Abstract
Despite the advancement in antiviral therapy, Hepatitis C remains a global health challenge and one of the leading causes of hepatitis related deaths worldwide. Hepatitis C virus, the causative agent, is a positive strand RNA virus that requires a liver specific microRNA called miR-122 for its replication. Unconventional to the canonical role of miRNAs in translation suppression by binding to 3′Untranslated Region (UTR) of messenger RNAs, miR-122 binds to two sites on the 5′UTR of viral genome and promotes viral propagation. In this review, we describe the unique relationship between the liver specific microRNA and HCV, the current knowledge on the mechanisms by which the virus uses miR-122 to promote the virus life cycle, and how miR-122 impacts viral tropism and pathogenesis. We will also discuss the use of anti-miR-122 therapy and its impact on viral evolution of miR-122-independent replication. This review further provides insight into how viruses manipulate host factors at the initial stage of infection to establish a successful infection.
1. Introduction
Hepatitis C Virus (HCV) is a blood borne pathogen and one of the leading causes of chronic liver disease worldwide. It has a global prevalence of 1%, corresponding to 71.1 million infections [1], and according to a report by World Health Organization (WHO), in 2016, approximately 1.34 million deaths were reported globally due to Hepatitis virus induced hepatic disorders [2]. HCV displays sequence variability leading to 7 different genotypes and an enormous list of subtypes [3]. Acute Hepatitis C infections are most often asymptomatic and 15–25% of the time are spontaneously resolved by the immune system [4]. However, most infections (75–85%) lead to chronic HCV, which in 15–25% of patients develops into liver cirrhosis, and in 1–5% and 2–5% into hepatocellular carcinoma (HCC) and end stage liver disease (ESLD), respectively [5,6]. However, unlike many chronic infections HCV can be cured. Until 2015, HCV treatments included a combination of pegylated interferons (pegIFN) alpha and Ribavirin (RBV) and induced a sustained virological response (SVR); the absence of detectable HCV RNA on blood testing 6 months after the completion of antiviral therapy, in 50–80% of patients depending on the virus genotype [7,8]. However, in addition to its poor efficacy, this therapy was expensive and induced severe adverse effects, including flu-like symptoms, hemolytic anemia, and psychiatric disturbances [9,10]. In 2015, the approval of several combinations of direct-acting antivirals (DAA) revolutionized HCV treatment by providing patients with oral therapy that was 95% effective at inducing SVR after 12–14 weeks of treatment with minimal adverse effect [11,12,13,14,15]. DAA therapy is being used to eliminate HCV in certain populations and regions, but an effective vaccine will be needed to control HCV on a global scale [15].
HCV is an enveloped virus with a positive sense RNA genome of ~9.6 kb in length. HCV infections start with virion entry, which is facilitated by attachment of the viral particles to several cell surface receptors and co-receptors followed by endocytosis [16]. The genome encodes a single polyprotein flanked by 5′ and 3′ untranslated regions (UTRs). The 5′UTR contains an Internal Ribosome Entry Sequence (IRES) that regulates translation of the viral polyprotein, a 3000 amino acid polypeptide that is proteolytically processed by host and viral proteases into 10 individual viral proteins; seven nonstructural proteins (p7, N2, NS3, NS4A, NS4B, NS5A, and NS5B) that facilitate polyprotein processing, viral RNA replication, and viral particle assembly, and three structural proteins (core, E1, and E2) that make up the virion particle [16]. The RNA genome replicates within replication complexes formed in association with infection-modified intracellular membranes via synthesis of a negative strand RNA genome intermediate [17]. A unique character of the HCV life-cycle is its reliance on a liver-specific microRNA, miR-122. miR-122 anneals to two sites on the 5′UTR upstream of the IRES and is required for viral RNA accumulation in infected cells [18]. The mechanism by which miR-122 promotes HCV propagation is not fully understood, but counter to the conventional role of microRNAs, which suppress translation and promote mRNA degradation, miR-122 promotes translation and stabilizes the viral genomic RNA and has also been reported to alter the viral RNA structures. It is speculated that HCV evolved to be dependent on miR-122 to limit virus replication to the liver [19], and miR-122 may also affect HCV pathogenesis with the viral genome acting as a miR-122 sponge affecting the normal cellular functions of miR-122 [20]. In this review, we will discuss how miR-122 promotes the HCV life cycle, how the unconventional relationship of miR-122 can be exploited to treat HCV infections, how the virus can escape the need for miR-122, and what we can learn from mutant viruses capable of miR-122-independent HCV replication.
2. HCV and miR-122
MicroRNAs (miRNA) are small non-coding RNAs of approximately 20–23 nucleotides that mediate post-transcriptional gene silencing by promoting mRNA degradation and repressing translation [21,22]. miR-122 is a liver-specific microRNA abundantly expressed in hepatocytes [approximately 660,000 copies per cell] which accounts for almost 72% of total miRNA pool in the liver, making it one of the most highly expressed miRNAs in any tissue [23]. The primary miR-122 (Pri-miR-122) is transcribed by RNA polymerase II from a single genomic locus on chromosome 18 [24] and like other miRNAs are processed by Drosha and Dicer into a mature miRNA that targets cellular mRNAs for translation suppression and degradation. The downstream functions of miR-122 are diverse and includes regulation of liver development, lipid and cholesterol metabolism, iron metabolism, and circadian rhythms [24,25,26,27,28,29]. miR-122 expression is typically observed to be lost in hepatocellular carcinoma (HCC), and miR-122 knockout mice developed persistent hepatosteatosis, fibrosis, and hepatocellular carcinoma (HCC), suggesting that miR-122 also has an important role as a tumor suppressor [30,31]. The liver specific expression pattern and sequence of mature miR-122 is highly conserved across the vertebrate lineage, suggesting a co-evolution of this microRNA with the emergence of the liver [32].
2.1. Annealing of miR-122
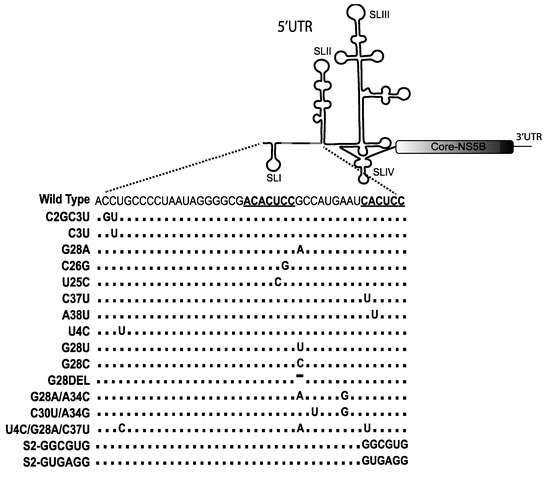
Unlike the canonical suppressive role of miRNAs, miR-122 promotes HCV replication and is essential for detectible HCV replication in cell culture [18,33,34,35,36,37]. miR-122 anneals directly to two homologous sites near the 5′ terminus of the HCV genome (Figure 1) [18,34,38,39]. The first miR-122 site (S1) includes a seed annealing site located adjacent to stem-loop I and auxiliary binding nucleotides at the extreme 5′ end of the viral genome such that miR-122 annealing generates a 3′ overhang on the viral genome 5′ terminus (Figure 1). The second miR-122 seed binding site (S2) is located at the base of stem-loop II and includes auxiliary binding near the S1 (Figure 1). Both seed and auxiliary annealing at both sites is required for efficient HCV replication [18,34,35]. Studies by Mortimier and Doudna, and Pang et al. suggested that miR-122 binds to site 2 with a higher affinity than site 1 [40,41], but there are contradicting reports of the importance of each binding site for replication promotion, with some studies suggested that site 1 binding is more important [33,42], and another study by Thibault et al. suggests an equal contribution of each site on viral replication [43]. In addition, a recent study showed that the specific binding pattern exhibited by miR-122 is not required for the mechanism by which miR-122 promotes replication since annealing of a single small RNA to nucleotides 23–35 promoted HCV replication as efficiently as annealing of two copies of miR-122 [44]. Since nucleotides 23–35 are also bound by miR-122 when annealed to both S1 and S2, this suggests that annealing to this region may be central to the mechanism of viral propagation by miR-122.
