Abstract
Dengue outbreaks have regularly been recorded in Lao People’s Democratic Republic (PDR) since the first detection of the disease in 1979. In 2012, an integrated arbovirus surveillance network was set up in Lao PDR and an entomological surveillance has been implemented since 2016 in Vientiane Capital. Here, we report a study combining epidemiological, phylogenetic, and entomological analyzes during the largest DENV-4 epidemic ever recorded in Lao PDR (2015–2019). Strikingly, from 2015 to 2019, we reported the DENV-4 emergence and spread at the country level after two large epidemics predominated by DENV-3 and DENV-1, respectively, in 2012–2013 and 2015. Our data revealed a significant difference in the median age of the patient infected by DENV-4 compared to the other serotypes. Phylogenetic analysis demonstrated the circulation of DENV-4 Genotype I at the country level since at least 2013. The entomological surveillance showed a predominance of Aedes aegypti compared to Aedes albopictus and high abundance of these vectors in dry and rainy seasons between 2016 and 2019, in Vientiane Capital. Overall, these results emphasized the importance of an integrated approach to evaluate factors, which could impact the circulation and the epidemiological profile of dengue viruses, especially in endemic countries like Lao PDR.
1. Introduction
In the context of globalization of trade and travel, the arboviruses’ epidemiology profiles have changed and their expansion is in constant progression []. Dengue fever is the most prevalent human arboviral disease in the world. Recent World Health Organization (WHO) statistics revealed an increase of the number of dengue cases reported from 505,430 cases in 2000 to 4.2 million in 2019, among which 70% of the burden is supported by Asia, and a modelling study estimated that 390 million people are infected by dengue virus per year [,,]. Even if these data should be interpreted cautiously, due to changes in declaration systems and the increased number of contributing countries, they do reflect an alarming evolution of dengue virus (DENV) epidemiology. In 2017, WHO estimated that, every year, more than 500,000 people, including a high proportion of children, experience severe clinical presentations that require hospitalization. The number of fatal dengue cases has increased from 960 in 2000 to 4032 in 2015 [].
Dengue fever is due to an infection of one of the four DENV serotypes transmitted to humans through the bite of an infected female mosquito of the Aedes genus []. In Lao People’s Democratic Republic (Lao PDR), the main vector in urban areas is Aedes aegypti [], whereas Aedes albopictus is considered a secondary vector, specifically in suburban, rural, and forested areas [,,].
DENV are single-stranded, positive-sense RNA viruses belonging to the Flavivirus genus, Flaviviridae family. Antigenic studies identified four distinct serotypes (DENV-1 to 4) []. Genetic studies confirmed the segregation of DENV strains into four main groups matching with the serotypes, but also into genotype subdivisions that are often characteristic of the geographical origin of the viral strains [,,,,]. Previous genetic studies have suggested that the four DENV serotypes emerged from sylvatic cycles in Asia [,,]. Dengue outbreaks caused by the four serotypes have regularly been recorded in Asia over the last decades []. DENV-4 was identified for the first time in the Philippines and in Thailand in 1953 []. Since then, DENV-4 has been reported yearly in the Indochinese peninsula, but with a global burden significantly lower compared to the three other DENV serotypes []. From a genetic point of view, DENV-4 isolates segregate in four different genotypes: Genotype I (Asia) is frequently reported in South East Asia []; Genotype II (Asia, South Pacific, and South America); Genotype III (Thailand); and Genotype IV (sylvatic strains from Malaysia) [,,,,].
Lao PDR is a land-locked country located in the middle of the Indochinese peninsula, surrounded by China (North), Cambodia (South), Thailand and Myanmar (West), and Vietnam (East). Lao PDR was a least-developed country of 7 million people in 2018, among which nearly 40% are under 20 years old (WHO and www.lsb.gov.la). The country is divided into 18 provinces. Since the first report of dengue hemorrhagic fever (DHF) cases in Lao PDR in 1979, DENV outbreaks have been regularly declared by the country and dengue fever has become a major national public health problem [,,]. All four dengue serotypes now circulate in rural and urban areas in Lao PDR [,,,,]. In 2008, a DENV-1 epidemic was described only in the North-West part of the country []. Then, in 2013, an extensive dengue outbreak with a predominance of DENV-3 caused 48,772 cases and 95 deaths at the country scale [].
Interestingly, DENV-4 has been rarely detected in Lao PDR. Previous studies reported a lower prevalence of this serotype compared to the others in 1990 in Vientiane Capital and in other provinces in 2008–2009 [,,]. Only sporadic cases have been detected in villages located around Vientiane Capital in 2006–2007 []. In 2012, an integrated arbovirus surveillance system, involving virologists and medical entomologists, was set up in Vientiane Capital by the Institut Pasteur du Laos and was gradually extended to different provinces in Lao PDR for the detection and monitoring of arbovirus epidemics [,,]. This surveillance system detected DENV-4 transmission in 2013 in Vientiane Capital’s suburban areas and subsequently its spread into the different districts of the city and then throughout the country.
This study aims to describe the DENV-4 switch from an endemic-sporadic circulation to a country-wide epidemic in Lao PDR in the context of multiple DENV serotypes co-circulation. A retrospective epidemiological study was conducted in Vientiane Capital to characterize the impact of the disease burden on the population. Furthermore, an entomological study was implemented in Vientiane Capital to estimate the population dynamics of the vectors at different time points between 2016 and 2019.
2. Results
2.1. Dengue Surveillance Activity between 2012 and 2019.
From 2012 to 2019, 15,152 samples were collected through the arbovirus surveillance system in 15 of the 18 provinces of Lao PDR (Table S1; Figure S1).
Of the 15,152 samples tested, 8771 (57.9%) were confirmed for a DENV infection by means of real-time RT-PCR and/or NS1 antigen detection. Among the confirmed cases, 8315 (54.9%) were found positives for DENV by RT-PCR and 448 (5%) cases were confirmed by NS1 antigen detection in plasma or in urine samples. The DENV serotypes were determined by real-time RT-PCR or sequencing after a viral culture for 3616 cases (43.5%).
