Abstract
We used whole genome sequencing (WGS) as an epidemiologic surveillance tool to elucidate the transmission dynamics of Shiga toxin-producing Escherichia coli (STEC) strains along the beef production chain in South Africa. Isolates were obtained from a cattle farm, abattoirs and retail outlets. Isolates were analysed using WGS on a MiSeq platform (Illumina, San Diego, CA, USA) and phylogenetic analysis was carried out. Of the 85 isolates, 39% (33) carried the stx gene and 61% (52) had lost the stx gene. The prevalence of stx subtypes was as follows; stx1a 55% (18/33), stx1b 52% (17/33), stx2a 55% (18/33), stx2b 27% (9/33), stx2dB 30% (10/33) and stx2d1A 15% (5/33). Thirty-five different serogenotypes were detected, of which 65% (56) were flagellar H-antigens and 34% (29) were both O-antigens and flagellar H-antigens. We identified 50 different sequence types (STs), and only nine of the isolates were assigned to three different clonal complexes. Core genome phylogenetic analysis revealed genetic relatedness, and isolates clustered mainly according to their STs and serogenotypes regardless of stx subtypes. This study provides evidence of horizontal transmission and recirculation of STEC strains in Gauteng province and demonstrates that every stage of the beef production chain plays a significant role in STEC entry into the food chain.
1. Introduction
Shiga toxin-producing Escherichia coli (STEC) is a food- and water-borne pathogen reported in numerous outbreaks worldwide [1]. STEC causes a broad spectrum of disease ranging from mild to severe bloody diarrhoea (haemorrhagic colitis; HC), and in some cases (5–10%) it can progress to haemolytic uraemic syndrome (HUS) [1,2]. The ability of STEC to cause human disease is influenced primarily by the production of Shiga-like toxins (stx), which are encoded by stx genes carried on bacteriophages [2,3]. The stx genes are classified into two major types, stx1 and stx2 [3]. Scheutz et al. [3] proposed a subtype classification of the two major stx variants: stx1 which consists of stx1a, stx1b, stx1c and stx1d and stx2 consisting of seven distinct variants, namely stx2a, stx2b, stx2c, stx2d, stx2e, stx2f and stx2g. Reports have shown that stx2a, stx2c and stx2d subtypes are associated with the development of HC and HUS [3]. A group of virulence factors encoded by a chromosomal region, described as the 35-kb locus of enterocyte effacement (LEE) pathogenicity island (PAI), confer the attaching and effacing (A/E) phenotype for STEC [4]. The STEC LEE comprises genes encoding intimin, an adhesion factor (eaeA), the translocated receptor of intimin (tir), the secreted proteins EspA, EspB and EspD and the type III secretion pathway [5]. Other potential and putative virulence factors of pathogenic E. coli, including a wide array of adhesins, toxins, siderophores and secretion systems, empower the organism to colonize the intestinal epithelium, evade or manipulate host defence mechanisms, provoke harmful inflammatory reactions within the host and inflict direct harm upon host cells and tissues [6].
Over 100 O-serotypes of the more than 470 known STEC serotypes have been associated with human disease [6]. STEC O157:H7 has gained notoriety in major foodborne outbreaks and sporadic cases worldwide including in the USA, Canada, Japan and the United Kingdom [7]. However, several non-O157 STEC strains have been frequently linked with HC and HUS, predominantly strains of several serogroups—O26, O45, O103, O111, O121 and O145, termed the “big six” [1]. Non-O157 STEC strains are reported to cause more infections than do O157:H7 strains in Europe [7], including the 2011 Germany and France outbreak of O104:H4 enteroaggregative STEC [8].
The hind gut of cattle is recognised as the main asymptomatic reservoir and has the capacity to shed STEC transiently [9]. The pathogen’s excretion rates and concentration in faeces contribute considerably to its epidemiology and transmission within herds and in humans [10]. If the pathogen load in cattle entering the abattoir is high, then the likelihood of carcass contamination in the beef production chain is increased [11]. As such, the cattle farm plays a vital role in the beef chain. Additionally, STEC strains have been associated with human disease through the consumption of undercooked beef or beef-based products [12,13], which have been contaminated by cattle faeces during slaughter or processing as a result of cross-contamination, mainly from the hide and occasionally from gut contents [14,15]. In addition to cattle farms and abattoirs, meat retail outlets play an important role in the transmission of STEC-contaminated raw beef and ready-to-eat (RTE) beef products [16]. Contamination could arise at several stages, such as during meat cutting and further processing, such as with mincemeat or sausages made from mincemeat. A few colonized livestock or contaminated carcasses could contaminate a large quantity of ground beef [17]. Consequently, the presence of STEC throughout the beef production chain is a potential public health hazard.
In South Africa, although a few studies have reported the prevalence and virulence profiles of bovine STEC isolates [18,19], little has been done using whole genome sequencing (WGS) as a subtyping method for bovine isolates recovered along the beef chain. This study aimed to apply WGS as a molecular subtyping method to assess the virulence potential, phylogenetic relationships and diversity of STEC isolates recovered along the beef chain in Gauteng, the most densely populated province of South Africa.