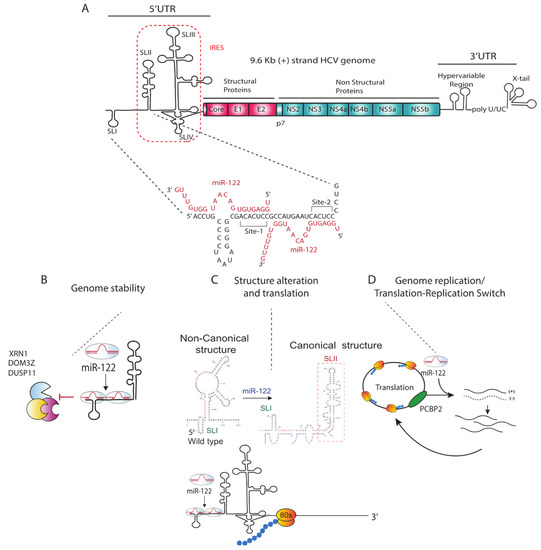
Figure 1.
The miR-122 annealing pattern and hypothetical mechanistic models of miR-122 promotion of HCV propagation. (A) HCV positive strand genomic RNA with miR-122 annealing pattern. miR-122 binds to seed and auxiliary binding sites on both binding site 1 (S1) and site 2 (S2). (B) miR-122 protects RNA genome against RNA degradation machinery. (C) miR-122 binding alters 5′UTR structure to a translation favourable structure (D) miR-122 acts as a switch between viral replication and translation.
In addition to the two seed binding sites on the 5′UTR, in silico analyses have suggested multiple alternative miR-122 binding sites on the viral genome, four in the NS5B coding region, and three (one very highly conserved) in the 3′UTR [44,45,46,47]. However, neither annealing of alternative small RNAs to these sites nor mutational analysis showed an impact on virus propagation, so the relevance of miR-122 annealing to these sites is still unknown [44,45].
2.2. miR-122-Protein Complexes and HCV Propagation
In addition to miR-122, HCV propagation is also dependent on host cellular proteins involved in miRNA biogenesis and translation suppression. Biogenesis of miRNAs is mediated by host proteins that include Drosha, which processes long pri-mRNAs into pre-miRNA hairpins, and Dicer that process the hairpins into mature miRNAs; so, unsurprisingly, Drosha and Dicer are also required for HCV replication, presumably to provide a supply of miR-122. In addition, several host proteins involved in miRNA translation suppression activity, including proteins that comprise the RNA Induced Silencing Complex, (RISC), such as Argonautes (Ago1, 2, 3, and 4), GW182, DiGeorge syndrome critical region 8 (DGCR8), and TAR RNA binding protein (TRBP) have also been shown to be involved in HCV replication promotion by miR-122 [36,37,48,49], suggesting a RISC-like protein complex is required for delivery of the miR-122 and perhaps the mechanism of replication promotion. The role of Ago2 has been studied in more detail and the Ago2:miR-122 complex binds to two sites (S1 and S2) on viral 5′UTR and is required for miR-122 promotion of virus replication [37,50,51,52]. Cells having a knockout of the Ago2 gene still supported viral replication, although to a lower level compared to the wild type cells, suggesting that other Ago isoforms (Ago 1, 3, and 4) are also capable of mediating miR-122-induced HCV propagation [51]. It has also been proposed by several groups that the Ago:miR-122 complex and not miR-122 alone likely promotes HCV propagation [50,51,53,54].
2.3. Other microRNAs Promoting HCV Replication
Recent findings by the Matsuura group suggest that other cellular miRNAs can bind to the viral 5′UTR and support viral replication in a miR-122-like and a non-miR-122-like manner [55]. The miRNAs identified to bind to viral 5′UTR at both miR-122 binding locations are miR-504-3p, miR-574-5p, and miR-1236-5p, and replication promotion requires at least six complementary nucleotides annealing with the viral RNA. miR-25-5p and miR-4730 were identified to bind to 7–8 nucleotides in a single site that bridge between sites 1 and 2 and promote viral replication. Altogether, these findings suggest that HCV can also usurp miRNAs other than miR-122 which could allow HCV to replicate in tissues other than the liver. Finally, at least four other miRNAs (miR-199a*, miR-196, let-7b, and miR-448) have been confirmed to bind to HCV genome, but these do not promote viral propagation [56]. These microRNAs function in a canonical manner to suppress viral translation, and let-7b can lead to cleavage and degradation of the viral genome [57]. Although these microRNAs are reported to inhibit viral propagation, their net negative effect appears to be superseded by promotion by miR-122 since knockdown of the microRNA pathway proteins leads to an overall reduction in viral propagation [19,37,50,52,56,58]
3. Mechanism of miR-122 Promotion of HCV Replication
Numerous studies have aimed to understand the mechanism by which miR-122 promotes virus replication and 3 primary mechanisms are proposed: (1) Protection of the viral genome from degradation, (2) Altering the structure of the 5′UTR to activate viral IRES translation, and (3) A direct role in virus replication (Figure 1).
3.1. miR-122 Protection of the Viral Genome from Degradation
The presence of a 7-methylguanylate cap at the 5′ end of eukaryotic mRNAs promotes translation, protects the 5′ end from cellular exonuclease degradation, and masks viral genomic RNA detection by cellular innate immune sensors. Viral RNAs obtain caps by various methods, including viral encoded capping enzymes (e.g., flavivirus and coronavirus) or by usurping caps from cellular mRNA (e.g., Influenza virus). However, the HCV genome, which is the template for both translation and replication, has an uncapped 5′ triphosphate end [50]. It was speculated that miR-122 annealing at the extreme 5′ end of the viral genome might act as an artificial cap to protect the triphosphate 5′ end of viral RNA from the cellular RNA degradation machinery or from detection by innate immune sensors [35,40,43,51,59,60,61,62]. While there is no evidence thus far that miR-122 protects against recognition by the innate immune sensors [62], studies by Shimakami et al. reported that transfection of miR-122 slowed the decay of a replication-defective viral RNAs, suggesting a role in protecting the viral genome from degradation [42,50]. Additionally, knockdown of exonucleases Xrn1 and Xrn2, which degrade RNA in a 5′ to 3′ direction, partially rescued HCV replication and further supported that miR-122 protects the viral genome from Xrn1 and, to a lesser extent, Xrn2 mediated degradation [43,60,61]. In a recent study, it was also observed that miR-122 protects the genome from cellular pyrophosphatases DOM3Z and DUSP11 [62,63]. DOM3Z is a cellular decapping and exoribonuclease protein with pyrophosphohydrolase and 5′-3′ exonuclease activity required for decapping and degradation of mRNA [64], and DUSP11 is another cellular 5’ pyrophosphatase required for regulating the cellular level of RNA transcripts from RNA pol I and III by hydrolyzing the di-and tri-phosphate from the 5′ end of the RNA [63]. DUSP11 has also been reported to directly act on the 5′ end of HCV RNA rendering it susceptible to Xrn1-mediated degradation [63]. Further, depletion of all three enzymes, Xrn1, DOM3Z, and DUSP11, rescued HCV replication in the absence of miR-122 [62], however, knockdown of Xrn1, DOM3Z, and DUSP11 did not reinstate HCV replication to an miR-122 dependent level and suggests that miR-122 mediated protection of viral RNA is not the only mechanism by which miR-122 promotes viral propagation [62,65].
3.2. Viral Translation Stimulation by miR-122-Induced Alteration of Genomic RNA and IRES Structure
MiR-122 annealing has been reported by several groups to stimulate HCV translation [33,37,52,53,58,66,67]. However, because of the difference between the magnitude of miR-122 induced viral translation (2 to 3 fold) and miR-122 enhancement of viral replication (1000 fold), the relevance of the impact of miR-122 induced viral translation on promotion of viral replication has been questioned. However, recent evidence suggests that stimulation of translation is a primary mechanism by which miR-122 promotes the HCV life-cycle. In a study by Kunden et al., small RNAs annealed to different locations on the 5′UTR were found to promote the HCV life-cycle with various efficiencies, and the efficiency of replication promotion having similar trends as their ability to stimulate transition suggesting the two functions are linked. In addition, miR-122 annealing has been hypothesized to alter the secondary structure of the 5′UTR and promote the formation of the thermodynamically stable canonical IRES structure [51,53,54]. In silico structure prediction and SHAPE analysis of 1–117 nt of viral 5′UTR in the absence of miR-122 predicts that it preferentially forms a non-canonical structure termed SLIIalt that would not support HCV IRES translation, and that annealing of miR-122 shifts the folding equilibrium toward formation of SLII, an RNA structure essential for IRES mediated translation [51,53,54,68]. Biophysical analyses of HCV IRES RNA structure and function indicated that SLII is essential for HCV IRES activation by inducing a conformational change in 40S ribosome and an interaction with the HCV IRES SLIV domain that facilitates the formation of the final 80S ribosomal complex that initiates translation, however, these biophysical analyses were done using HCV RNAs that lacked the 5′ region to which miR-122 binds, thus detailed biophysical analyses of the impact of miR-122 on the IRES structure remain to be confirmed. (Figure 1).