Overall, 11,858 samples (78.4%) were collected in Vientiane Capital, notwithstanding the progressive extension of the surveillance system to other Lao provinces from 2015 and onwards. In 2012 and 2013, DENV serotype identification was mainly performed for isolates collected in Vientiane Capital, which represented, respectively, 80% and 87% of the samples tested. In 2014, only 134 suspected cases were investigated. The dramatically low number of samples tested by the arbovirus surveillance network was consistent with the poor syndromic activity recorded at the country level by the National Center for Laboratory and Epidemiology in charge of the centralization of the mandatory clinical declaration system (Figure 1). Furthermore, since 2012, the laboratory-based survey seemed to follow the same dengue epidemiological profile as the national syndromic surveillance system (Figure 1). A total of 14 (10.4%) were confirmed by DENV RT-PCR in 2014 and the DENV serotype could be determined for 11 samples (78.6%) (Figure 2).
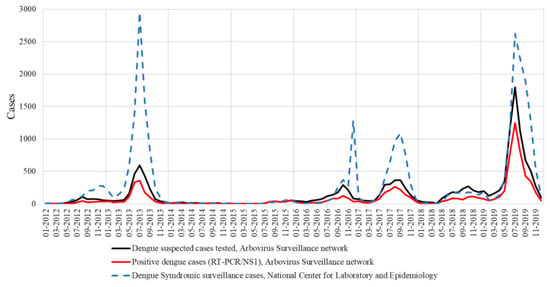
Figure 1.
National syndromic surveillance system versus laboratory-based survey in Lao PDR.
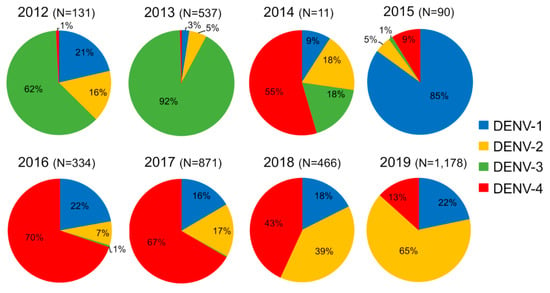
Figure 2.
Dengue serotype distribution between 2012 and 2019 from samples collected by the Institut Pasteur du Laos arbovirus surveillance network. In parentheses, number of samples tested per year.
Among the cases confirmed by RT-PCR in Vientiane Capital, 85.7% and 62.4% were respectively serotyped in 2016 and in 2018 (Table S1). The proportion of samples serotyped varied from year to year (from 28.2% in 2019 to 76.6% in 2014), but the proportion of samples from Vientiane Capital remained constant over time (between 50% and 65%) (Table S1).
In 2012 and 2013, all four DENV serotypes were detected, but DENV-3 serotype was predominant (62% in 2012 and 92% in 2013) (Figure 2). This serotype gradually decreased and vanished in December 2013 []. Since then, only 2 DENV-3 sporadic cases were collected in Saravane in 2016 and 3 cases were detected in Vientiane Capital in 2017. In 2015, DENV-1 was at the origin of 85% of the confirmed dengue cases at the country level and this serotype continued to circulate at a low level in Lao PDR until 2019.
Between 2012 and 2015, DENV-4 samples represented less than 9% of the total cases (Figure 2) and were only recorded in the sub-urban districts of Vientiane Capital and in Vientiane Province, which are mainly rural areas. In 2014, clusters of DENV-4 cases were identified in Vientiane Capital. At the end of 2015, the dengue epidemiological profile changed in Vientiane capital with the identification of grouped cases of DENV-4 in urban districts of Vientiane Capital followed by a rapid increase of the proportion of DENV-4 cases in Vientiane Capital from 9% in 2015 to 70% and 67%, respectively, in 2016 and 2017. The outbreak peaked between June and August 2017 (Figure 3). The same trend was observed at the country level (Figure 3). In 2018, the proportion of DENV-4 samples decreased to 43% and DENV-2 became the predominant serotype at the end of the year. In 2019, at the country level, DENV-4 still represented 13% of the cases serotyped (13.2% in Vientiane Capital) with a co-circulation of DENV-1 and DENV-2 (Figure 2).
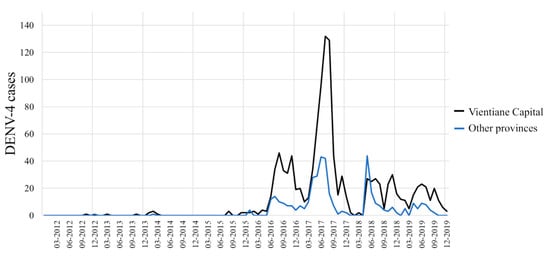
Figure 3.
Dengue serotype 4 distribution between 2012 and 2019 per month from samples collected by the Institut Pasteur du Laos arbovirus surveillance network in Vientiane Capital and in other Lao provinces.
2.2. Epidemiological Analysis of the Patients Infected by DENV-4
Demographic and clinical characteristics of the patients reported by the surveillance network are presented in Table 1. Patients infected with DENV-4 serotype were significantly older (mean age 27.4 vs. 24.5, p < 0.0001) than patients infected with other DENV serotypes (Table 1 and Figure 4). Sex ratio, days to diagnosis, and severity were no different for DENV-4 when compared to the other serotypes.

Table 1.
Demographic and clinical characteristics of the 15,152 patients investigated by the Institut Pasteurs du Laos arbovirus surveillance system between 2012 and 2019.
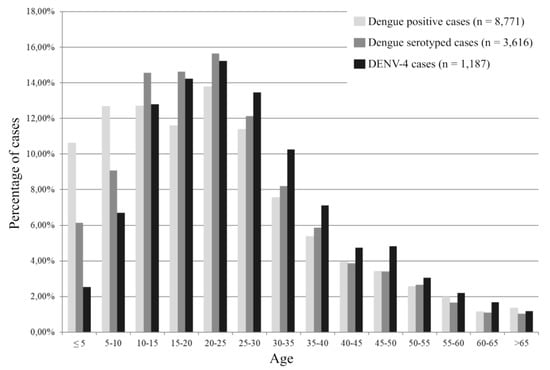
Figure 4.