2. Materials and Methods
2.1. Sources of Isolates
The STEC isolates in this study were recovered from three sources as previously described in Onyeka et al. [20,21,22].
2.1.1. Cattle Feedlots
Isolates (n = 30) were recovered from a longitudinal study conducted between September 2016 and February 2017, which determined the presence of shedders and super-shedders in a feedlot cattle operation in northern Gauteng, South Africa. Faecal samples were collected by rectal grab from randomly selected individual animals [23].
2.1.2. Abattoir and Retail Outlets
During November 2015 to November 2016, a random cross-sectional survey investigated STEC prevalence and molecular characteristics on beef carcasses and in beef products in Gauteng. For the abattoir study, 12 abattoirs located in Gauteng North (5), Gauteng East (4) and Gauteng West (3) were selected for the survey. Individual animals and carcasses were tagged and tracked in simple or continuous slaughter lines, and samples were obtained at different point locations in processing plants. From the abattoir study, 28 isolates were recovered. In addition, 7 isolates were recovered from tagged cattle followed from the farm to the abattoir in February for slaughter. The samples used in this study comprised carcass swabs in swab rinse kit solution (SRK), swabs from perineum hide swabs, swabs from walls and floor, faeces, rinsates and abattoir effluents [24].
For the retail study, 31 retail outlets (large chain supermarkets and butcheries) located across northern Gauteng were sampled by purchasing five different raw beef and ready-to-eat beef products. The samples comprised raw beef including brisket, mincemeat and boerewors and beef-based RTE products including biltong and cold meat and were sampled from the retail outlets during four seasons: summer, autumn, winter and spring. A total of 21 isolates were recovered from the retail outlets [20].
2.1.3. Isolation of STEC Strains
Only enrichment broth cultures that were PCR-positive for stx1, stx2 or both were considered positive for STEC and were cultured to isolate STEC strains. To isolate O157 STEC the procedure consisted of immunomagnetic separation (IMS) assays using Dynabeads® anti-E. coli O157 (Thermo Fisher Scientific, Waltham, USA), as recommended by the manufacturer. The immune-concentrated bacterial suspensions were then inoculated onto sorbitol with MacConkey agar (SMAC) supplemented with potassium tellurite 2.5 mg/L and cefixime 0.05 mg/L (Himedia Laboratories Pvt., Thane, India). Likewise, 10 μL of enriched broth sample was streaked on a chromogenic agar, CHROMagar O157 (CHROMagar Microbiology, Paris, France) supplemented with potassium tellurite 2.5 mg/L and cefixime 0.05 mg/L (Himedia Laboratories Pvt., Thane, India). Subsequently, the plates were incubated for 24–30 h at 37 °C, and up to seven suspect colonies with different phenotypes were picked from each plate and tested by latex agglutination (Welcolex® E. coli O157 Rapid latex agglutination test, Remel, Leicestershire, UK). Enriched control strain—E. coli ATCC 43888 (O157:H7)—was also inoculated for phenotypic control and assessment.
To isolate non-O157 STEC, 10 μL of enriched broth sample was streaked on MacConkey agar containing crystal violet and salt and onto CHROMagar STEC (CHROMagar Microbiology, Paris, France). The plates were incubated for 24–30 h at 37 °C, and representative suspect colonies were subcultured on nutrient agar plates for further biochemical testing. For further biochemical confirmation, isolates were randomly selected and confirmed as E. coli using the bioMérieux Vitek 2 Compact system (BioMérieux, Marcy l’Étoile, France).
2.1.4. Multiplex PCR to Identify Virulence
A DNA template of STEC isolates was prepared using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The DNA templates were investigated for the presence of stx1, stx2, eaeA and hlyA genes using mPCR as described by Paton and Paton [25] and Lindsey et al. [26], with slight modifications. All the isolates positive for the presence of toxin genes were stored at −20 °C until subjected to analysis.
2.1.5. Validation of mPCR
The assay conditions were optimized using molecular control strains obtained from the National Institute for Communicable Diseases—Centre for Enteric Diseases, South Africa (2014-2015 VTEC EQA—E. coli RR18-3022 O157, eaeA, stx1a, stx2a) and the enrichment control strain E. coli ATCC 43888 (O157:H7) stx1. The mPCR was validated by Sanger sequencing of PCR products.
2.2. Whole Genome Sequencing and Analysis
The Nextera XT DNA library prep kit was used to prepare paired-end libraries for 85 genomic DNA isolates, followed by 2 × 300-bp sequencing on a MiSeq platform (Illumina, Inc., San Diego, CA, USA) aiming at a coverage of at least 100-fold. The resultant paired-end reads were checked for quality control (QC) of average Q-score > 20 and trimmed using FASTP version 0.19.5 [27]. The sequence reads were de novo assembled using SKESA version 2.4.0 [28]. Gene annotation of all contiguous sequences (contigs) was carried out using Prokka [29]. Multilocus sequence typing (MLST) using the seven conserved housekeeping genes of E. coli scheme 1 was determined using Seemann T, mlst Github (https://github.com/tseemann/mlst, accessed on 20 March 2023). ABRicate [30] and subsequently ECtyper [31], was used for in silico serotyping of E. coli. Comprehensive antibiotic resistance database (CARD) was used for antimicrobial resistance genes [32], and E. coli virulence factors were determined using known virulence factors obtained from the virulence factor database (VFDB) [33].