A recent report proposed a dynamic model for the mechanism of Ago:miR-122 promotion of HCV that involved sequential annealing to the two sites on the 5′UTR. The Ago:miR-122 complex was modeled to first bind to site 2, and it serves as an RNA chaperone to re-fold the RNA into the functional SLII conformation of the active IRES, which is then followed by the subsequent binding of a second Ago:miR-122 complex to site 1, promoting viral genome protection by cellular endonucleases and pyrophosphatases. Binding of Ago2 at site 1 was also modeled to stabilize the complex through interactions between Ago and SLII and then release the Ago:miR-122 auxiliary interactions at Site 2 [68] (Figure 1). However, a recent study by Kunden et al. showed that annealing of a single Ago2:small RNA complex at nucleotide position 27–45, a region that spans the annealing site 1 seed and the site 2 auxiliary, both stabilize the genome and promote virus replication to levels equivalent or more than that of miR-122. That a single small RNA:Ago complex can promote genome stabilization, virus translation, and the virus life cycle suggests that the complex annealing pattern, 5′ end annealing, and the dynamics of Ago2:miR-122 binding is not essential for the mechanism of miR-122 promotion of HCV [44].
3.3. A Direct Role for miR-122 in Promoting Genome Amplification
It has been hypothesized for many years that miR-122 has a direct role in HCV genome replication or in regulating the transition from genome translation to genome replication. A study by Masaki et al. showed that miR-122 induced an increase in viral replication that preceded an increase in viral protein synthesis, suggesting a direct role for miR-122 in promoting viral genomic RNA replication that did not rely on increased viral protein synthesis [69]. The study also found that miR-122-stimulated viral replication ceased following siRNA depletion of host poly[rC] binding protein 2 [PCBP2]. PCBP2 interacts with the HCV 5′UTR to promote genome circularization and genome replication [70], and Masaki et al. also showed that miR-122 displaced PCBP2 from HCV RNA in cell-free pull-down experiments and thus speculated that miR-122 binding might enhance the replicating pool of viral RNA by displacing PCBP2 [69]. Additionally, consistent with this notion was reduced polysome association of HCV RNA upon miR-122 transfection, suggesting a role as a molecular switch, activating replication of viral genomes [69,71]. Alternatively, miR-122 binding to the HCV RNA may promote genome synthesis and activate replication by displacing the positive strand from the newly synthesized negative strand replication intermediate to increase accessibility of the 3′ end for initiation of positive strand genomes. Thus, miR-122 is hypothesized to play a role at early stages of the virus life cycle, through enhancement in viral translation and genome stability but may also have a direct role in inducing genome replication.
4. Clinical Significance of miR-122 in HCV Infections
4.1. miR-122, HCV Liver Tropism and Pathogenesis
HCV has evolved to infect the liver and is a major factor affecting HCV′s liver-specificality. That HCV usurps miR-122 may also contribute to liver induced pathogenesis by acting as a miR-122 sponge and de-repressing cellular mRNAs targeted by miR-122 [20]. Genome-wide high-throughput sequencing of Ago2 crosslinked and immunoprecipitation (HITS-CLIP) analysis in HCV infected and naïve cells revealed a significant reduction in global AGO binding of miR-122 targeted mRNA and de-repression of miR-122-targeted mRNAs. Further, microarray data from liver biopsies of HCV-infected and naive patients showed a significant de-repression of similar miR-122 target mRNAs and confirmed the HCV sponge effect in vivo. The sponge effect by the viral RNA on miR-122 affected cell proliferation and survival, collagen production, and hepatic stellate cell activation, which could influence liver inflammation [20]. Since miR-122 is a known tumor suppressor, miR-122 sequestration during an HCV infection may also contribute to HCC development in chronic infection [30,31,72,73].
4.2. miR-122 as a Biomarker for Chronic Hepatitis C Infection and Liver Diseases
Circulating microRNAs in human peripheral blood have been increasingly regarded as potential indicators of a variety of physiological and pathological conditions, including liver injury induced by hepatotoxic agents and viral hepatitis [74,75,76]. Circulating microRNAs are secreted in different body fluids through exosomes and macrovesicles, making them potential and relatively non-invasive biomarkers for the detection of different stages of a disease progression [77]. Circulating miR-122 has been studied as promising circulating biomarkers for liver disease conditions because its expression is dysregulated in liver diseases. Since microRNAs are stable in human plasma/serum and miR-122 being a tissue-specific miRNA, its abnormal presence in the serum can indicate liver injuries [20,30,75,78,79]. Assessing circulating miR-122 levels has shown promise in diagnosing liver pathogenesis in various diseases such HBV, HIV, and/or HCV-associated chronic viral hepatitis, and as a biomarker for chronic liver infection [75,80,81].
5. Other Viruses That Rely on miR-122 or Other miRNAs
5.1. Non-Primate Hepacivirus and miR-122: Similarities, Differences, and Evolution
In addition to HCV, genomes of other viruses in the genus hepacivirus likely anneal to miR-122. Recent studies have identified several novel hepaciviruses including equine hepacivirus, or non-primate hepacivirus (NPHV), Norway rat hepacivirus (NrHV) or rodent hepacivirus-nr-1 (RHV-nr-1), bat hepaciviruses; bovine hepaciviruses (BovHepV), and Guerza hepacivirus, and they all exhibit liver tropism and have at least one miR-122 binding site on their genome [74,82,83,84,85,86,87,88]. Studies have confirmed miR-122 stimulation of translation in NPHV and BovHepV, and miR-122 sequestration inhibits NPHV accumulation in cell culture [74,89]. GB virus B (GBV-B), a relative of HCV isolated from laboratory tamarins, also has two miR-122 binding sites on its 5′UTR RNA and is dependent on miR-122 and Ago2 abundance for viral propagation [90]. The presence of the miR-122 binding site in hepaciviruses of different species suggests evolution from a common liver-tropic ancestor [19] and a conserved mechanism of action. Altogether, it can be proposed that throughout host–virus coevolution, hepaciviruses may have used miR-122 as a common strategy to establish liver tropism.
5.2. Let-7 and MiR-17 Promotion of Bovine Viral Diarrhea Virus
The dependency of viruses on miRNAs is rare but is shared by one other virus from the Flaviviridae family. A study by Scheel et al. reported that Bovine viral diarrhea virus (BVDV) an animal pathogen from pestivirus genus, also requires microRNAs let-7 and miR-17 for their propagation [91,92]. However, unlike miR-122 and HCV, these microRNAs bind to the 3′UTR of the BVDV genome to promote translation and replication. The discovery of other viruses dependent on host microRNA provides another example of the unusual relationship between the host and virus, which might provide insight into common mechanisms of action and a broader picture of host-pathogen co-evolution.
7. Mechanism of miR-122-Independent Replication
7.1. Replication Promotion by Other microRNAs
Cell culture models of miR-122-independent HCV replication have been used to study mechanisms by which the virus escapes the need for miR-122. One possibility is that HCV variants reported to replicate in the absence of miR-122 use other cellular miRNAs, such as those identified by Ono et al. to anneal to the 5′UTR [55]. However, several studies on miR-122-independent replication also showed virus replication in Dicer and Drosha knockout cells, which are devoid of miRNAs that rely on the canonical miR-122 biogenesis pathways. HCV-S2-GGCGUG which has been reported previously for replicating in the absence of miR-122 was capable of replicating in Dicer KO Huh 7.5 cells, and several mutants were shown to replicate in Drosha knockout cells [19,65]. Additionally, another study showed that Ago2 interaction with HCV genome was not detected for miR-122-independent replication of G28A, suggesting that miR-122 independent replication of HCV is not because of the binding of other microRNAs to the 5′UTR of the viral RNA and no other microRNA compensate for miR-122-independent replication of G28A HCV [71]. However, these studies do not remove the possibility of the involvement of small cellular RNAs that do not rely on Dicer or Drosha or other Ago isoforms.