Age distribution of patients investigated by the Institut Pasteur du Laos arbovirus surveillance network.
Among the 52 DENV fatal cases investigated by the arbovirus surveillance network between 2012 and 2019, 9 were infected by DENV-4. These nine cases were identified between 2016 and 2019. In Vientiane Capital, four cases were detected in September 2016, August 2017, January 2019, and December 2019. In Attapeu province, 2 DENV-4 fatal cases were identified in July and September 2017. Two fatal cases were also recorded in June 2018 in Xayaboury and Champassak provinces and 1 case in Saravane province in April 2019 (Figure S1).
2.3. Phylogenetic Analysis of DENV-4
A panel of 45 samples, i.e., plasma (n = 29) or cell supernatant (n = 16), from DENV-4 cases collected between 2013 and 2019, were investigated for the phylogenetic analysis (Table 2). This panel included samples from different Lao provinces, cases imported from foreign Asian countries, patients with different degrees of dengue severity, and fatal cases. The sequence analysis based on a fragment spanning 1485 nucleotides of the envelop gene revealed that DENV-4 isolates grouped in two different genotypes (i.e., Genotype I and Genotype II) (Figure 5).

Table 2.
References of Lao DENV-4 isolates.

Figure 5.
Maximum likelihood phylogenetic tree of DENV-4 sequences from Lao PDR. The tree was constructed on the envelope protein gene (1485 nt). Only the bootstrap values > 70 are shown. Scale bar indicates the nucleotide substitution per site. The Lao strains sequenced in this study are indicated in red and the triangles highlight the strains from fatal cases. The Lao strain previously described is indicated in blue. Black triangles identify isolates obtained from fatal cases.
Genotype II was identified in samples from European tourists traveling in Southeast Asia (Thailand and Malaysia) in 2014 and who developed a dengue-like syndrome during their stay in Vientiane. These isolates clustered with DENV-4 strains identified in Malaysia in 2009 and 2014 (>99% identity) (Figure 5) [].
All autochthonous DENV-4 sequences belonged to the Genotype I. Among the Genotype I, the Lao strains isolated between 2013 and 2019 displayed a maximum nucleotide distance of 2.7% from each other. Interestingly, our series of isolates showed 3.1% to 4.1% of divergence with a Lao strain (KY849762) isolated in 2009 in Saravane province (Figure 5) []. When compared to DENV-4 reference strains collected in other Asian countries from 2011 to 2016, less than 2.5% of nucleotide divergence was found with the Lao DENV-4 isolates (Figure 5). In this study, eight DENV-4 isolates from fatal cases could be analyzed. These isolates belonged to the Genotype I but segregated in different phylogenetic clusters formed by the different Lao isolates characterized since 2013 (Figure 5). Globally, the phylogenetic analysis revealed the active circulation of DENV-4 Genotype I since at least 2013 in Lao PDR. Furthermore, several clusters were detected and could emphasize several introduction events in the country from neighboring countries such as Thailand (Figure 5).
2.4. Entomological Survey on the Aedes sp. Vectors in Vientiane Capital
The results of the abundance of the adult dengue vectors collected between 2016 and 2019 are presented in Figure 6. A total of 1276 specimens were collected and Aedes aegypti was the most abundant species, representing more than 86% of the total. Aedes albopictus represented 14% of the total. The percentage of Ae. albopictus varied between 3.2% in 2018 to 57.8% in 2016 when the study only started in May. In 2017 and 2019, the percentages were 12.6% and 9.9%, respectively. Every year, the vectors were more abundant during the rainy season between May and August, but the mosquito populations remained throughout the dry season albeit at a low level. Of note, no collections were made during the month of April; and in 2016, the collection started in May only.
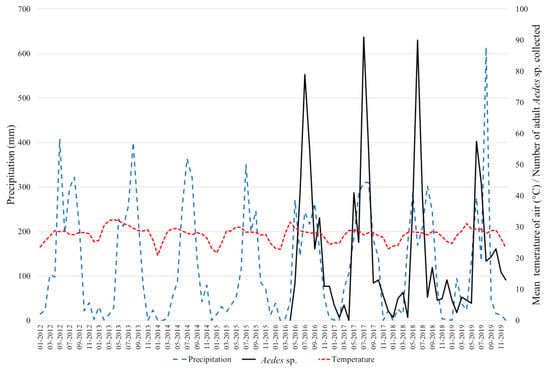
Figure 6.
Monthly rainfall (mm; in blue) and mean temperature in the air (°C; in red) recorded between 2012 and 2019 in Vientiane Capital (17°58′12″ N, 102°34′14″ E). Number of Aedes aegypti and Aedes albopictus adult mosquitoes collected monthly between 2016 and 2019 in Vientiane Capital (in black).
3. Discussion
Since the first report on dengue in Lao PDR, limited data have been published on DENV serotypes/genotypes circulating at the country level []. Nevertheless, recent studies suggest a complex and dynamic dengue virus circulation countrywide, but without reference to DENV-4 [,,,]. This study is a first step in filling this gap by providing genetic and epidemiologic information on the first DENV-4 epidemic recorded in Lao PDR. The progressive increase of DENV-4 burden occurred in a context of the co-circulation of different DENV serotypes previously described and maintained over the eight years of surveillance (i.e., 2012–2019) [,,]. Since 2009, DENV-4 circulated sporadically in the country [,]. Sporadic cases or small clusters of grouped cases were observed in Vientiane Capital between February 2014 and August 2015. Subsequently, the proportion of DENV-4 cases gradually increased among the samples tested by the arbovirus surveillance network in Vientiane Capital and became predominant in 2016 at the country level until October 2018. Since then, DENV-2 has overcome DENV-4 in Vientiane Capital and at the country level []. However, DENV-4 still circulated at a significant level in 2019 (13%; Figure 2). These data reveal a circulation of DENV-4 during at least six years (2014–2019) in Lao PDR and could be correlated with an insufficient immunity protection of the Lao population against this serotype as suggested by the impact of DENV-4 in the different age groups.
