Abstract
The intracellular pathogens of the genus Brucella are phylogenetically close to Ochrobactrum, a diverse group of free-living bacteria with a few species occasionally infecting medically compromised patients. A group of taxonomists recently included all Ochrobactrum organisms in the genus Brucella based on global genome analyses and alleged equivalences with genera such as Mycobacterium. Here, we demonstrate that such equivalencies are incorrect because they overlook the complexities of pathogenicity. By summarizing Brucella and Ochrobactrum divergences in lifestyle, structure, physiology, population, closed versus open pangenomes, genomic traits, and pathogenicity, we show that when they are adequately understood, they are highly relevant in taxonomy and not unidimensional quantitative characters. Thus, the Ochrobactrum and Brucella differences are not limited to their assignments to different “risk-groups”, a biologically (and hence, taxonomically) oversimplified description that, moreover, does not support ignoring the nomen periculosum rule, as proposed. Since the epidemiology, prophylaxis, diagnosis, and treatment are thoroughly unrelated, merging free-living Ochrobactrum organisms with highly pathogenic Brucella organisms brings evident risks for veterinarians, medical doctors, and public health authorities who confront brucellosis, a significant zoonosis worldwide. Therefore, from taxonomical and practical standpoints, the Brucella and Ochrobactrum genera must be maintained apart. Consequently, we urge researchers, culture collections, and databases to keep their canonical nomenclature.
1. Introduction
The Brucella organisms are among the first recognized zoonotic bacteria, becoming classic examples in the history of infectious diseases. However, since these bacteria do not show conspicuous traits upon culture and because the disease they cause (brucellosis) lacks pathognomonic signs and symptoms, it has taken over 100 years to realize the close resemblance of strains isolated from different hosts and to define the Brucella genus; investigation in which the peculiar pathogenicity and epidemiology of these bacteria were key [1]. Taxonomically, minimum standards for the Brucella genus were defined in 1975 emphasizing virulence and intracellular pathogenicity as required traits for including species in this genus [2]. About a decade later, a group of investigators showed that these pathogens were close to some soil bacteria that occasionally caused opportunistic nosocomial infections, known as the CDC group Vd or Alcaligenes/Achromobacter cluster [3,4,5], subsequently described as the Ochrobactrum genus [6]. It soon became evident that the Brucella and Ochrobactrum organisms belong to the Class 2 Alphaproteobacteria, thus closely related to plant pathogens and endosymbionts such as Agrobacterium and Sinorhizobium species [4,5,7]. Therefore, both genera are part of the so-called Rhizobiales, from the taxonomical perspective within Brucellaceae, a sister group of Bartonellaceae [8]. From the beginning, the Brucella and Ochrobactrum species have been maintained in two different genera based on relevant phenotypic, genotypic, biological, and epidemiological differences. A group of taxonomists recently joined these two groups in a single genus, “Brucella”, based on a BLAST Distance Phylogeny approach (a cladistic methodology that uses only genes identified as orthologs using bioinformatic tools) and a supposed equivalence with some genera of pathogenic bacteria [9]. The arguments put forward to justify this claim follow (for simplicity, we omitted the citations):
“The overall genomic divergence of the Brucella-Ochrobactrum clade was lower than in many clades harboring a single genus only. Brucella differs from Ochrobactrum regarding its pathogenic lifestyle, which may be reflected in the smaller genome size of Brucella. However, Ochrobactrum species are also known from clinical specimens, including its type species, and a more pronounced genome size reduction of pathogenic species nested within a partially non-pathogenic genus was observed elsewhere, as, e.g., in Mycobacterium leprae. Mycobacterium can also serve as an example for a genus harboring distinct risk groups, much like Burkholderia and Yersinia. Hence, the difference between Brucella and Ochrobactrum regarding their risk-group assignment could hardly be used as an argument against their inclusion in the same genus. Known phenotypic differences, if any, appeared to be restricted to autapomorphies of Brucella that may well be linked to its evolutionary adaptation to pathogenesis. Despite the differences in genome size, the gene-content analysis provided more support for the combined Brucella–Ochrobactrum clade than for the subclades”.[9] (Reproduced under Creative Commons CC-BY license)
Here, we argue that including Ochrobactrum in the genus Brucella following such genomic analysis and supposed equivalences with other genera of pathogens is incorrect. After briefly reviewing the different criteria used by biologists to delineate the genus, we analyze the Ochrobactrum versus Brucella case and show that a genus concept that emphasizes cladistics disregarding typology (i.e., in-depth genomic analysis, structure and physiology, population structure, ecology, and lifestyle) is insufficient, a deficiency particularly clear in the case of pathogens. We conclude that a correct taxonomical approach needs to include a proper understanding of pathogenicity. Moreover, concerning the highly infectious and vicious Brucella organisms, we argue that any taxonomical approach requires the multiple dimensions of pathogenicity and the practical implications of virulence. Consequently, we concluded that Ochrobactrum and Brucella organisms must be maintained apart in two different genera.