7.2. 5′UTR Mutations Stabilize the Viral Genome
A method by which miR-122 is reported to promote HCV replication is through stabilization of the viral genome (Figure 3). Based on this, Chahal et al. suggested that mutations to the auxiliary region of miR-122 binding site 1 (C2G, C3U, U4C) located near the genomic 5′ terminus protect the genome from degradation, and this notion was supported by in vitro stability assays showing that the mutant RNAs were resistant to degradation by Xrn1 [68]. However, small RNAs that anneal to nucleotides 19–37 and not to the 5′ terminus also stabilized the genome and promoted HCV replication, indicating that miR-122 5′ end annealing is not essential for genome stabilization [44].
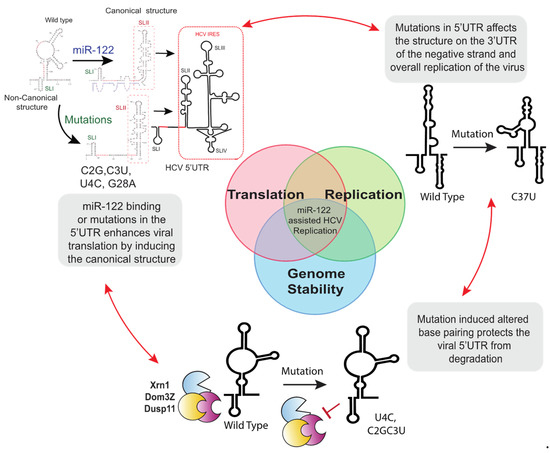
Figure 3.
A model for the mechanism of miR-122 independent replication. miR-122-independent replication is an interplay between genome translation, replication, and stability. See text for details.
7.3. 5′UTR Mutations Modulate the 5′UTR RNA Structure and Stimulate Virus Translation
Another proposed mechanism of action of miR-122 is as a chaperon that alters viral 5′UTR RNA structures, refolding SLII into the canonical structure of an active viral IRES (Figure 3) [51,53,54]. Computational and SHAPE analysis of the 5′UTR viral RNA of mutants replicating independently of miR-122 predicts that their 5′UTRs have a higher propensity to form a structure similar to the canonical SLII structure formed by wild type virus after miR-122 annealing. Thus, it was proposed that 5′ RNA structure may be dynamic and that mutations that allow for miR-122-independent replication shift the folding equilibrium toward that of the active IRES even in the absence of miR-122 [51]. In support of this, there was a correlation between the mutation induced enhancement of translation and the efficiency of miR-122-independent replication for the HCV mutants capable of miR-122-independent replication [53,65]. Further, knocking down of RNA degrading enzymes rescued replication of several mutants, some to miR-122-dependent levels, suggesting that the roles of miR-122 can be compensated by enhancing virus translation and stabilizing the viral genome. Experiments also demonstrated that enhanced translation rescued viral replication by approximately 100-fold, whereas enhanced stability rescued viral replication approximately 10-fold, and suggested that the major role for miR-122 is translation stimulation with stabilization being important but less potent [65].
7.4. Removal of a Negative Regulator of Viral Propagation
It is also possible that the HCV 5′ terminal region functions as a switchable regulatory element whose primary function is to make the virus reliant on miR-122 (Figure 4). This hypothesis is based on miR-122 regulation of a related hapacivirus, GB virus B (GBV-B). GBV-B also requires miR-122 for efficient viral RNA propagation but was found to be capable of replication independently of miR-122 after simply removing the miR-122 binding region [90]. The deletion mutant lacked much of the 5′UTR, including both miR-122 binding sites (∆4-29), and it was first isolated by growing the GBV-B replicons in Hep3B cells, human hepatoma cells that lack miR-122 expression (Figure 4) [90,116]. A similar deletion of the miR-122 binding region in HCV was not tolerated (unpublished data), presumably because the complementary sequence forms RNA structures on the 3′UTR of the negative strange that are essential for genome replication [117]. However, that GBV-B can tolerate removal of the miR-122 binding sites, suggesting this region is dispensable and that its inclusion makes virus replication rely on miR-122, perhaps to regulate virus tropism.
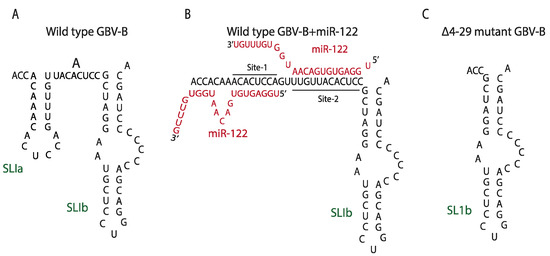
Figure 4.
Predicted structures of 5′UTR of HCV and GBV-B with or without miR-122. (A) Wild type GBV-B without miR-122 (1–62 nt), (B) Wild type GBV-B with miR-122 (1–62 nt), (C) ∆4-29 mutant GBV-B without miR-122.
8. Dynamics of miR-122-Independent Replication and miR-122’s Impact on Different Stages of the Virus Lifecycle
Models of miR-122-independent replication have allowed researchers to study how the HCV replication cycle differs when miR-122 is present or absent. Our recent publication showed that miR-122-independent replication in cultured cells is manifested as efficient replication in a smaller number of cells than when miR-122 is present; but, it also showed that once virus replication is established the level of virus protein expression in each infected cell is similar whether miR-122 is present or absent [102]. This suggests that that miR-122 is required to establish an infection but is dispensable after an infection has been established. This notion is also supported by a study showing efficient miR-122-independent replication of HCV replicons [71]. In support of miR-122 being dispensable for an ongoing infection, no detectable miR-122 has been observed within the HCV replicase complex in vivo and the addition of miR-122 or miR-122-antisense inhibitors has no effect on HCV RNA synthesis within membrane-bound replicase complexes isolated ex vivo from HCV infected cells and [118]. However, miR-122 supplementation and antagonism showed a small positive influence on ongoing viral replication when measured by luciferase expression. This suggested a minor supportive role in an ongoing infection. This could be a direct role of miR-122 on genome replication as discussed previously, but we also hypothesized that miR-122 anneals to newly synthesized HCV genomes that exit replication complexes to stabilize and promote translation to augment their ability to establish new replication complexes within an infected cell or after division of an infected cell [102]. Together, this study proposed a model in which miR-122 is essential at the initial stage of infection to allow the virus to reach a protein synthesis threshold required to establish replication complexes and an infection and then may use the same mechanism to assist in the formation of new replication complexes. However, how and when miR-122 dissociates from the viral genome in this model is unknown.
Model for the Role of miR-122 in the HCV Life-Cycle: A Host Factor That Allows HCV to Overcome Infection Barriers
Establishment of a successful viral infection is determined by the outcome of competition between viral translation/replication kinetics and host antiviral response. Host cells defend against viral infection using strategies that include translation inhibition and viral genome degradation to create a barrier to the initiation of an infection. In turn, viruses have evolved many mechanisms to block the activation of the antiviral pathways to maintain virus translation and genome stability [119,120]. However, a recent publication suggests that the dynamics of virus translation and replication can also overcome the antiviral barrier. For example, to establish a picornavirus infection requires that the virus produce sufficient protein to successfully switch from genome translation to genome replication, but in 15–20% of picornavirus infected cells the incoming viral RNA fails to translate sufficiently to initiate the switch, likely due to antiviral conditions in the cell. However, the genomes that fail to switch to replication will try again, reinitiating translation to further attempt to establish an infection [121]. Similarly, we propose that miR-122 allows HCV to overcome a barrier to infection, specifically RNA degradation by host enzymes and poor translation efficiency. In the absence of miR-122, translation efficiency is poor and the genomic RNA is subject to degradation; this barrier blocks the establishment of an infection. In the presence of miR-122, the RNA is stabilized, translation is efficient, and an infection is established in most cells. However, with mutant HCV genomes having enhanced genome stability or translation efficiency without miR-122, the barrier is overcome in a small number of cells stochastically and an infection is established. Similarly, knockdown of RNA degradation enzymes also reduces the barrier and enhances HCV infection success in the absence of miR-122, in some cases to levels similar to that with miR-122 (Figure 5) [44,53,62,65].