This study highlighted the significant difference in the age of the patients infected with DENV. The syndromic patients infected with DENV-4 were significantly older than the patients infected with other serotypes. The Lao population is young (~40% under 20 years old) (WHO and www.lsb.gov.la) and this data emphasize the risk of dengue emergence in Lao PDR due to a high percentage of the population immunologically naive for DENV. In this study, the mean age (27.4) of the patients infected by DENV-4 appeared to be consistent with studies from other DENV-endemic countries [,]. However, the fact that syndromic DENV-4 patients were older could suggest the recent emergence of this serotype and/or a low DENV-4 circulation in Lao PDR at least since 2009 []. These findings demonstrate that a large proportion of the Lao population is susceptible to DENV-4 and highlights the need to study the immune status of populations living in dengue-endemic countries in order to predict and prevent future DENV outbreaks as previously done in Lao PDR [] and in Asia in general [,,]. Furthermore, this study highlights the need to perform serotype-specific studies of the immune status of the population.
Here, we describe the pre-epidemic circulation of DENV-4 in Lao PDR and the specific features of this serotype during the outbreak in Vientiane Capital and at the country level in the context of DENV serotypes co-circulation. Clusters of DENV-4 infections were identified sporadically several years before the outbreak started. Indeed, serotype-specific surveillance networks are able to finely monitor the profile of DENV circulating and detect early warning signals. Strengthening the prediction capacity of DENV outbreaks by combining syndromic surveillance systems with a laboratory surveillance system has been demonstrated in Lao PDR ([]; present study). Even if a systematic exhaustive investigation of suspected cases was clearly unrealistic, an appropriate sample size allowed for the drawing of an accurate picture of DENV serotypes’ history in a country over the last eight years. In this way, the actual disease burden of dengue could be calculated, including at the serotype level. The amplitude over time and the burden by serotype and by age groups could be used as proxies to estimate the level of herd immunity in the Lao population. Interestingly, the local lab-based surveillance system in Vientiane city followed the trends of the syndromic surveillance data. Based on this approach, mathematical models could be developed to determine parameters including costs of long-term sustainable, combined surveillance systems.
The disease severity of dengue fever was not associated with the predominance of DENV-4 in Lao PDR. Indeed, no significant difference in the disease severity was found between the DENV serotypes in the confirmed samples collected by the arbovirus surveillance network. This finding is in agreement with previous observations, such as during a DENV-4 outbreak in Brazil, which failed to show any link between this serotype and dengue severe symptoms []. Furthermore, in Lao PDR, the fatal cases due to a DENV-4 infection were recorded during the entire outbreak and no link could be established with the number of positive DENV-4 cases. Indeed, in our study, the same number of fatal cases was recorded in 2017 and 2019, respectively, during the peak of the outbreak and at the end of the outbreak. Moreover, dengue fatal cases were observed during the active DENV transmission period, as well as when dengue cases were recorded at a low level.
Previous phylogenetic studies showed the recent circulation of two DENV-4 genotypes (Genotype I and II) in Southeast Asia [,]. Both genotypes were detected in Lao PDR, but in two different and independent types of patients, i.e., Lao native autochthonous cases and imported cases by foreigners. All autochthonous cases recorded at the country level investigated here belonged to DENV-4 Genotype I. The first evidence of the presence of DENV-4 Genotype I in Lao PDR came from a strain that was first identified in Saravane Province isolated in 2009 []. Our serial investigation suggests that this genotype has spread and circulated at least since 2013 at the country level. The genetic difference (2.7% of nucleotide divergence) between the former strain and the recent isolates could be the result of several introductions of DENV-4 Genotype I in Lao PDR since 2009 from different Asian countries. The composition of the different clusters and the circulation of the Lao isolates over several years are a signature of successful consecutive emergences of those independent introduction events. DENV-4 Genotype II was only detected in two imported cases from Thailand/Malaysia in 2014 (Figure 5). Interestingly, the predominant circulation of the DENV-4 Genotype I in Lao PDR contrasted with some results obtained in South East Asia. Indeed, in the Philippines, a genotype turnover from Genotype I to Genotype II was observed before 2013, and this seemed to match with a high DENV-4 circulation in the country between 2012 and 2016 []. The results found in the Philippines were consistent with those from Malaysia in 2001 that showed the circulation of the DENV-4 Genotype II []. However, both introductions of DENV-4 Genotype II were recorded in February 2014, the dry season in Lao PDR (October–April) and a decrease of vector density. This context, associated with the abnormal lack of rainfalls until August 2015, could explain the non-emergence of DENV-4 Genotype II in Vientiane Capital. However, these dramatically unfavorable conditions for DENV transmission did not hamper the maintenance of a sufficient inoculum of a mixt population of DENV-4 Genotype I isolates at the origin of the rapid spread-off since September 2015. Hence, the genotype selection could impact the epidemiology of DENV emergence and expansion of the virus. Indeed, as previously described in India, China, or the WHO Pacific region, a DENV genotype switch could increase the risk of dengue outbreak [,,]. This selection of DENV genotype could be influenced by several factors, such as serotype-specific herd immunity, viremia titer in humans, and the ability of local mosquito populations, such as Aedes aegypti, to transmit the virus under the influence of environmental and climatic conditions [,,,,,]. These factors need to be investigated for DENV serotypes and genotypes, which circulated in a specific geographical context to improve the knowledge on DENV epidemiology and to prevent DENV outbreaks [].
The entomological results demonstrate the presence of both Ae. aegypti and Ae. albopictus in Vientiane Capital with an increase of vector density during the rainfall season as previously described in Asia. The dynamic of DENV transmission in human population follows the trend of the dynamic of Aedes vectors []. Moreover, predominance of Ae. aegypti was highlighted since 2016 in Vientiane Capital. Vector competence of Ae. aegypti from Lao PDR was investigated for DENV-1 and showed that 50% of the females orally exposed to the virus could transmit DENV-1 []. These data indicate a high efficiency of transmission of this serotype by Ae. aegypti from Lao PDR compared to other Ae. aegypti populations from the Pacific region (3–37% transmission efficiency), from French Guiana (<10% transmission efficiency), and from Florida (33% transmission efficiency) in the USA [,,]. DENV transmission ability appeared to be a specific interaction between the mosquito population and a virus strain [,]. Vazeille et al. [] suggested a possible competition between serotypes at the midgut level in co-infected mosquitoes leading to a drastically different transmission potential and, in this case, favoring the competitive displacement of DENV-1 by DENV-4. Thus, the ability of Lao vectors for DENV-4 must be investigated to determine how this factor could influence the onset of new dengue epidemics in Lao PDR.