2.3. Phylogenetic Analysis of STEC Isolates
A rapid, large-scale prokaryote pan-genome analysis pipeline (Roary) was used to determine genetic relationships among the STEC genomes [34], and randomized accelerated maximum-likelihood (RAxML) analysis [35] was used to reconstruct a maximum-likelihood phylogenetic tree based on core genome single nucleotide polymorphisms (SNPs). To extract predicted coding regions from Prokka-annotated assemblies and convert them to protein sequences, the core genome alignment module in Roary was employed [32]. BlastP (https://blast.ncbi.nlm.nih.gov, accessed on 15 March 2023) was used to compare all protein sequences with one another. Proteins that had alignment similarity of ≥70% and were present in at least 90% of the isolates were defined as the core genome. RAxML [17] was used to create a bootstrapped maximum-likelihood phylogenetic tree from the resulting core genome alignment and visualized and annotated in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 30 March 2023). See Supplementary Table S1 for information on the 85 isolates used in this study, NCBI accession numbers and corresponding URLs.
3. Results
3.1. Isolates
Of the 85 PCR-confirmed STEC isolates submitted for WGS, only 39% (33) harboured the stx gene, and 61% (52) had lost the stx gene. Of the 52, 85% (44) harboured the eae gene, and 15% (8) lacked the eae gene but harboured other adherence genes (Supplementary Table S2). The prevalence of stx subtypes was as follows: stx1a 55% (18/33), stx1b 52% (17/33), stx2a 55% (18/33), stx2b 27% (9/33), stx2dB 30% (10/33) and stx2d1A 15% (5/33) (Table 1). Thirty-five different serogenotypes, with two novel serogenotypes, were found among the 85 isolates, of which 65% (56/85) were flagellar H-antigens with O-antigen untypeable, and 34% (29/85) were both O-antigens and flagellar H-antigens including O8:H19. The stx2d toxin defined by Scheutz et al. [3], namely stx2d1A, was found in five isolates, whereas stx2dB was found in ten isolates. Fourteen different stx subtype combinations were found among the 33 isolates (Table 1).

Table 1.
Molecular and epidemiological data associated with 33 Shiga toxin-producing Escherichia coli isolates recovered from the beef production chain in Gauteng, South Africa.
3.2. Multilocus Sequence Typing
We identified 50 different sequence types (STs), including five isolates of novel STs and three of unknown STs, among the 85 isolates. The most frequent STs were ST306 (5/85; 6%) and Novel (5/85; 6%). Only nine of the isolates were assigned to three different clonal complexes (ST that matched the central genotype at five or six loci), the remaining STs identified in this study could not be assigned to any clonal complex (Table 2).

Table 2.
MLST clonal complexes found in 85 Shiga toxin-producing Escherichia coli isolates recovered along the beef chain in Gauteng, South Africa.
3.3. Virulence Genes
A total of 552 putative virulence genes were determined (Supplementary Table S2). The genes included adherence, secretory (type II/III/IV/VI secretory system/effectors) and toxin (heat-labile/stable enterotoxin, cytolethal distending toxins, colicin, exotoxin cytotoxic necrotizing factor, haemolysin and subtilase cytotoxin), among others (Figure 1). From the 85 isolates, the prevalence of the LEE encoded genes was as follows: eae 19% (16), EspA 20% (17), EspB 19% (16) and EspD 20% (17). The prevalence of plasmid-encoded virulence-associated genes was as follows: espP 26% (22), katp 11% (9), subA 6% (5) and saa 4% (3). Others prevalences included the autotransporter proteins ehaA 62% (53) and ehaB 74% (63), a heme uptake-related gene chuA 6% (5) and the haemolysin gene hlyA 31% (26). Furthermore, among the 52 stx-negative isolates, the virulence factors eae, tir and chuA were identified in the beef chain in the farm 12% (2) and retail 20% (3) isolates, each. Catalase-peroxidase (katP) was found in isolates from the farm 18% (3) and retail shops 20% (3) in the beef chain. Only one isolate (perineal-PdJ2-4) harboured cytolethal distending toxins (cdtIIIA, cdtIIIB and cdtIIIC). Additionally, we identified a selection of virulence genes associated with a high risk of diarrhoea and severe disease in humans [19], including aatA 6% (5/85), cif 7% (6/85), escV 11.8% (10/85), EspA 20% (17/85), nleA 9.4% (8/85), nleB 13% (11/85) and tccp 3.5% (3/85).
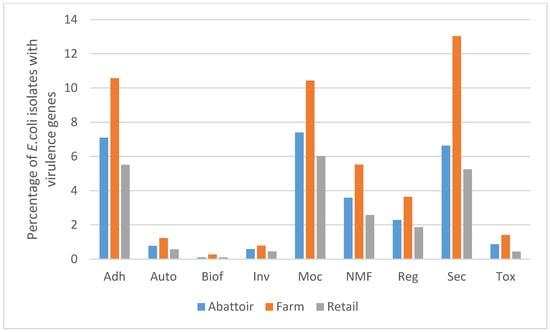
Figure 1.