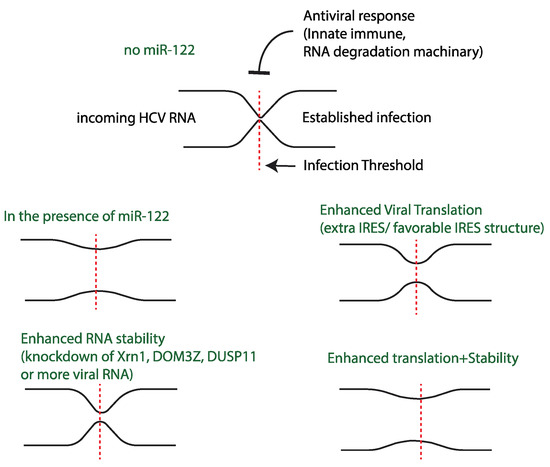
Figure 5.
Model for miR-122 promotion of HCV: Schematic representation of infection bottleneck of HCV in the presence of different infection limiting variables.
Recent studies have revealed that barriers to infection can also be surmounted by infection of a cell by multiple virions since en bloc transmission of viruses within extracellular vesicles containing multiple viral particles containing multiple viral genomes are more infectious than individual free virions [122,123]. The average number of HCV genomic RNA copies is 1 to 8 per cell in vivo [124]; 100 fold lower RNA levels than tissue culture cells expressing HCV replicons or infectious virus [125]. Since a very low copy number of viral genomes are found in hepatocytes, the presence of miR-122 may be even more important for infection initiation in vivo. Thus, we suggest that miR-122 regulates the dynamics of infection initiation and the establishment of a successful infection.
While our data and model suggest that miR-122 does not have a major role following the initiation of an infection, there is a small positive impact during an ongoing infection. We hypothesize that this is due to the stabilization and translation stimulation of newly synthesized genomes as they initiate new replication complexes within a cell, or following division of an infected cell. In infected patients, chronic HCV infections have been suggested to be maintained by a continuous cycle of viral infection and clearance in a small subpopulation of hepatocytes in the liver [124], and we propose that miR-122 supports the dynamics of this cycle and explains why miR-122 antagonists have a potent effect on chronic infections.
9. Conclusions and Future Directions
The liver specific microRNA, miR-122, is essential for efficient HCV replication. It stabilizes the viral genomic RNA and stimulates translation by inducing viral genomic RNA structure modifications; miR-122 limits HCV tropism to the liver where it allows the virus to overcome barriers to infection, and it supports the dynamic infection of hepatocytes during a chronic liver infection. However, gaps in knowledge remain. Biophysical details of the RNA structures formed with and without miR-122 are still lacking, as are the details regarding the viral and host RISC and RNA binding proteins involved. A direct role for miR-122 in regulating the switch between virus translation and replication has been proposed for many years and while there appears an indirect influence on genome replication through enhanced translation and genome stabilization, whether there is a direct impact on the switch to genome replication is still unresolved. Further, details of the impact of miR-122 on the replication and tropism of other hepacivirus that also anneal miR-122 will allow a better understanding of how and why they evolved to be regulated by miR-122, and understanding the mechanisms of HCV genome structure–function will provide insight into the RNA genome structure and function of other viruses.
Author Contributions
Conceptualization, M.P., M.A.P. and J.A.W.; article preparation and editing, M.P., M.A.P. and J.A.W.; review of final manuscript, M.P., M.A.P. and J.A.W.; supervision, J.A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) [MOP-133458], Natural Science and Research council of Canada (NSERC) [RGPIN-2018-06335} and the University of Saskatchewan (CoMBRIDGE) to J.A.W. M.P. was funded by a University of Saskatchewan Graduate Teaching Fellowship. M.A.P. is funded by a CanHepC Ph.D. Scholarship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Blach, S.; Zeuzem, S.; Manns, M.; Altraif, I.; Duberg, A.-S.; Muljono, D.H.; Waked, I.; Alavian, S.M.; Lee, M.-H.; Negro, F.; et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: A modelling study. Lancet Gastroenterol. Hepatol. 2017, 2, 161–176. [Google Scholar]
- World Health Organization; Global Hepatitis Programme. Global Hepatitis Report. 2017. Available online: http://apps.who.int/iris/bitstream/10665/255016/1/9789241565455-eng.pdf?ua=1 (accessed on 6 July 2022).
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2013, 59, 318–327. [Google Scholar]
- Chung, R.T. Acute Hepatitis C Virus Infection. Clin. Infect. Dis. 2005, 41 (Suppl. S1), S14–S17. [Google Scholar]
- Chen, S.L.; Morgan, T.R. The Natural History of Hepatitis C Virus (HCV) Infection. Int. J. Med. Sci. 2006, 3, 47–52. [Google Scholar] [CrossRef]
- Hoofnagle, J.H. Course and outcome of hepatitis C. Hepatology 2002, 36 (Suppl. S1), S21–S29. [Google Scholar]
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Goodman, Z.D.; Koury, K.; Ling, M.-H.; Albrecht, J.K. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar]
- Tsubota, A.; Fujise, K.; Namiki, Y.; Tada, N. Peginterferon and ribavirin treatment for hepatitis C virus infection. World J. Gastroenterol. 2011, 17, 419–432. [Google Scholar] [CrossRef]
- Thomas, E.; Ghany, M.G.; Liang, T.J. The application and mechanism of action of ribavirin in therapy of hepatitis C. Antivir. Chem. Chemother. 2012, 23, 1–12. [Google Scholar]
- Dixit, N.M.; Perelson, A.S. The metabolism, pharmacokinetics and mechanisms of antiviral activity of ribavirin against hepatitis C virus. Cell. Mol. Life Sci. CMLS 2006, 63, 832–842. [Google Scholar]
- Lin, K.; Perni, R.B.; Kwong, A.D.; Lin, C. VX-950, a Novel Hepatitis C Virus (HCV) NS3-4A Protease Inhibitor, Exhibits Potent Antiviral Activities in HCV Replicon Cells. Antimicrob. Agents Chemother. 2006, 50, 1813–1822. [Google Scholar]
- Fridell, R.A.; Qiu, D.; Wang, C.; Valera, L.; Gao, M. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 2010, 54, 3641–3650. [Google Scholar] [PubMed]
- Vermehren, J.; Sarrazin, C. New HCV therapies on the horizon. Clin. Microbiol. Infect. 2011, 17, 122–134. [Google Scholar] [PubMed]
- Asselah, T. Sofosbuvir for the treatment of hepatitis C virus. Expert Opin. Pharmacother. 2014, 15, 121–130. [Google Scholar] [CrossRef]
- Das, D.; Pandya, M. Recent Advancement of Direct-acting Antiviral Agents (DAAs) in Hepatitis C Therapy. Mini Rev. Med. Chem. 2018, 18, 584–596. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [PubMed]
- Bartenschlager, R.; Cosset, F.L.; Lohmann, V. Hepatitis C virus replication cycle. J. Hepatol. 2010, 53, 583–585. [Google Scholar]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar]
- Yu, Y.; Scheel, T.K.H.; Luna, J.M.; Chung, H.; Nishiuchi, E.; Scull, M.A.; Echeverría, N.; Ricardo-Lax, I.; Kapoor, A.; Lipkin, I.W.; et al. miRNA independent hepacivirus variants suggest a strong evolutionary pressure to maintain miR-122 dependence. PLOS Pathog. 2017, 13, e1006694. [Google Scholar]