DENV emergence and spread are complex mechanisms, especially in endemic countries like Lao PDR. This study shows the need to combine results obtained by the DENV laboratory-based surveillance system and epidemiologic studies, combined with entomological data, along with more research oriented approaches, such as virus genetic, viral kinetic, or virus/vector interaction findings, to determine the impact of these factors on DENV transmission in each specific geographical context.
4. Materials and Methods
4.1. Human Samples Collection
In 2012, an arbovirus surveillance system was set up and coordinated by the Institut Pasteur du Laos. It was first implemented in Vientiane Capital and then progressively extended to hospitals in 18 provinces, with Vientiane Capital’s hospital network being the most active. From 2012 to 2019, human samples were collected through this surveillance hospital network. Suspected dengue fever cases were defined according to the WHO’s definition (fever onset ≥ 38 °C for less than 7 days with at least one of the following accompanying symptoms: headache; myalgia; arthralgia; retro-orbital pain; digestive troubles or hemorrhaging) []. After obtaining informed consent, clinicians filled a standardized clinical report form (CRF). Cases were classified according to the WHO criteria (1997), defining the severity as Dengue fever (DF), Dengue hemorrhagic fever (DHF), or Dengue shock syndrome (DSS). Venous blood samples (5 mL) were taken from patients. Samples were stored at 4 °C during transportation to the Institut Pasteur du Laos for analysis and serotypes and genotypes investigation.
4.2. Ethical Statement
Ethics approval was obtained from the Lao National Ethics Committee for Health Research (N°2018.116). Oral and written informed consent were obtained from all participants, or a parent or legal guardian.
4.3. Dengue Virus Screening
Samples collected by the arbovirus surveillance system were screened by RT-PCR as previously described [] with a pan-dengue real-time RT-PCR [] and the DENV serotypes were determined with specific real-time RT-PCR [].
4.4. Gene E Sequencing Analysis
Viral genomic RNA was extracted from 45 human plasma using a Nucleospin DX or Nucleospin 96 core kit purification kit (Macherey-Nagel) according to the manufacturer’s instructions (Table 2).
Gene E sequencing (1485nt) was performed using primers FGT1, FGT2, and F3 (Table 3). First Stand cDNA were generated using a Maxima H Minus First Stand cDNA Synthesis kit (Thermo Scientific, Waltham, MA, USA) and the PCR was performed using a Phusion Flash High-Fidelity PCR Master Kit (New England Biolabs® Inc, Waltham, MA, USA). Amplified fragments were purified using ExoSAP-ITTM PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Purified fragments were sequenced using a BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystem, Waltham, MA, USA) on a Genetic Analyzer 3500xL (Applied Biosystem, Waltham, MA, USA).

Table 3.
List and positions of primers used for DENV-4 complete envelope gene RT-PCR and sequencing.
Sequences were analyzed using Chromas software (www.technelysium.com.au) and aligned with the multiple sequence alignment software Clustal W integrated in BioEdit version 7.0.5.3 software (Manchester, United Kingdom) [,]. For the phylogenetic analysis, a maximum likelihood tree was constructed using MEGA version 7 (www.megasoftware.net), with a kimura-2 parameter model with a bootstrap of 1000 replication [] as previously described for dengue virus phylogenetic analysis [,,,,]).
4.5. Epidemiological Analysis
Demographic and clinical data were extracted from the surveillance network’s de-identified database. Continuous variables were summarized using mean and standard deviation (SD), and categorical variables were summarized using frequencies and percentages. Continuous data were compared using parametric Student test and rates using the Chi-square and Fisher exact test. Statistical significance was set at the 5% level. Analyses were conducted using STATA software (version 14.0; StatCorp LP, College Station, TX, USA).
4.6. Mosquito Surveillance
Five mosquito sentinel sites in five different villages of Vientiane Capital were chosen for the mosquito surveillance and the GPS locations are presented in Table 4. The vectors abundance study took place during both dry and rainy seasons between 2016 and 2019. BG sentinel traps® (Biogents, Regensburg, Germany) were used to follow the dynamics of mosquito population abundance in the city (4 traps per locations, except at Kao-Gnot with two traps). After every weekly collection, the adult mosquitoes were brought back to the laboratory for morphological identification.

Table 4.
Mosquito collection locations in Vientiane Capital.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/9/728/s1, Figure S1: Map of Lao PDR, Table S1: Geographic origins of the DENV samples serotyped by the Institut Pasteur du Laos arbovirus surveillance system.
Author Contributions
Conceptualization: E.C., V.P., S.M., and M.G.; methodology, E.C., V.P., S.M., and M.G.; validation, E.C., V.P., S.M., and M.G.; formal Analysis, E.C., V.P., and S.M.; investigation, E.C., V.P., S.V., P.B., T.A.C., S.S., and S.M.; resources, S.S., S.V., T.A.C., T.X., P.P., P.B., S.K., and S.M.; writing—original draft preparation, E.C., V.P., S.S., S.M., and M.G.; writing—review and editing, E.C., V.P., J.P., P.T.B., M.C., S.M., and M.G.; visualization, E.C., V.P., and S.M.; supervision, M.G. and E.C.; project administration, M.G.; funding acquisition, M.G., P.T.B., O.T., and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Agence Française de Développement grant n° CZZ 2146 01 (Ecomore2 project) and by UNITEDengue and Global Partnership Programme, Canada (ASEAN-GPP Grant Phase 3—Laboratory Capacity Development for diagnostics of Emerging Dangerous Pathogens).