Bar-chart showing the percentage of 85 E.coli isolates from beef abattoirs, feedlot and retail outlets (raw beef and ready-to-eat beef products) that possessed virulence genes with their manually annotated functions in Gauteng, South Africa. Adh: adherence, Auto: autotransporter, Biof: biofilm, Inv: invasive, MoC: motility/chemotaxis, NMF: nutritional/metabolic factor, Reg: regulatory, Sec: secretory, Tox: toxin.
3.4. Antimicrobial Resistance Genes
We detected 66 genes of which multidrug (MDR) efflux pump genes were the most prevalent 55% (36), including the acriflavine efflux system AcrAB-TolC (acrA—100%; acrB—96.5%; TolC—100%) and regulators such as cpxA (98.8) and gadX (95%) (Table 3). Of notable mention is the presence of E. coli ampicillin class C (AmpC) β-lactamase genes, detected in 97.6% (83/85) of the isolates. Interestingly, we observed a low prevalence of antimicrobial resistance genes in the WOAH-OIE [23] classified list of “veterinary critically important antimicrobial agents” in cattle, which included aminoglycosides-modifying enzymes [24], nucleotidyltransferases encoded by aadA (5%), aadA2 (2%), aadA3 (4%) and aadA4 (1%) and phosphotransferases aph (6)-Id (9.4%) and aph (3″)-Ib (9.4%) which mediate resistance against kanamycin. Others were Fosfomycin-modifying enzymes such as metalloenzyme FosA7 (4.7%), nonfluorinated/fluorinated phenicols genes cmIA6 (1%) and floR (4%) and β-lactamases TEM-1 (4%) and TEM-150 (14%) (Table 3 and Supplementary Table S3).

Table 3.
Occurrence of 66 genes that code for antimicrobial resistance in 85 STEC isolates from the beef production chain in Gauteng, South Africa.
3.5. Phylogenetic Analysis
The phylogenetic tree was built with only 82 isolates; the three isolates with unknown ST were excluded from the tree. The 82 isolates contained 4760 genes, of which 32.81% were the core genes (shared by all 82 isolates), used in constructing the tree. Core genome phylogenetic analysis revealed that isolates clustered mainly according to their STs and serogenotypes regardless of stx subtype. The 82 isolates were categorised into 12 clades, partly based on their STs and serogenotypes. Figure 2 shows the distribution of the 12 clades, with clades that contain similar STs being highlighted. The clade formed by ST515 belonging to serotype H29 showed a close relationship with isolates from cattle faeces (abattoir, Gauteng east), biltong (retail outlet, Gauteng north) and cattle faeces (feedlot, Gauteng north). The clade containing ST730 and ST361 showed intra-farm transmission. In addition, similar patterns of genetic relatedness were shown in isolates with ST306 (five cattle intra-farm) and ST4017 (inter-abattoir in Gauteng west and Gauteng north). However, we observed an outgroup clade including isolates from abattoir hide (ST95) and from three cattle from the farm (ST6353, ST11 and ST6546).
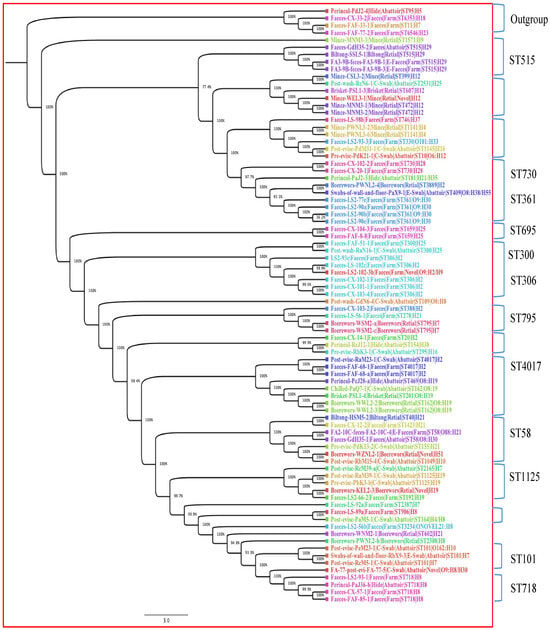
Figure 2.
Phylogenetic tree of 82 STEC isolates based on 4760 genes (defined by core genome) from different stages in the beef production chain in Gauteng, South Africa. The node percentages are the bootstrap values from 100 replicates representing the confidence estimates of the tree topology. The scale bar indicates 10% nucleotide sequence divergence. The colours represent isolates belonging to the same sequence types.
4. Discussion
In this study, we explored the potential of WGS as an epidemiologic surveillance tool to elucidate the molecular characteristics and transmission dynamics of STEC along the beef production chain (the farm-to-fork approach) in South Africa. The subtyping of stx genes revealed that only 39% (33) of the 85 isolates harboured the stx gene, and 61% (52) had lost the stx gene, a phenomenon termed ‘STEC lost Shiga toxin’ [36], given that our previous studies had confirmed these as STEC isolates [20,21,22]. The loss of the stx genes might have occurred during the initial subcultivation step or during subculturing of preserved frozen cultures to obtain genomic DNA [36,37,38]. Several studies have indicated a correlation between the loss of stx genes and the serotype or the specific subtype of stx, which are less stable in non-O157 strains [36,37,38]. Our data support these observations, since all 85 isolates were non-O157 STEC. Consequently, great caution must be exercised in the aetiological diagnosis of HC and HUS, given the possibility of a loss of stx genes.