- Luna, J.M.; Scheel, T.K.H.; Danino, T.; Shaw, K.S.; Mele, A.; Fak, J.J.; Nishiuchi, E.; Takacs, C.N.; Catanese, M.T.; de Jong, Y.P.; et al. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell 2015, 160, 1099–1110. [Google Scholar]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Chang, J.; Nicolas, E.; Marks, D.; Sander, C.; Lerro, A.; Buendia, M.A.; Xu, C.; Mason, W.S.; Moloshok, T.; Bort, R.; et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004, 1, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjärn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008, 36, 1153–1162. [Google Scholar] [CrossRef]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Gatfield, D.; Esau, C.C.; Green, C.B. MicroRNA-122 Modulates the Rhythmic Expression Profile of the Circadian Deadenylase Nocturnin in Mouse Liver. PLoS ONE 2010, 5, e11264. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Vujic Spasić, M.; Altamura, S.; Elmén, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmüller, U.; et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef]
- Tsai, W.C.; Hsu, S.D.; Hsu, C.S.; Lai, T.C.; Chen, S.J.; Shen, R.; Huang, Y.; Chen, H.C.; Lee, C.H.; Tsai, T.F.; et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Investig. 2012, 122, 2884–2897. [Google Scholar] [CrossRef]
- Hsu, S.-H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; La Perle, K.; Chivukula, R.R.; et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883. [Google Scholar] [CrossRef]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H.A. MicroRNA Expression in Zebrafish Embryonic Development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Jangra, R.K.; Yi, M.; Lemon, S.M. Regulation of Hepatitis C Virus Translation and Infectious Virus Production by the MicroRNA miR-122. J. Virol. 2010, 84, 6615–6625. [Google Scholar] [CrossRef]
- Jopling, C.L. Regulation of hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 2008, 36, 1220–1223. [Google Scholar] [CrossRef]
- Machlin, E.S.; Sarnow, P.; Sagan, S.M. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. USA 2011, 108, 3193. [Google Scholar] [CrossRef] [PubMed]
- Randall, G.; Panis, M.; Cooper, J.D.; Tellinghuisen, T.L.; Sukhodolets, K.E.; Pfeffer, S.; Landthaler, M.; Landgraf, P.; Kan, S.; Lindenbach, B.D.; et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 2007, 104, 12884–12889. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.A.; Zhang, C.; Huys, A.; Richardson, C.D. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J. Virol. 2011, 85, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Norman, K.L.; Sarnow, P. Positive and Negative Modulation of Viral and Cellular mRNAs by Liver-specific MicroRNA miR-122. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Schütz, S.; Sarnow, P. Position-dependent Function for a Tandem MicroRNA miR-122 Binding Site Located in the Hepatitis C Virus RNA Genome. Cell Host Microbe 2008, 4, 77–85. [Google Scholar] [CrossRef]
- Mortimer, S.A.; Doudna, J.A. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013, 41, 4230–4424. [Google Scholar] [CrossRef]
- Pang, P.S.; Pham, E.A.; Elazar, M.; Patel, S.G.; Eckart, M.R.; Glenn, J.S. Structural Map of a MicroRNA-122: Hepatitis C Virus Complex. J. Virol. 2012, 86, 1250–1254. [Google Scholar] [CrossRef]
- Shimakami, T.; Yamane, D.; Welsch, C.; Hensley, L.; Jangra, R.K.; Lemon, S.M. Base pairing between hepatitis C virus RNA and microRNA 122 3’ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 2012, 86, 7372–7383. [Google Scholar] [CrossRef] [PubMed]
- Thibault, P.A.; Huys, A.; Amador-Cañizares, Y.; Gailius, J.E.; Pinel, D.E.; Wilson, J.A. Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5′ Untranslated Region. J. Virol. 2015, 89, 6294–6311. [Google Scholar] [CrossRef] [PubMed]
- Kunden, R.D.; Ghezelbash, S.; Khan, J.Q.; Wilson, J.A. Location specific annealing of miR-122 and other small RNAs defines an Hepatitis C Virus 5′ UTR regulatory element with distinct impacts on virus translation and genome stability. Nucleic Acids Res. 2020, 48, 9235–9249. [Google Scholar] [CrossRef] [PubMed]
- Bernier, A.; Sagan, S.M. Beyond sites 1 and 2, miR-122 target sites in the HCV genome have negligible contributions to HCV RNA accumulation in cell culture. J. Gen. Virol. 2019, 100, 217–226. [Google Scholar] [CrossRef]
- Gerresheim, G.K.; Dünnes, N.; Nieder-Röhrmann, A.; Shalamova, L.A.; Fricke, M.; Hofacker, I.; Zu Siederdissen, C.H.; Marz, M.; Niepmann, M. microRNA-122 target sites in the hepatitis C virus RNA NS5B coding region and 3′ untranslated region: Function in replication and influence of RNA secondary structure. Experientia 2017, 74, 747–760. [Google Scholar] [CrossRef]
- Nasheri, N.; Singaravelu, R.; Goodmurphy, M.; Lyn, R.K.; Pezacki, J.P. Competing roles of microRNA-122 recognition elements in hepatitis C virus RNA. Virology 2011, 410, 336–344. [Google Scholar] [CrossRef]
- Sarnow, P.; Sagan, S.M. Unraveling the Mysterious Interactions Between Hepatitis C Virus RNA and Liver-Specific MicroRNA-122. Annu. Rev. Virol. 2016, 3, 309–332. [Google Scholar] [CrossRef]
- Zhang, C.; Huys, A.; Thibault, P.A.; Wilson, J.A. Requirements for human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology 2012, 433, 479–488. [Google Scholar] [CrossRef]
- Shimakami, T.; Yamane, D.; Jangra, R.K.; Kempf, B.J.; Spaniel, C.; Barton, D.J.; Lemon, S.M. Stabilization of hepatitis C virus RNA by an Ago2–miR-122 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 941–946. [Google Scholar] [CrossRef]
- Amador-Cañizares, Y.; Panigrahi, M.; Huys, A.; Kunden, R.; Adams, H.M.; Schinold, M.J.; A Wilson, J. miR-122, small RNA annealing and sequence mutations alter the predicted structure of the Hepatitis C virus 5′ UTR RNA to stabilize and promote viral RNA accumulation. Nucleic Acids Res. 2018, 46, 9776–9792. [Google Scholar] [CrossRef]
- Conrad, K.D.; Giering, F.; Erfurth, C.; Neumann, A.; Fehr, C.; Meister, G.; Niepmann, M. microRNA-122 Dependent Binding of Ago2 Protein to Hepatitis C Virus RNA Is Associated with Enhanced RNA Stability and Translation Stimulation. PLoS ONE 2013, 8, e56272. [Google Scholar]
- Schult, P.; Roth, H.; Adams, R.L.; Mas, C.; Imbert, L.; Orlik, C.; Ruggieri, A.; Pyle, A.M.; Lohmann, V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018, 9, 2613. [Google Scholar] [CrossRef] [PubMed]
- Chahal, J.; Gebert, L.; Gan, H.H.; Camacho, E.; Gunsalus, K.C.; Macrae, I.J.; Sagan, S.M. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5′ terminus. Nucleic Acids Res. 2019, 47, 5307–5324. [Google Scholar] [CrossRef]
- Ono, C.; Fukuhara, T.; Li, S.; Wang, J.; Sato, A.; Izumi, T.; Fauzyah, Y.; Yamamoto, T.; Morioka, Y.; Dokholyan, N.V.; et al. Various miRNAs compensate the role of miR-122 on HCV replication. PLoS Pathog. 2020, 16, e1008308. [Google Scholar] [CrossRef]
- Conrad, K.D.; Niepmann, M. The role of microRNAs in hepatitis C virus RNA replication. Arch. Virol. 2014, 159, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-C.; Yeh, Y.-J.; Tseng, C.-P.; Hsu, S.-D.; Chang, Y.-L.; Sakamoto, N.; Huang, H.-D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. 2012, 69, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.E.; Lewis, A.P.; Jopling, C.L. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011, 39, 7716–7729. [Google Scholar] [CrossRef]
- Li, Y.; Masaki, T.; Yamane, D.; McGivern, D.R.; Lemon, S.M. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 2013, 110, 1881. [Google Scholar] [CrossRef]
