S.M.; Williams, K.P. A third lineage with two-piece tmRNA. Nucleic Acids Res. 2004, 32, 4531–4538. [Google Scholar] [CrossRef]
- Gaudin, C.; Zhou, X.; Williams, K.P.; Felden, B. Two-piece tmRNA in cyanobacteria and its structural analysis. Nucleic Acids Res. 2002, 30, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.D.; Sauer, R.T. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007, 76, 101–124. [Google Scholar] [CrossRef]
- Karzai, A.W.; Susskind, M.M.; Sauer, R. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 1999, 18, 3793–3799. [Google Scholar] [CrossRef]
- Hanawa-Suetsugu, K.; Takagi, M.; Inokuchi, H.; Himeno, H.; Muto, A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002, 30, 1620–1629. [Google Scholar] [CrossRef]
- Shimizu, Y.; Ueda, T. The role of SmpB protein in trans-translation. FEBS Lett. 2002, 514, 74–77. [Google Scholar] [CrossRef]
- Wower, J.; Zwieb, C.W.; Hoffman, D.W.; Wower, I.K. SmpB: A protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry 2002, 41, 8826–8836. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, M.; Hubbard, T.J.; Proctor, M.; Freund, S.M.; Murzin, A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid–binding fold. Cell 1997, 88, 235–242. [Google Scholar] [CrossRef]
- Dong, G.; Nowakowski, J.; Hoffman, D.W. Structure of small protein B: The protein component of the tmRNA–SmpB system for ribosome rescue. EMBO J. 2002, 21, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Someya, T.; Nameki, N.; Hosoi, H.; Suzuki, S.; Hatanaka, H.; Fujii, M.; Terada, T.; Shirouzu, M.; Inoue, Y.; Shibata, T.; et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 2003, 535, 94–100. [Google Scholar] [CrossRef]
- Gutmann, S.; Haebel, P.W.; Metzinger, L.; Sutter, M.; Felden, B.; Ban, N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 2003, 424, 699–703. [Google Scholar] [CrossRef]
- Bessho, Y.; Shibata, R.; Sekine, S.; Murayama, K.; Higashijima, K.; Hori-Takemoto, C.; Shirouzu, M.; Kuramitsu, S.; Yokoyama, S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc. Natl. Acad. Sci. USA 2007, 104, 8293–8298. [Google Scholar] [CrossRef]
- Kaur, S.; Gillet, R.; Li, W.; Gursky, R.; Frank, J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc. Natl. Acad. Sci. USA 2006, 103, 16484–16489. [Google Scholar] [CrossRef] [PubMed]
- Kurita, D.; Sasaki, R.; Muto, A.; Himeno, H. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res. 2007, 35, 7248–7255. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nonin-Lecomte, S.; Germain-Amiot, N.; Gillet, R.; Hallier, M.; Ponchon, L.; Dardel, F.; Felden, B. Ribosome hijacking: A role for small protein B during trans -translation. EMBO Rep. 2009, 10, 160–165. [Google Scholar] [CrossRef]
- Rae, C.D.; Gordiyenko, Y.; Ramakrishnan, V. How a circularized tmRNA moves through the ribosome. Science 2019, 363, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Guyomar, C.; D’Urso, G.; Chat, S.; Giudice, E.; Gillet, R. Structures of tmRNA and SmpB as they transit through the ribosome. Nat. Commun. 2021, 12, 4909. [Google Scholar] [CrossRef]
- Neubauer, C.; Gillet, R.; Kelley, A.C.; Ramakrishnan, V. Decoding in the absence of a codon by tmRNA and SmpB in the Ribosome. Science 2012, 335, 1366–1369. [Google Scholar] [CrossRef]
- Kurita, D.; Muto, A.; Himeno, H. Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA 2010, 16, 980–990. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, G.; Guyomar, C.; Chat, S.; Giudice, E.; Gillet, R. Insights into the ribosomal trans-translation rescue system: Lessons from recent structural studies. FEBS J. 2022; in press. [Google Scholar]