Acknowledgments
We thank Lee Ching Ng, Director, Environmental Health Institute (EHI), National Environment Agency, Chanditha Hapuarachchi, and Carmen Koo for their sequencing technical assistance. We also thank the Institut Pasteur du Laos staff for the mosquito collections and identification (Phoutmany Thammavong, Phonesavanh Luangamath, Kaithong Lakeomany, Somphat Nilaxay, and Vaekey Vungkyly).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cao-Lormeau, V.-M. Tropical Islands as New Hubs for Emerging Arboviruses. Emerg. Infect. Dis. 2016, 22, 913–915. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Dengue and Severe Dengue Update 23 June 2020. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 14 July 2020).
- Gubler, D. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002, 10, 100–103. [Google Scholar] [CrossRef]
- Marcombe, S.; Fustec, B.; Cattel, J.; Chonephetsarath, S.; Thammavong, P.; Phommavanh, N.; David, J.-P.; Corbel, V.; Sutherland, I.W.; Hertz, J.C.; et al. Distribution of insecticide resistance and mechanisms involved in the arbovirus vector Aedes aegypti in Laos and implication for vector control. Barker CM, editor. PLoS Negl. Trop. Dis. 2019, 13, e0007852. [Google Scholar] [CrossRef]
- Hawley, W.A. The biology of Aedes albopictus. J. Am. Mosq. Control. Assoc. Suppl. 1988, 1, 1–39. [Google Scholar]
- Tangena, J.-A.; Marcombe, S.; Thammavong, P.; Chonephetsarath, S.; Somphong, B.; Sayteng, K.; Grandadam, M.; Sutherland, I.W.; Lindsay, S.W.; Brey, P.T. Bionomics and insecticide resistance of the arboviral vector Aedes albopictus in northern Lao PDR. PLoS ONE 2018, 13, e0206387. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Bustos-Arriaga, J.; Green, A.; James, S.; Lau, L.; Montoya, M.; Wang, C.; VanBlargan, L.A.; et al. Dengue viruses cluster antigenically but not as discrete serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef]
- Henchal, E.A.; Putnak, J.R. The dengue viruses. Clin. Microbiol. Rev. 1990, 3, 376–396. [Google Scholar] [CrossRef]
- Weaver, S.; Vasilakis, N. Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 2009, 9, 523–540. [Google Scholar] [CrossRef]
- Gubler, D. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.-J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Rico-Hesse, R. Microevolution and virulence of dengue viruses. Adv. Exp. Med. Biol. 2003, 59, 315–341. [Google Scholar]
- Rico-Hesse, R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 1990, 174, 479–493. [Google Scholar] [CrossRef]
- Wang, E.; Ni, H.; Xu, R.; Barrett, A.D.T.; Watowich, S.J.; Gubler, D.J.; Weaver, S. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 2000, 74, 3227–3234. [Google Scholar] [CrossRef]
- Messina, J.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Bhatt, S.; Katzelnick, L.; Howes, R.E.; Battle, K.E.; et al. Global spread of dengue virus types: Mapping the 70 year history. Trends Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef]
- Hamel, R.; Surasombatpattana, P.; Wichit, S.; Dauvé, A.; Donato, C.; Pompon, J.F.; Vijaykrishna, D.; Liegeois, F.; Vargas, R.M.; Luplertlop, N.; et al. Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand. PLoS ONE 2019, 14, e0221179. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Gubler, D.J.; Trent, D.W. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 1997, 78, 2279–2284. [Google Scholar] [CrossRef]
- Heringer, M.; Souza, T.M.A.; Lima, M.D.R.Q.; Nunes, P.C.G.; Faria, N.R.D.C.; De Bruycker-Nogueira, F.; Chouin-Carneiro, T.; Nogueira, R.M.R.; Dos Santos, F.B. Dengue type 4 in Rio de Janeiro, Brazil: Case characterization following its introduction in an endemic region. BMC Infect. Dis. 2017, 17, 410. [Google Scholar] [CrossRef]
- Klungthong, C.; Zhang, C.; Mammen, M.P.; Ubol, S.; Holmes, E.C. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 2004, 329, 168–179. [Google Scholar] [CrossRef]
- Fukunaga, T.; Phommasack, B.; Bounlu, K.; Saito, M.; Tadano, M.; Makino, Y.; Kanemura, K.; Arakaki, S.; Shinjo, M.; Insisiengmay, S. Epidemiological situation of dengue infection in Lao P.D.R. Trop. Med. 1994, 35, 219–227. [Google Scholar]
- Khampapongpane, B.; Lewis, H.C.; Ketmayoon, P.; Phonekeo, D.; Somoulay, V.; Khamsing, A.; Phengxay, M.; Sisouk, T.; Sisouk, P.; Bryant, J.E. National dengue surveillance in the Lao People’s Democratic Republic, 2006–2012: Epidemiological and laboratory findings. West. Pac. Surveill Response J. 2014, 5, 7–13. [Google Scholar]
- Soukaloun, D. Dengue infection in Lao PDR. Southeast Asian J. Trop. Med. Public Health. 2014, 45, 113–119. [Google Scholar] [PubMed]
- Bounlu, K.; Tadano, M.; Makino, Y.; Arakaki, S.; Kanemura, K.; Fukunaga, T. A seroepidemiological study of dengue and Japanese encephalitis virus infections in Vientiane, Lao PDR. Jpn. J. Trop. Med. Hyg. 1992, 20, 149–156. [Google Scholar] [CrossRef]