- Sedano, C.D.; Sarnow, P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe 2014, 16, 257–264. [Google Scholar] [CrossRef]
- Li, Y.; Yamane, D.; Lemon, S.M. Dissecting the Roles of the 5′ Exoribonucleases Xrn1 and Xrn2 in Restricting Hepatitis C Virus Replication. J. Virol. 2015, 89, 4857–4865. [Google Scholar] [CrossRef]
- Amador-Cañizares, Y.; Bernier, A.; Wilson, J.A.; Sagan, S.M. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5’ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018, 46, 5139–5158. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Lam, V.L.; Chirayil, R.P.; Randall, G.; Sullivan, C.S. RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proc. Natl. Acad. Sci. USA 2018, 115, 8197–8202. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Chang, J.H.; Kilic, T.; Tong, L.; Kiledjian, M. A mammalian pre-mRNA 5’ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013, 50, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, M.; Palmer, M.A.; Wilson, J.A. Enhanced virus translation enables miR-122-independent Hepatitis C Virus propagation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schüttler, C.G.; Fehr, C.; Jünemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Niepmann, M.; Shalamova, L.A.; Gerresheim, G.K.; Rossbach, O. Signals Involved in Regulation of Hepatitis C Virus RNA Genome Translation and Replication. Front. Microbiol. 2018, 9, 395. [Google Scholar] [CrossRef]
- Chahal, J.; Gebert, L.F.R.; Camargo, C.; MacRae, I.J.; Sagan, S.M. miR-122–based therapies select for three distinct resistance mechanisms based on alterations in RNA structure. Proc. Natl. Acad. Sci. USA 2021, 118, e2103671118. [Google Scholar] [CrossRef]
- Masaki, T.; Arend, K.C.; Li, Y.; Yamane, D.; McGivern, D.R.; Kato, T.; Wakita, T.; Moorman, N.J.; Lemon, S.M. miR-122 Stimulates Hepatitis C Virus RNA Synthesis by Altering the Balance of Viral RNAs Engaged in Replication versus Translation. Cell Host Microbe 2015, 17, 217–228. [Google Scholar] [CrossRef]
- Wang, L.; Jeng, K.-S.; Lai, M.M.C. Poly(C)-Binding Protein 2 Interacts with Sequences Required for Viral Replication in the Hepatitis C Virus (HCV) 5′ Untranslated Region and Directs HCV RNA Replication through Circularizing the Viral Genome. J. Virol. 2011, 85, 7954–7964. [Google Scholar] [CrossRef]
- Ono, C.; Fukuhara, T.; Motooka, D.; Nakamura, S.; Okuzaki, D.; Yamamoto, S.; Tamura, T.; Mori, H.; Sato, A.; Uemura, K.; et al. Characterization of miR-122-independent propagation of HCV. PLOS Pathog. 2017, 13, e1006374. [Google Scholar] [CrossRef]
- Bai, S.; Nasser, M.W.; Wang, B.; Hsu, S.-H.; Datta, J.; Kutay, H.; Yadav, A.; Nuovo, G.; Kumar, P.; Ghoshal, K. MicroRNA-122 Inhibits Tumorigenic Properties of Hepatocellular Carcinoma Cells and Sensitizes These Cells to Sorafenib. J. Biol. Chem. 2009, 284, 32015–32027. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Pfeffer, S.; Baumert, T.F. miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. J. Hepatol. 2013, 58, 821–823. [Google Scholar] [CrossRef] [PubMed]
- Scheel, T.K.; Kapoor, A.; Nishiuchi, E.; Brock, K.V.; Yu, Y.; Andrus, L.; Gu, M.; Renshaw, R.W.; Dubovi, E.J.; McDonough, S.P.; et al. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc. Natl. Acad. Sci. USA 2015, 112, 2192–2197. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Dai, F.; Shi, B.; Chen, L.; Zhang, X.; Zang, G.; Zhang, J.; Chen, X.; Qian, F.; et al. Comparison of Circulating, Hepatocyte Specific Messenger RNA and microRNA as Biomarkers for Chronic Hepatitis B and C. PLoS ONE 2014, 9, e92112. [Google Scholar] [CrossRef]
- Pinzani, P.; Salvianti, F.; Pazzagli, M.; Orlando, C. Circulating nucleic acids in cancer and pregnancy. Methods 2010, 50, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Thakral, S.; Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef]
- Murray, D.D.; Suzuki, K.; Law, M.; Trebicka, J.; Neuhaus Nordwall, J.; Johnson, M.; Vjecha, M.J.; Kelleher, A.D.; Emery, S. Circulating miR-122 and miR-200a as biomarkers for fatal liver disease in ART-treated, HIV-1-infected individuals. Sci. Rep. 2017, 7, 10934. [Google Scholar] [CrossRef]
- Zhou, X.; Fang, S.; Wang, M.; Xiong, A.; Zheng, C.; Wang, J.; Yin, C. Diagnostic value of circulating miRNA-122 for hepatitis B virus and/or hepatitis C virus-associated chronic viral hepatitis. Biosci. Rep. 2019, 39, BSR20190900. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Dubovi, E.J.; Simmonds, P.; Medina, J.L.; Henriquez, J.A.; Mishra, N.; Wagner, J.; Tokarz, R.; Cullen, J.M.; Iadarola, M.J.; et al. Serology-Enabled Discovery of Genetically Diverse Hepaciviruses in a New Host. J. Virol. 2012, 86, 6171–6178. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Lukashev, A.N.; Gmyl, A.; Coutard, B.; Adam, A.; Ritz, D.; Leijten, L.M.; Van Riel, D.; et al. Evidence for Novel Hepaciviruses in Rodents. PLOS Pathog. 2013, 9, e1003438. [Google Scholar]
- Kapoor, A.; Simmonds, P.; Scheel, T.K.H.; Hjelle, B.; Cullen, J.M.; Burbelo, P.D.; Chauhan, L.V.; Duraisamy, R.; Leon, M.S.; Jain, K.; et al. Identification of Rodent Homologs of Hepatitis C Virus and Pegiviruses. mBio 2013, 4, e00216-13. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Simmonds, P.; Gerold, G.; Qaisar, N.; Jain, K.; Henriquez, J.A.; Firth, C.; Hirschberg, D.L.; Rice, C.M.; Shields, S.; et al. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. USA 2011, 108, 11608–11613. [Google Scholar]
- Lattimer, J.; Stewart, H.; Locker, N.; Tuplin, A.; Stonehouse, N.J.; Harris, M. Structure–function analysis of the equine hepacivirus 5′ untranslated region highlights the conservation of translational mechanisms across the hepaciviruses. J. Gen. Virol. 2019, 100, 1501–1514. [Google Scholar] [CrossRef]
- Lu, G.; Ou, J.; Zhao, J.; Li, S. Presence of a Novel Subtype of Bovine Hepacivirus in China and Expanded Classification of Bovine Hepacivirus Strains Worldwide into 7 Subtypes. Viruses 2019, 11, 843. [Google Scholar] [CrossRef] [Green Version]
- Quan, P.-L.; Firth, C.; Conte, J.M.; Williams, S.H.; Zambrana-Torrelio, C.M.; Anthony, S.J.; Ellison, J.A.; Gilbert, A.T.; Kuzmin, I.V.; Niezgoda, M.; et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 2013, 110, 8194–8199. [Google Scholar] [CrossRef]
- Baron, A.L.; Schoeniger, A.; Becher, P.; Baechlein, C. Mutational Analysis of the Bovine Hepacivirus Internal Ribosome Entry Site. J. Virol. 2018, 92, e01974-17. [Google Scholar] [CrossRef]
- Sagan, S.M.; Sarnow, P.; Wilson, J.A. Modulation of GB virus B RNA abundance by microRNA-122: Dependence on and escape from microRNA-122 restriction. J. Virol. 2013, 87, 7338–7347. [Google Scholar] [CrossRef]
- Kokkonos, K.G.; Fossat, N.; Nielsen, L.; Holm, C.; Hepkema, W.; Bukh, J.; Scheel, T.K.H. Evolutionary selection of pestivirus variants with altered or no microRNA dependency. Nucleic Acids Res. 2020, 48, 5555–5571. [Google Scholar]
- Scheel, T.; Luna, J.; Liniger, M.; Nishiuchi, E.; Rozen-Gagnon, K.; Shlomai, A.; Auray, G.; Gerber, M.; Fak, J.; Keller, I.; et al. A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host Microbe 2016, 19, 409–423. [Google Scholar] [PubMed]