Epidemiologic studies and cytotoxicity assays have revealed that the different subtypes may be associated with varying degrees of virulence or severity [1,2,39]. In this study, we detected stx2d genes (stx2d1A and stx2dB) and combinations of stx1a, stx1b and eae (8 isolates) and stx2a, stx2dB and eae (2 isolates), which have the potential to cause HC and HUS in humans [1,39,40].
In addition to the stx genes, we observed genes encoding 81 type III secretion systems (T3SSs). These are major virulence genes that contribute to the severity of STEC disease [32]. The presence of T3SSs in our isolates is of public health importance, as this presence in cattle populations, abattoirs and meat products in South Africa increases the risk for zoonotic, environmental and foodborne transmission of the most virulent strains [41].
From the 85 isolates, we found 35 serogenotypes of which 65% (56/85) were O-serogroup untypeable (ONT). Among the 56 flagellar antigens we identified H2, H7, H8, H12, H16, H19, H21, H25 and H28, which have been associated with pathogenic STEC [40,42]. Additionally, STEC ONT:H7 in this study harboured the highest number of virulence-associated genes linked with severe clinical symptoms (stx2dB, stx2a, subA, eae, espP, hlyA, katP, tir, chuA and astA). Other isolates which had more virulence genes included ONT:H25 (stx2dB, stx2a, eae, espP, hlyA, katp, tir and astA), ONT:H2 (stx2dB, stx2a, espP, saa, tir and astA), ONT:H8 (stx2d1A, stx2a, espP, hlyA, katp and astA) and O8:H19 (stx2dB, stx2a, subA, espP, saa and hlyA). Our results confirm that pathogenic E. coli in the beef production chain in Gauteng, South Africa comprises a genetically heterogeneous family of bacteria. Notably, O8:H19 (five isolates), ONT:H8 (six isolates) and ONT:H21(six isolates) have been linked with human disease in South Africa [41]. Furthermore, in the Netherlands and Germany, O8:H19 has been associated with HUS, while O8:H8 has been associated with mild infection [43]. STEC O8:H19 have been recovered from healthy cattle across the globe, including Europe [43], China [44] and Mexico [40].
In South Africa and other southern African countries, the importance of STEC has been highlighted by numerous clinical cases of diarrhoea in children and adults between 2006-2013 in which a diverse range of STEC serogroups (O4, O5, O8:H19, ONT:H8, ONT:H21,O21, O26, O84, O111, O113, O117 and O157) were implicated [45].
This study revealed a high prevalence of E. coli ampicillin class C (AmpC) β-lactamase genes, detected in 98% (83/85) of the isolates, clinically known to confer resistance to penicillin-like and cephalosporin-class antibiotics [46]. Our result is comparable with the findings of Iweriebor et al. [47], who reported AmpC beta-lactamases (penicillin and cephalosporin resistance) in 90% of isolates originating from two dairy cattle farms in South Africa. Additionally, multidrug efflux pumps serve as a primary defence mechanism in all bacteria, reducing the intracellular concentration of antimicrobials. A single multidrug efflux pump can expel various antibiotics, thereby contributing to bacterial pathogenicity and multidrug resistance [48]. For example, the AcrAB-TolC observed in this study is a house-keeping efflux pump which is involved in the extrusion of a wide spectrum of antibiotics including macrolides, linezolid, novobiocin, rifampin, fusidic acid, chloramphenicol, fluoroquinolone, tetracycline, nalidixic acid and β-lactam antibiotics among others [49,50]. Hence, the observed prevalence of 55% (36/66) of MDR efflux pump genes is also notable, considering that the WHO [51] has included AMR among the “top 10 threats for global health”. This trend of resistance, along with the MDR profiles, could stem from the isolate source as it originates from livestock, mainly cattle, and could be attributable to the magnitude and scale of AMR presence and persistence in the study area. South Africa and other industrializing economies such as China, Brazil, India and Russia are regarded as hotspots for antimicrobial resistance due to intensive livestock production and the concomitant increase in antibiotic use in animal husbandry [52,53].
Interestingly, the cgMLST revealed a high genomic diversity of strains, with only nine isolates grouped into three clonal complexes (Table 2). The three abattoir samples originating from three different geographic locations (Pre-evis-PdK13-2-Gauteng north, Post-evis-RbM15-4-Gauteng west and Faeces-GdH35-1-Gauteng east) were indistinguishable, which suggests that either there is recirculation of the same strain through horizontal transmission across the province, or less likely, that the cattle slaughtered in these abattoirs were sourced from the same farm [9]. In addition, the two clonal complexes comprising strains from two different abattoirs and one retail outlet (Brisket-PSL1-retail/Chilled-PaQ7-1-abattoir/Perineal-PcJ28-a-abattoir and Post-evis-PdM31-1-abattoir/Pre-evis-PdK21-1-abattoir/Mince-PWNL3-2-retail) originating from the same geographic location (Gauteng north) suggest horizontal transmission and strain recirculation in Gauteng north. Recirculation of STEC could occur from carrier cattle such as super-shedders, from faecal environmental contamination including wastewater irrigations and indirectly via humans and other vectors acting as vehicles of recirculation in a geographic region [9].