2. The Concept of Genus
After Linnaeus, biologists used a binomial nomenclature to which the name for an organism was based on two terms, the genus and the species, and this naming convention has been maintained in modern taxonomy. While there have been numerous debates regarding the species concept in sexual and asexual organisms, the genus and other higher taxonomical taxa have received less attention because they are much more elusive than the species [10], the entities active in nature. Since the genus and other higher taxonomical hierarchies result from averaging and factorizing common characteristics of known extinct and extant species within a group, they are undoubtedly artificial constructs of the human mind intended to bring a practical order in our observations of biological diversity [11,12]. Accordingly, the taxonomic effort is not, sensu stricto, a scientific endeavor but an epistemological one that includes two dialectically inseparable procedures: the analytical and the typological processes.
2.1. The Analytical Process
The analytical process uses quantitatively measurable characteristics of the subjects organized in ranks, commonly depicted as cladograms or phylogenetic trees, which are hypotheses on the evolutionary course of the species. It must be borne in mind that, whereas this analysis is of fundamental importance in evolutionary studies, these representations are based on numerical values and thus, depend on the parameters quantified. Not surprisingly, in genomic sequence comparisons, the distance between two groups with different genome sizes (the case of Ochrobactrum and Brucella, see below) may be enlarged or narrowed depending on whether the tree is constructed based on SNPs, core genomes, pangenomes, or nonsynonymous or synonymous sites, among several options [13]. Nonetheless, the punctuated ancestor-descendant relationships depicted through dichotomic nodes and branches in a tree are representations based on similarities presented as numerical values. Still, the interpretation of these values follows circular reasoning (the principle of circulus in probando) because a group of organisms descended from a common ancestor (premise) are closely related (conclusion), and they are closely related (premise) because they descended from a common ancestor (conclusion). In addition, drawing a cut-off line in a given branch for defining a cluster of organisms and including them in a taxon, such as a genus, is a decision that cannot be taken solely on quantitative parameters and thus, is usually based on assessments made by other methods [14,15,16]. Moreover, the branching points are not fixed features of the possible trees because we do not know the characteristics of the still undiscovered or extinct individuals not incorporated in the analysis. Consequently, the clustering or the division of groups (including the genus) reflects perspectives that, although obtained after analyzing multiple characteristics, are thus strictly speaking anthropocentric.
2.2. The Typological Process
The typological process follows a methodology that conceptually separates items in various dimensions by exploring common and exclusive properties to identify an “ideal type.” Therefore, this process is an intellectual construction that stresses specific properties of different realities that, although critically important, are not necessarily linked to quantitative characters (as those used in cladistics). Thus, the typological process is a flexible heuristic system that adapts to decision-making while working with complex data [17]. Typologies are crucial because they provide efficient and practical bases for comparisons and a framework for giving operative and predictive names, two essential conditions of taxonomy [12] that are particularly relevant in the case of pathogenic bacteria.