- van der Ree, M.H.; de Vree, J.M.; Stelma, F.; Willemse, S.; van der Valk, M.; Rietdijk, S.; Molenkamp, R.; Schinkel, J.; van Nuenen, A.C.; Beuers, U.; et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial. Lancet 2017, 389, 709–717. [Google Scholar]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; Van Der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [PubMed]
- Zeisel, M.B.; Baumert, T.F. Clinical development of hepatitis C virus host-targeting agents. Lancet 2017, 389, 674–675. [Google Scholar] [CrossRef]
- Li, Y.-P.; Gottwein, J.M.; Scheel, T.K.; Jensen, T.B.; Bukh, J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc. Natl. Acad. Sci. USA 2011, 108, 4991–4996. [Google Scholar] [PubMed]
- Liu, F.; Shimakami, T.; Murai, K.; Shirasaki, T.; Funaki, M.; Honda, M.; Murakami, S.; Yi, M.; Tang, H.; Kaneko, S. Efficient Suppression of Hepatitis C Virus Replication by Combination Treatment with miR-122 Antagonism and Direct-acting Antivirals in Cell Culture Systems. Sci. Rep. 2016, 6, 30939. [Google Scholar]
- Mata, M.; Neben, S.; Majzoub, K.; Carette, J.; Ramanathan, M.; Khavari, P.A.; Sarnow, P. Impact of a patient-derived hepatitis C viral RNA genome with a mutated microRNA binding site. PLOS Pathog. 2019, 15, e1007467. [Google Scholar]
- Ottosen, S.; Parsley, T.B.; Yang, L.; Zeh, K.; van Doorn, L.J.; van der Veer, E.; Raney, A.K.; Hodges, M.R.; Patick, A.K. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 2015, 59, 599–608. [Google Scholar]
- van der Ree, M.H.; van der Meer, A.J.; de Bruijne, J.; Maan, R.; van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J.; Rodriguez-Torres, M.; Kupcova, V.; et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antivir. Res. 2014, 111, 53–59. [Google Scholar]
- Thibault, P.A.; Huys, A.; Dhillon, P.; Wilson, J.A. MicroRNA-122-dependent and -independent replication of Hepatitis C Virus in Hep3B human hepatoma cells. Virology 2013, 436, 179–190. [Google Scholar]
- Panigrahi, M.; Thibault, P.A.; Wilson, J.A. MicroRNA 122 Affects both the Initiation and the Maintenance of Hepatitis C Virus Infections. J. Virol. 2022, 96, e0190321. [Google Scholar] [PubMed]
- Israelow, B.; Mullokandov, G.; Agudo, J.; Sourisseau, M.; Bashir, A.; Maldonado, A.Y.; Dar, A.C.; Brown, B.D.; Evans, M.J. Hepatitis C virus genetics affects miR-122 requirements and response to miR-122 inhibitors. Nat. Commun. 2014, 5, 5408. [Google Scholar] [PubMed]
- Hopcraft, S.E.; Azarm, K.D.; Israelow, B.; Lévêque, N.; Schwarz, M.C.; Hsu, T.-H.; Chambers, M.T.; Sourisseau, M.; Semler, B.L.; Evans, M.J. Viral Determinants of miR-122-Independent Hepatitis C Virus Replication. mSphere 2016, 1, e00009-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackard, J.T.; Kemmer, N.; Sherman, K.E. Extrahepatic replication of HCV: Insights into clinical manifestations and biological consequences. Hepatology 2006, 44, 15–22. [Google Scholar]
- Agnello, V.; De Rosa, F.G. Extrahepatic disease manifestations of HCV infection: Some current issues. J. Hepatol. 2004, 40, 341–352. [Google Scholar]
- Cacoub, P.; Poynard, T.; Ghillani, P.; Charlotte, F.; Olivi, M.; Piette, J.C.; Opolon, P. Extrahepatic manifestations of chronic hepatitis C. Arthritis Rheumatol. 1999, 42, 2204–2212. [Google Scholar]
- Husa, P. Extrahepatic manifestations of HCV infection. Vnitr Lek. 2016, 62 (Suppl. S2), 18–22. [Google Scholar]
- Mayo, M.J. Extrahepatic manifestations of hepatitis C infection. Am. J. Med Sci. 2003, 325, 135–148. [Google Scholar]
- Arcaini, L.; Vallisa, D.; Rattotti, S.; Ferretti, V.V.; Ferreri, A.J.M.; Bernuzzi, P.; Merli, M.; Varettoni, M.; Chiappella, A.; Ambrosetti, A.; et al. Antiviral treatment in patients with indolent B-cell lymphomas associated with HCV infection: A study of the Fondazione Italiana Linfomi. Ann. Oncol. 2014, 25, 1404–1410. [Google Scholar] [CrossRef]
- Chen, C.L.; Huang, J.Y.; Wang, C.H.; Tahara, S.M.; Zhou, L.; Kondo, Y.; Schechter, J.; Su, L.; Lai, M.M.; Wakita, T.; et al. Hepatitis C virus has a genetically determined lymphotropism through co-receptor B7.2. Nat. Commun. 2017, 8, 13882. [Google Scholar]
- Dai, B.; Chen, A.Y.; Corkum, C.P.; Peroutka, R.J.; Landon, A.; Houng, S.; Muniandy, P.A.; Zhang, Y.; Lehrmann, E.; Mazan-Mamczarz, K.; et al. Hepatitis C virus upregulates B-cell receptor signaling: A novel mechanism for HCV-associated B-cell lymphoproliferative disorders. Oncogene 2015, 35, 2979–2990. [Google Scholar] [PubMed]
- Lunel, F.; Musset, L.; Cacoub, P.; Frangeul, L.; Cresta, P.; Perrin, M.; Grippon, P.; Hoang, C.; Piette, J.; Huraux, J.-M.; et al. Cryoglobulinemia in chronic liver diseases: Role of hepatitis C virus and liver damage. Gastroenterology 1994, 106, 1291–1300. [Google Scholar] [PubMed]
- Sarhan, M.A.; Pham, T.N.Q.; Chen, A.Y.; Michalak, T.I. Hepatitis C Virus Infection of Human T Lymphocytes Is Mediated by CD5. J. Virol. 2012, 86, 3723–3735. [Google Scholar] [PubMed]
- Bukong, T.N.; Momen-Heravi, F.; Kodys, K.; Bala, S.; Szabo, G. Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90. PLoS Pathog. 2014, 10, e1004424. [Google Scholar]
- De Tomassi, A.; Pizzuti, M.; Traboni, C. Hep3B Human Hepatoma Cells Support Replication of the Wild-Type and a 5′-End Deletion Mutant GB Virus B Replicon. J. Virol. 2003, 77, 11875–11881. [Google Scholar]
- Friebe, P.; Bartenschlager, R. Role of RNA Structures in Genome Terminal Sequences of the Hepatitis C Virus for Replication and Assembly. J. Virol. 2009, 83, 11989–11995. [Google Scholar]
- Villanueva, R.A.; Jangra, R.K.; Yi, M.; Pyles, R.; Bourne, N.; Lemon, S.M. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antivir. Res. 2010, 88, 119–123. [Google Scholar]
- Walsh, D.; Mathews, M.B.; Mohr, I. Tinkering with Translation: Protein Synthesis in Virus-Infected Cells. Cold Spring Harb. Perspect. Biol. 2012, 5, a012351. [Google Scholar] [CrossRef]
- Beachboard, D.C.; Horner, S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 2016, 32, 113–119. [Google Scholar]
- Boersma, S.; Rabouw, H.H.; Bruurs, L.J.M.; Pavlovič, T.; van Vliet, A.L.W.; Beumer, J.; Clevers, H.; van Kuppeveld, F.J.M.; Tanenbaum, M.E. Translation and Replication Dynamics of Single RNA Viruses. Cell 2020, 183, 1930–1945. [Google Scholar]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.-L.; Mutsafi, Y.; De Jésus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I.; et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe 2018, 24, 208–220.e8. [Google Scholar] [PubMed]
- Kerviel, A.; Zhang, M.; Altan-Bonnet, N. A New Infectious Unit: Extracellular Vesicles Carrying Virus Populations. Annu. Rev. Cell Dev. Biol. 2021, 37, 171–197. [Google Scholar] [PubMed]
- Stiffler, J.D.; Nguyen, M.; Sohn, J.A.; Liu, C.; Kaplan, D.; Seeger, C. Focal Distribution of Hepatitis C Virus RNA in Infected Livers. Lindenbach B, editor. PLoS ONE 2009, 4, e6661. [Google Scholar]
- Zhu, Q.; Guo, J.T.; Seeger, C. Replication of Hepatitis C Virus Subgenomes in Nonhepatic Epithelial and Mouse Hepatoma Cells. J. Virol. 2003, 77, 9204–9210. [Google Scholar] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).