- Fei, X.; Bell, T.A.; Barkow, S.R.; Baker, T.A.; Sauer, R.T. Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. eLife 2020, 9, e61496. [Google Scholar] [CrossRef] [PubMed]
- Karzai, A.W.; Roche, E.D.; Sauer, R.T. The SsrA–SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 2000, 7, 449–455. [Google Scholar] [CrossRef]
- Richards, J.; Mehta, P.; Karzai, A.W. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 2006, 62, 1700–1712. [Google Scholar] [CrossRef]
- Cheng, Z.-F.; Deutscher, M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002, 277, 21624–21629. [Google Scholar] [CrossRef] [PubMed]
- Matos, R.G.; Pobre, V.; Reis, F.P.; Malecki, M.; Andrade, J.M.; Arraiano, C.M. Structure and degradation mechanisms of 3′ to 5′ exoribonucleases. In Ribonucleases. Nucleic Acids and Molecular Biology; Nicholson, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 192–222. [Google Scholar] [CrossRef]
- Saramago, M.; Bárria, C.; Dos Santos, R.F.; Silva, I.; Pobre, V.; Domingues, S.; Andrade, J.; Viegas, S.; Arraiano, C.M. The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 2014, 18, 105–115. [Google Scholar] [CrossRef]
- Ramadoss, N.S.; Alumasa, J.N.; Cheng, L.; Wang, Y.; Li, S.; Chambers, B.S.; Chang, H.; Chatterjee, A.K.; Brinker, A.; Engels, I.H.; et al. Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. USA 2013, 110, 10282–10287. [Google Scholar] [CrossRef]
- Huang, Y.; Alumasa, J.N.; Callaghan, L.T.; Baugh, R.S.; Rae, C.D.; Keiler, K.C.; McGillivray, S.M. A Small-molecule inhibitor of trans-translation synergistically interacts with cathelicidin antimicrobial peptides to impair survival of Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e02362-18. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, J.; Vioque, A. Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA 2001, 7, 1708–1716. [Google Scholar]
- Vioque, A.; de la Cruz, J. Trans-translation and protein synthesis inhibitors. FEMS Microbiol. Lett. 2003, 218, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Feaga, H.A.; Viollier, P.; Keiler, K.C. Release of nonstop ribosomes is essential. mBio 2014, 5, e01916-14. [Google Scholar] [CrossRef]
- Giudice, E.; Macé, K.; Gillet, R. Trans-translation exposed: Understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 2014, 5, 113. [Google Scholar] [CrossRef]
- Andini, N.; Nash, K.A. Expression of tmRNA in mycobacteria is increased by antimicrobial agents that target the ribosome. FEMS Microbiol. Lett. 2011, 322, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Luidalepp, H.; Hallier, M.; Felden, B.; Tenson, T. tmRNA decreases the bactericidal activity of aminoglycosides and the susceptibility to inhibitors of cell wall synthesis. RNA Biol. 2005, 2, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ji, L.; Shi, W.; Xie, J.; Zhang, Y. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2477–2481. [Google Scholar] [CrossRef] [PubMed]
- Alumasa, J.N.; Manzanillo, P.S.; Peterson, N.D.; Lundrigan, T.; Baughn, A.D.; Cox, J.S.; Keiler, K.C. Ribosome rescue inhibitors kill actively growing and nonreplicating persister Mycobacterium tuberculosis cells. ACS Infect. Dis. 2017, 3, 634–644. [Google Scholar] [CrossRef]
- Aron, Z.D.; Mehrani, A.; Hoffer, E.D.; Connolly, K.L.; Srinivas, P.; Torhan, M.C.; Alumasa, J.N.; Cabrera, M.; Hosangadi, D.; Barbor, J.S.; et al. Trans-translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo. Nat. Commun. 2021, 12, 1779. [Google Scholar] [CrossRef]