The phylogenetic tree revealed that a common ancestor might exist for strains of the same sequence type in the beef production chain in South Africa. An outgroup clade comprising an isolate (Perineal swab-PdJ2-4-Gauteng north) from an abattoir hide (ST95) and three faecal isolates (Cx33-2, FaF33-1 and FaF77-2) from cattle from the feedlot (ST6353, ST11 and ST6546) was also present. The isolates belonging to these sequence types, notably ST95, had been confirmed as STEC in previous studies [20,21,22]. Interestingly, ST95 in this study also harboured four genes (UTI89_C3190, UTI89_C3191, UTI89_C3194 and UTI89_C3202) associated with uropathogenic E. coli (UPEC) (Supplementary Table S2) and could be related to the clonal lineage of one of the predominant clonal extraintestinal pathogenic E. coli (ExPEC) groups (ST131, ST69, ST95 and ST73) incriminated in human infections globally, including in the United Kingdom, Spain and France. ST95 ranked second among the most prevalent clonal ExPEC groups recovered from patients with bloodstream infections (BSI) [54]. Furthermore, the STEC O157:H7 strains predominantly belonged to ST11, which has previously been associated with diarrhoea and HUS [55].
Given the difficulty of isolating STEC from food and environmental samples, including faeces, we consider culture-based methods as a major limitation in this study. For further studies, we recommend using metagenomics to study transmission dynamics. With metagenomics, field samples can be sequenced directly, thus bypassing culture-based limitations while simultaneously increasing the opportunity for the discovery of novel pathogens [56]. Additionally, the O-antigen in most of the strains of E. coli could not be O-serotyped; we identify this as a limitation of the study, which requires further investigation.
In conclusion, this study provided evidence of genetic diversity in STEC strains throughout the beef production chain. The detection of stx2d (stx2d1A and stx2dB) and serotype O8:H19 that may cause severe disease including HC and HUS in humans is notable. The high prevalence of MDR efflux pump and AmpC genes constitutes a potential source of resistance genes in the southern African region, with negative impact on food security and public health. Knowledge about the prevalence of these resistance genes is crucial to curtail their dissemination in Africa. The three clonal complexes are strong evidence of horizontal transmission and recirculation of STEC strains throughout the beef production chain in Gauteng province, South Africa.
To our knowledge, this is the first study to comprehensively characterise STEC isolates recovered from the beef production chain in an area and provide evidence of horizontal transmission using WGS data. These data are valuable for hazard identification and risk assessment and for the development of intervention strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13090732/s1 Table S1: Information on 85 isolates used in this study from the beef production chain in Gauteng, South Africa; Table S2: Occurrence of 552 VFDB annotated genes in the 85 STEC genome assemblies.; Table S3: Occurrence of 66 genes that code for antimicrobial resistance (AMR) in 85 STEC isolates from the beef production chain in Gauteng, South Africa
Author Contributions
Conceptualization, A.A.A. and P.N.T.; Methodology, L.O.O., A.I. and K.H.K.; Software, A.I. and M.A.; Formal analysis, L.O.O., A.I., M.A. and P.N.T.; Investigation, L.O.O.; Resources, A.A.A., A.I. and K.H.K.; Data curation, M.A. and P.N.T.; Writing—original draft, L.O.O.; Writing—review & editing, L.O.O., A.A.A., A.I., M.A., K.H.K. and P.N.T.; Supervision, A.A.A., K.H.K. and P.N.T.; Project administration, P.N.T.; Funding acquisition, L.O.O., A.A.A. and P.N.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by Red Meat Research and Development South Africa (Grant numbers NAS2015-0116, VET2018-0005) and the University of Pretoria.
Institutional Review Board Statement
Ethical approval for the study was obtained from the University of Pretoria Animal Ethics Committee (S4285-15, V019-15). Permission was obtained from the Veterinary Public Health section of the Gauteng Department of Agriculture and Rural Development (GDARD).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in [National Center for Biotechnology Information] at https://account.ncbi.nlm.nih.gov/?back_url=https%3A%2F%2Fdataview.ncbi.nlm.nih.gov%2F, reference number [PRJNA706921], accessed on 20 July 2024.