- Makino, Y.; Saito, M.; Phommasack, B.; Vongxay, P.; Kanemura, K.; Pothawan, T.; Bounsou; Insisiengmay, S.; Sompaw; Fukunaga, T. Arbovirus Infections in Pilot Areas in Laos. Trop. Med. 1995, 36, 131–139. [Google Scholar]
- Guo, X.; Zhao, Q.; Wu, C.; Zuo, S.; Zhang, X.; Jia, N.; Liu, J.; Zhou, H.-N.; Zhang, J.-S. First isolation of dengue virus from Lao PDR in a Chinese traveler. Virol. J. 2013, 10, 70. [Google Scholar] [CrossRef]
- Dubot-Pérès, A.; Vongphrachanh, P.; Denny, J.; Phetsouvanh, R.; Linthavong, S.; Sengkeopraseuth, B.; Khasing, A.; Xaythideth, V.; Moore, C.E.; Vongsouvath, M.; et al. An Epidemic of Dengue-1 in a Remote Village in Rural Laos. PLoS Negl. Trop. Dis. 2013, 7, e2360. [Google Scholar] [CrossRef]
- Lao, M.; Caro, V.; Thiberge, J.-M.; Bounmany, P.; Vongpayloth, K.; Buchy, P.; Duong, V.; Vanhlasy, C.; Hospied, J.-M.; Thongsna, M.; et al. Co-Circulation of Dengue Virus Type 3 Genotypes in Vientiane Capital, Lao PDR. PLoS ONE 2014, 9, e115569. [Google Scholar] [CrossRef]
- Castonguay-Vanier, J.; Klitting, R.; Sengvilaipaseuth, O.; Piorkowski, G.; Baronti, C.; Sibounheuang, B.; Vongsouvath, M.; Chanthongthip, A.; Thongpaseuth, S.; Mayxay, M.; et al. Molecular epidemiology of dengue viruses in three provinces of Lao PDR, 2006–2010. PLoS Neglected Trop. Dis. 2018, 12, e0006203. [Google Scholar] [CrossRef]
- Somlor, S.; Vongpayloth, K.; Diancourt, L.; Buchy, P.; Duong, V.; Phonekeo, D.; Ketmayoon, P.; Vongphrachanh, P.; Brey, P.T.; Caro, V.; et al. Chikungunya virus emergence in the Lao PDR, 2012–2013. PLoS ONE 2017, 12, e0189879. [Google Scholar] [CrossRef]
- Yang, C.-F.; Chang, S.-F.; Hsu, T.-C.; Su, C.-L.; Wang, T.-C.; Lin, S.-H.; Yang, S.-L.; Lin, C.-C.; Shu, P.-Y. Molecular characterization and phylogenetic analysis of dengue viruses imported into Taiwan during 2011–2016. PLoS Neglected Trop. Dis. 2018, 12, e0006773. [Google Scholar] [CrossRef] [PubMed]
- Calvez, E.; Somlor, S.; Viengphouthong, S.; Balière, C.; Bounmany, P.; Keosenhom, S.; Caro, V.; Grandadam, M. Rapid genotyping protocol to improve dengue virus serotype 2 survey in Lao PDR. PLoS ONE 2020, 15, e0237384. [Google Scholar] [CrossRef] [PubMed]
- Lestari, C.S.W.; Yohan, B.; Yunita, A.; Meutiawati, F.; Hayati, R.F.; Trimarsanto, H.; Sasmono, R. Phylogenetic and evolutionary analyses of dengue viruses isolated in Jakarta, Indonesia. Virus Genes 2017, 53, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Tiraki, D.; Diwan, A.; Lalwani, S.K.; Modak, M.; Mishra, A.C.; Arankalle, V.A. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE 2018, 13, e0192672. [Google Scholar] [CrossRef] [PubMed]
- Vongpunsawad, S.; Intharasongkroh, D.; Thongmee, T.; Poovorawan, Y. Seroprevalence of antibodies to dengue and chikungunya viruses in Thailand. PLoS ONE 2017, 12, e0180560. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-J.; Liu, C.-K.; Tsai, W.-Y.; Liu, L.-T.; Tyson, J.; Tsai, C.-Y.; Lin, P.-C.; Wang, W.-K. Seroprevalence of dengue virus in two districts of Kaohsiung City after the largest dengue outbreak in Taiwan since World War II. PLoS Neglected Trop. Dis. 2018, 12, e0006879. [Google Scholar] [CrossRef] [PubMed]
- Fox-Lewis, A.; Hopkins, J.; Sar, P.; Sao, S.; Pheaktra, N.; Day, N.P.J.; Blacksell, S.D.; Turner, P. Seroprevalence of Dengue Virus and Rickettsial Infections in Cambodian Children. Am. J. Trop. Med. Hyg. 2019, 100, 635–638. [Google Scholar] [CrossRef]
- Luz, M.A.D.V.; Nabeshima, T.; Moi, M.L.; Dimamay, M.T.A.; Pangilinan, L.-A.S.; Dimamay, M.P.S.; Matias, R.R.; Mapua, C.A.; Buerano, C.C.; De Guzman, F.; et al. An Epidemic of Dengue Virus Serotype-4 during the 2015–2017: The Emergence of a Novel Genotype IIa of DENV-4 in the Philippines. Jpn. J. Infect. Dis. 2019, 72, 413–419. [Google Scholar] [CrossRef]
- Abubakar, S.; Wong, P.-F.; Chan, Y.F. Emergence of dengue virus type 4 genotype IIA in Malaysia. J. Gen. Virol. 2002, 83, 2437–2442. [Google Scholar] [CrossRef]
- Ahamed, S.F.; Rosario, V.; Britto, C.; Dias, M.; Nayak, K.; Chandele, A.; Murali-Krishna, K.; Shet, A. Emergence of new genotypes and lineages of dengue viruses during the 2012–15 epidemics in southern India. Int. J. Infect. Dis. 2019, 84, S34–S43. [Google Scholar] [CrossRef]
- Jiang, L.; Jing, Q.L.; Liu, Y.; Cao, Y.M.; Su, W.Z.; Biao, D.; Yang, Z.C. Molecular characterization and genotype shift of dengue virus strains between 2001 and 2014 in Guangzhou. Epidemiol. Infect. 2016, 145, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Dupont-Rouzeyrol, M.; Aubry, M.; O’Connor, O.; Roche, C.; Gourinat, A.-C.; Guigon, A.; Pyke, A.; Grangeon, J.-P.; Nilles, E.J.; Chanteau, S.; et al. Epidemiological and molecular features of dengue virus type-1 in New Caledonia, South Pacific, 2001–2013. Virol. J. 2014, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Hang, V.T.T.; Holmes, E.C.; Veasna, D.; Quy, N.T.; Hien, T.T.; Quail, M.; Churcher, C.; Parkhill, J.; Cardosa, J.; Farrar, J.; et al. Emergence of the Asian 1 Genotype of Dengue Virus Serotype 2 in Viet Nam: In Vivo Fitness Advantage and Lineage Replacement in South-East Asia. PLoS Negl. Trop. Dis. 2010, 4, e757. [Google Scholar] [CrossRef]