- Senges, C.H.R.; Stepanek, J.J.; Wenzel, M.; Raatschen, N.; Ay, Ü.; Märtens, Y.; Prochnow, P.; Hernández, M.V.; Yayci, A.; Schubert, B.; et al. Comparison of proteomic responses as global approach to antibiotic mechanism of action elucidation. Antimicrob. Agents Chemother. 2020, 65, e01373-20. [Google Scholar] [CrossRef]
- Macé, K.; Demay, F.; Guyomar, C.; Georgeault, S.; Giudice, E.; Goude, R.; Trautwetter, A.; Ermel, G.; Blanco, C.; Gillet, R. A genetic tool to quantify trans-translation activity in vivo. J. Mol. Biol. 2017, 429, 3617–3625. [Google Scholar] [CrossRef] [PubMed]
- Brunel, R.; Descours, G.; Durieux, I.; Doublet, P.; Jarraud, S.; Charpentier, X. KKL-35 exhibits potent antibiotic activity against Legionella species independently of trans-translation inhibition. Antimicrob. Agents Chemother. 2018, 62, e01459-17. [Google Scholar] [CrossRef]
- Guyomar, C.; Thépaut, M.; Nonin-Lecomte, S.; Méreau, A.; Goude, R.; Gillet, R. Reassembling green fluorescent protein for in vitro evaluation of trans-translation. Nucleic Acids Res. 2020, 48, e22. [Google Scholar] [CrossRef]
- Tresse, C.; Radigue, R.; Gomes Von Borowski, R.; Thepaut, M.; Le, H.H.; Demay, F.; Georgeault, S.; Dhalluin, A.; Trautwetter, A.; Ermel, G.; et al. Synthesis and evaluation of 1,3,4-oxadiazole derivatives for development as broad-spectrum antibiotics. Bioorganic Med. Chem. 2019, 27, 115097. [Google Scholar] [CrossRef]
- Cole, S.T. Microbiology. Pyrazinamide—Old TB drug finds new target. Science 2011, 333, 1583–1584. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E.; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Bi, J.; Cai, Q.; Liao, X.; Li, W.; Guo, C.; Zhang, Q.; Lin, T.; Zhao, Y.; et al. Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide. Mol. Microbiol. 2015, 95, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Dillon, N.A.; Peterson, N.D.; Feaga, H.A.; Keiler, K.C.; Baughn, A.D. Anti-tubercular activity of pyrazinamide is independent of trans-translation and RpsA. Sci. Rep. 2017, 7, 6135. [Google Scholar] [CrossRef] [PubMed]
- Vallejos-Sánchez, K.; Lopez, J.M.; Antiparra, R.; Toscano, E.; Saavedra, H.; Kirwan, D.E.; Amzel, L.M.; Gilman, R.H.; Maruenda, H.; Sheen, P.; et al. Mycobacterium tuberculosis ribosomal protein S1 (RpsA) and variants with truncated C-terminal end show absence of interaction with pyrazinoic acid. Sci. Rep. 2020, 10, 8356. [Google Scholar] [CrossRef] [PubMed]
- Reverdatto, S.; Burz, D.S.; Shekhtman, A. Peptide aptamers: Development and applications. Curr. Top. Med. Chem. 2015, 15, 1082–1101. [Google Scholar] [CrossRef]
- Liu, P.; Huang, D.; Hu, X.; Tang, Y.; Ma, X.; Yan, R.; Han, Q.; Guo, J.; Zhang, Y.; Sun, Q.; et al. Targeting inhibition of SmpB by peptide aptamer attenuates the virulence to protect zebrafish against Aeromonas veronii infection. Front. Microbiol. 2017, 8, 1766. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhou, Z.; Huang, H.; Ren, Y.; Zhang, Y.; Li, G.; Zhou, Z.; Wang, L. Complete genome sequence of Aeromonas veronii Strain B565. J. Bacteriol. 2011, 193, 3389–3390. [Google Scholar] [CrossRef]
- Liu, P.; Chen, Y.; Wang, D.; Tang, Y.; Tang, H.; Song, H.; Sun, Q.; Zhang, Y.; Liu, Z. Genetic selection of peptide aptamers that interact and inhibit both small protein B and alternative ribosome-rescue factor A of Aeromonas veronii C4. Front. Microbiol. 2016, 7, 1228. [Google Scholar] [CrossRef]
- Thépaut, M.; Campos-Silva, R.; Renard, E.; Barloy-Hubler, F.; Ennifar, E.; Boujard, D.; Gillet, R. Safe and easy in vitro evaluation of tmRNA-SmpB-mediated trans-translation from ESKAPE pathogenic bacteria. RNA 2021, 27, 1390–1399. [Google Scholar] [CrossRef]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).