Acknowledgments
We are grateful for the support we received from the staff of the Sequencing Core Facility, National Institute for Communicable Diseases (CED-NICD), Johannesburg, South Africa.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO/WHO STEC Expert Group. Hazard identification and characterization: Criteria for categorizing Shiga toxin–producing Escherichia coli on a risk basis. J. Food Prot. 2019, 82, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Melton-Celsa, A.R. Shiga toxin (Stx) classification, structure, and function. Microbiol. Spectr. 2014, 2, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B. The locus of enterocyte effacement pathogenicity island of Shiga toxin-producing Escherichia coli O157:H7 and other attaching and effacing E. Coli. Jpn. J. Med. Sci. Biol. 1998, 51 (Suppl. S1), S101–S107. [Google Scholar] [CrossRef]
- Mora, A.; Herrrera, A.; López, C.; Dahbi, G.; Mamani, R.; Pita, J.M.; Alonso, M.P.; Llovo, J.; Bernárdez, M.I.; Blanco, J.E.; et al. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104: H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 2011, 14, 121–141. [Google Scholar] [CrossRef]
- Blanco, M.; Blanco, J.E.; Mora, A.; Dahbi, G.; Alonso, M.P.; González, E.A.; Bernárdez, M.I.; Blanco, J. Serotypes, Virulence Genes, and Intimin Types of Shiga Toxin (Verotoxin)-Producing Escherichia coli Isolates from Cattle in Spain and Identification of a New Intimin Variant Gene (eae-ξ). J. Clin. Microbiol. 2004, 42, 645–651. [Google Scholar] [CrossRef]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli—A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Loneragan, G.H.; Carr, M.A.; Nisbet, D.J. Shiga toxin-producing Escherichia coli (STEC) ecology in cattle and management based options for reducing fecal shedding. Agric. Food Anal. Bacteriol. 2013, 3, 39–69. [Google Scholar]
- Munns, K.D.; Selinger, L.B.; Stanford, K.; Guan, L.; Callaway, T.R.; McAllister, T.A. Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog. Dis. 2015, 12, 89–103. [Google Scholar] [CrossRef]
- Collins, J.D. Slaughtering and processing of livestock. Agricu Mech. Autom. 2009, 2, 342. [Google Scholar] [CrossRef]
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 2010, 140, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.; Chinen, I.; Miliwebsky, E.; Masana, M. Risk factors for Shiga toxin-producing Escherichia coli-associated human diseases. In Enterohemorrhagic Escherichia coli and Other Shiga Toxin-Producing; Wiley: Hoboken, NJ, USA, 2015; pp. 359–380. [Google Scholar] [CrossRef]
- Fremaux, B.; Prigent-Combaret, C.; Vernozy-Rozand, C. Long-term survival of Shiga toxin-producing Escherichia coli in cattle effluents and environment: An updated review. Vet. Microbiol. 2008, 132, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, J.M.; Doherty, A.M.; Sheridan, J.J.; Thomson-Carter, F.M.; Garvey, P.; McGuire, L.; Blair, I.; McDowell, D. The prevalence and spread of Escherichia coli O157:H7 at a commercial beef abattoir. J. Appl. Microbiol. 2003, 95, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; MacLeod, E.T.; El Bayomi, R.M.; Mohsen, R.A.; Nassar, A.H. Molecular characterization of Escherichia coli O157:H7 and non-O157 shiga toxin producing E. coli from retail meat and humans. Zagazig Vet. J. 2017, 45, 250–261. [Google Scholar] [CrossRef]
- Duffy, G.; Cummins, E.; Nally, P.; O’Brien, S.; Butler, F. A review of quantitative microbial risk assessment in the management of Escherichia coli O157:H7 on beef. Meat Sci. 2006, 74, 76–88. [Google Scholar] [CrossRef]
- Bumunang, E.W.; McAllister, T.A.; Zaheer, R.; Ortega Polo, R.; Stanford, K.; King, R.; Niu, Y.D.; Ateba, C.N. Characterization of non-O157 Escherichia coli from cattle faecal samples in the North-West Province of South Africa. Microorganisms 2019, 7, 272. [Google Scholar] [CrossRef]
- Karama, M.; Mainga, A.O.; Cenci-Goga, B.T.; Malahlela, M.; El-Ashram, S.; Kalake, A. Molecular profiling and antimicrobial resistance of Shiga toxin-producing Escherichia coli O26, O45, O103, O121, O145 and O157 isolates from cattle on cow-calf operations in South Africa. Sci. Rep. 2019, 9, 11930. [Google Scholar] [CrossRef]
- Onyeka, L.O.; Adesiyun, A.A.; Keddy, K.H.; Madoroba, E.; Manqele, A.; Thompson, P.N. Shiga toxin–producing Escherichia coli contamination of raw beef and beef-based ready-to-eat products at retail outlets in Pretoria, South Africa. J. Food Protect. 2020, 83, 476–484. [Google Scholar] [CrossRef]
- Onyeka, L.O.; Adesiyun, A.A.; Keddy, K.H.; Manqele, A.; Madoroba, E.; Thompson, P.N. Prevalence, risk factors and molecular characteristics of Shiga toxin-producing Escherichia coli in beef abattoirs in Gauteng, South Africa. Food Control. 2021, 123, 107746. [Google Scholar] [CrossRef]
- Onyeka, L.O.; Adesiyun, A.A.; Keddy, K.H.; Manqele, A.; Madoroba, E.; Thompson, P.N. Prevalence and patterns of fecal shedding of Shiga toxin–producing Escherichia coli by cattle at a commercial feedlot in South Africa. J. Food Safety. 2022, 42, e12961. [Google Scholar] [CrossRef]
- WOAH-OIE List of Antimicrobial Agents of Veterinary Importance 2021. Available online: https://www.woah.org/app/uploads/2021/06/a-oie-list-antimicrobials-june2021.pdf (accessed on 15 August 2024).