- Quiner, C.A.; Parameswaran, P.; Ciota, A.T.; Ehrbar, D.J.; Dodson, B.L.; Schlesinger, S.; Kramer, L.D.; Harris, E. Increased Replicative Fitness of a Dengue Virus 2 Clade in Native Mosquitoes: Potential Contribution to a Clade Replacement Event in Nicaragua. J. Virol. 2014, 88, 13125–13134. [Google Scholar] [CrossRef]
- Lambrechts, L.; Fansiri, T.; Pongsiri, A.; Thaisomboonsuk, B.; Klungthong, C.; Richardson, J.H.; Ponlawat, A.; Jarman, R.G.; Scott, T.W. Dengue-1 Virus Clade Replacement in Thailand Associated with Enhanced Mosquito Transmission. J. Virol. 2011, 86, 1853–1861. [Google Scholar] [CrossRef]
- Hanley, K.A.; Nelson, J.T.; Schirtzinger, E.E.; Whitehead, S.S.; Hanson, C.T. Superior infectivity for mosquito vectors contributes to competitive displacement among strains of dengue virus. BMC Ecol. 2008, 8, 1. [Google Scholar] [CrossRef]
- Fansiri, T.; Fontaine, A.; Diancourt, L.; Caro, V.; Thaisomboonsuk, B.; Richardson, J.H.; Jarman, R.G.; Ponlawat, A.; Lambrechts, L. Genetic Mapping of Specific Interactions between Aedes aegypti Mosquitoes and Dengue Viruses. PLoS Genet. 2013, 9, e1003621. [Google Scholar] [CrossRef]
- Lambrechts, L. Quantitative genetics of Aedes aegypti vector competence for dengue viruses: Towards a new paradigm? Trends Parasitol. 2011, 27, 111–114. [Google Scholar] [CrossRef]
- Cologna, R.; Armstrong, P.M.; Rico-Hesse, R. Selection for Virulent Dengue Viruses Occurs in Humans and Mosquitoes. J. Virol. 2005, 79, 853–859. [Google Scholar] [CrossRef]
- Wai, K.T.; Arunachalam, N.; Tana, S.; Espino, F.; Kittayapong, P.; Abeyewickreme, W.; Hapangama, D.; Tyagi, B.K.; Htun, P.T.; Koyadun, S.; et al. Estimating dengue vector abundance in the wet and dry season: Implications for targeted vector control in urban and peri-urban Asia. Pathog. Glob. Health 2012, 106, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Miot, E.F.; Calvez, E.; Aubry, F.; Dabo, S.; Grandadam, M.; Marcombe, S.; Oke, C.; Logan, J.G.; Brey, P.T.; Lambrechts, L. Risk of arbovirus emergence via bridge vectors: Case study of the sylvatic mosquito Aedes malayensis in the Nakai district, Laos. Sci. Rep. 2020, 10, 7750–7759. [Google Scholar] [CrossRef] [PubMed]
- Calvez, E.; Guillaumot, L.; Girault, D.; Richard, V.; O’Connor, O.; Paoaafaite, T.; Teurlai, M.; Pocquet, N.; Cao-Lormeau, V.-M.; Dupont-Rouzeyrol, M. Dengue-1 virus and vector competence of Aedes aegypti (Diptera: Culicidae) populations from New Caledonia. Parasites Vectors 2017, 10, 381. [Google Scholar] [CrossRef] [PubMed]
- Vazeille, M.; Gaborit, P.; Mousson, L.; Girod, R.; Failloux, A.-B. Competitive advantage of a dengue 4 virus when co-infecting the mosquito Aedes aegypti with a dengue 1 virus. BMC Infect. Dis. 2016, 16, 318. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Anderson, S.L.; Alto, B.W. Vector competence of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) for dengue virus in the Florida Keys. J. Med. Entomol. 2012, 49, 942–946. [Google Scholar] [CrossRef]
- Lambrechts, L.; Chevillon, C.; Albright, R.G.; Thaisomboonsuk, B.K.; Richardson, J.H.; Jarman, R.G.; Scott, T.W. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol. Boil. 2009, 9, 160. [Google Scholar] [CrossRef]
- Warrilow, D.; Northill, J.A.; Pyke, A.; Smith, G.A. Single rapid TaqMan fluorogenic probe based PCR assay that detects all four dengue serotypes. J. Med. Virol. 2002, 66, 524–528. [Google Scholar] [CrossRef]
- Ito, M.; Takasaki, T.; Yamada, K.-I.; Nerome, R.; Tajima, S.; Kurane, I. Development and Evaluation of Fluorogenic TaqMan Reverse Transcriptase PCR Assays for Detection of Dengue Virus Types 1 to 4. J. Clin. Microbiol. 2004, 42, 5935–5937. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Afreen, N.; Naqvi, I.H.; Broor, S.; Ahmed, A.E.; Kazim, S.N.; Dohare, R.; Kumar, M.; Parveen, S. Evolutionary Analysis of Dengue Serotype 2 Viruses Using Phylogenetic and Bayesian Methods from New Delhi, India. PLoS Neglected Trop. Dis. 2016, 10, e0004511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, K.-S.; Lai, Y.-L.; Lo, S.; Barkham, T.; Aw, P.; Ooi, P.-L.; Tai, J.-C.; Hibberd, M.L.; Johansson, P.; Khoo, S.-P.; et al. Dengue Virus Surveillance for Early Warning, Singapore. Emerg. Infect. Dis. 2010, 16, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Ali, I. The Complete Genome Phylogeny of Geographically Distinct Dengue Virus Serotype 2 Isolates (1944–2013) Supports Further Groupings within the Cosmopolitan Genotype. PLoS ONE 2015, 10, e0138900. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).