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updates. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Paton, J.C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Pouseele, H.; Chen, J.C.; Strockbine, N.A.; Carleton, H.A. Implementation of whole genome sequencing (WGS) for identification and characterization of Shiga toxin-producing Escherichia coli (STEC) in the United States. Front. Microbiol. 2016, 7, 766. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome biology. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O: H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar] [CrossRef]
- Bessonov, K.; Laing, C.; Robertson, J.; Yong, I.; Ziebell, K.; Gannon, V.P.; Nichani, A. ECTyper: In silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom. 2021, 7, 000728. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Joris, M.A.; Verstraete, K.; Reu, K.D.; Zutter, L.D. Loss of vtx genes after the first subcultivation step of verocytotoxigenic Escherichia coli O157 and non-O157 during isolation from naturally contaminated fecal samples. Toxins 2011, 3, 672–677. [Google Scholar] [CrossRef]
- Mellmann, A.; Lu, S.; Karch, H.; Xu, J.G.; Harmsen, D.; Schmidt, M.A.; Bielaszewska, M. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157: NM. Appl. Environ. Microbiol. 2008, 74, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Senthakumaran, T.; Brandal, L.T.; Lindstedt, B.A.; Jørgensen, S.B.; Charnock, C.; Tunsjø, H.S. Implications of stx loss for clinical diagnostics of Shiga toxin-producing Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2361–2370. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Prager, R.; Kock, R.; Mellmann, A.; Zhang, W.; Tschäpe, H.; Tarr, P.I.; Karch, H. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 2007, 73, 3144–3150. [Google Scholar] [CrossRef] [PubMed]
- Amézquita-López, B.A.; Quiñones, B.; Lee, B.G.; Chaidez, C. Virulence profiling of Shiga toxin-producing Escherichia coli recovered from domestic farm animals in Northwestern Mexico. Front. Cell Infect. Microbiol. 2014, 4, 7. [Google Scholar] [CrossRef]
- Coombes, B.K.; Wickham, M.E.; Mascarenhas, M.; Gruenheid, S.; Finlay, B.B.; Karmali, M.A. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 2008, 74, 2153–2160. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Fratamico, P.M.; Bono, J.L.; Baranzoni, G.M.; Chen, C.Y. Genome sequencing and comparative genomics provides insights on the evolutionary dynamics and pathogenic potential of different H-serotypes of Shiga toxin-producing Escherichia coli O104. BMC Microbiol. 2015, 15, 83. [Google Scholar] [CrossRef]
- Friesema, I.H.; Keijzer-Veen, M.G.; Koppejan, M.; Schipper, H.S.; van Griethuysen, A.J.; Heck, M.E.; van Pelt, W. Hemolytic uremic syndrome associated with Escherichia coli O8: H19 and Shiga toxin 2f gene. Emerg. Infect. Dis. 2015, 21, 168. [Google Scholar] [CrossRef]
- Fan, R.; Shao, K.; Yang, X.; Bai, X.; Fu, S.; Sun, H.; Xu, Y.; Wang, H.; Li, Q.; Hu, B.; et al. High prevalence of non-O157 Shiga toxin-producing Escherichia coli in beef cattle detected by combining four selective agars. BMC Microbiol. 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Karama, M.; Cenci-Goga, B.T.; Malahlela, M.; Smith, A.M.; Keddy, K.H.; El-Ashram, S.; Kabiru, L.M.; Kalake, A. Virulence characteristics and antimicrobial resistance profiles of shiga toxin-producing Escherichia coli isolates from humans in South Africa: 2006–2013. Toxins 2019, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.A.; Kudva, I.T. Antibiotic-resistant Shiga toxin-producing Escherichia coli: An overview of prevalence and intervention strategies. Zoonoses Public. Health. 2019, 66, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Iwu, C.J.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015, 15, 213. [Google Scholar] [CrossRef]
- Piddock, L.J. Multidrug-resistance efflux pumps? not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Nikaido, H. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 1. [Google Scholar]
- Nishino, K.; Yamasaki, S.; Nakashima, R.; Zwama, M.; Hayashi-Nishino, M. Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 2021, 12, 737288. [Google Scholar] [CrossRef]
- WHO Ten Threaths to Global Health in 2019. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 15 August 2024).
- Grace, D.; Lindahl, J.F.; Nguyen-Viet, H.; Kakkar, M. Antimicrobial use in developing countries. In Proceedings of the World Veterinary Association (WVA)/World Medical Association (WMA) Global Conference on One Health, Madrid, Spain, 21–22 May 2015; ILRI:: Nairobi, Kenya, 2015. Available online: https://cgspace.cgiar.org/handle/10568/67030 (accessed on 8 August 2024).
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sciences. 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.C.; Stanford, K.; McAllister, T.A.; Thomas, J.; Reuter, T. Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle faeces. J. Microbiol. Methods 2014, 105, 22–30. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).