Ideally, the analytical and typological essences coincide in a dialectic relationship. However, while acquiring the former (e.g., DNA sequence comparisons) is a straightforward two-dimensional process often amenable to quantification, the typological essence (e.g., ecology, lifestyle, pathogenicity, etc.) includes more than two dimensions and, therefore, requires qualitative assessments. For example, the biological species concept of the famous evolutionist Ernst Mayr (1904–2005) stated that, within a defined cluster (through the analytical process) of living organisms with common characteristics, species are those “groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups” [18]. This famous Mayr’s declaration, mainly used to define most species throughout the plant and animal kingdoms, stressed the typological essence of reproduction as the epistemological conceptualization of the species as a taxon on which evolution occurred through gene flow by interacting phenotypes. The typological characteristics are even more relevant when defining two different genera since they display different “types” precluding reproduction. As expected, in other live systems such as the prokaryotes, where conjugation can be entirely precluded or occur between the same species, between different species, or among species of different genera, the typological principles are more difficult to grasp; however, they cannot be ignored because they are at the core of the diversity of life that taxonomy aims to represent.
2.3. The Analytical and Typological Processes Define the Genus
The integration of the analytical and typological processes is reflected in different proportions in the two main ways biologists interpret the genus, i.e., cladistics and evolutionary systematics [19]. The cladistic definition relies heavily or solely on the quantitative analytical process and proposes that a genus is a group of species more closely related among them than with the species of another genus, implying that it must be monophyletic. However, this definition makes no provision that paraphyletic subsets in a monophyletic group can be diverging evolutionary units occupying widely different ecological niches. Furthermore, as discussed above, defining monophyly using quantitative approaches (i.e., strict cladistics) is not straightforward and cannot avoid subjective considerations. A 94.5% or lower sequence identity for two 16S rRNA genes has been proposed as evidence for distinct genera. However, this represents a practical convention to introduce order in the taxonomy of uncultured archaea and bacteria, not a taxonomically objective truth [16]. In fact, the authors of this proposal explicitly acknowledge that such a threshold is a minimum value that does not preclude the formation of separate genera with higher sequence identities if supported by other phenotypic, genetic, or environmental data (the case of Ochrobactrum and Brucella, see below) [16].
Moreover, it has been emphasized that such a “lower cut-off window” of 16S rRNA gene sequence similarity, while reasonable for the above-cited practical purpose, was based on the evaluation of genera previously delineated by a broad spectrum of methods [15,16]. Clearly, these cautions in applying “lower cut-off windows” illustrate why integrating the analytical and typological process is necessary and support the second interpretation of the genus, i.e., the systematic evolutionary definition. This definition postulates that a genus is a group of species of common ancestry (or a single species for monospecific genera) that occupies an ecological situation different from that occupied by the species of another genus. Following this definition, the genus can be monophyletic or paraphyletic (when the latter is a subset of a monophyletic group), thus, bypassing the problem of strictly defining the branching nodes that occurs in cladistics. Emphasizing ecology (and, therefore, its structural and physiological bases) also introduces typology and thus, becomes operative and predictive. The need to incorporate ecology in taxonomy is obvious, and it is entirely relevant in the case of those bacteria (commensal or pathogens) that colonize and thrive in a given host. In addition, the systematic evolutionary definition considers the evolutionary hypotheses derived from cladistic analyses and puts them in a biologically and taxonomically meaningful perspective. Specifically, in the context of our discussion of Brucella and Ochrobactrum, it is necessary to consider hypotheses on how bacterial pathogens emerge and evolve in nature.
3. Pathogenicity and Its Taxonomical Implications: The Brucella and Ochrobactrum Case
Below, we discuss why the equivalence arguments proposed by Hördt et al. [9] to merge Ochrobactrum and Brucella in a single genus are incorrect and why, when the analytical and typological approaches are appropriately applied, the Brucella and the Ochrobactrum organisms are clearly identified as widely different groups each belonging to a different genus. Their overall characteristics are summarized in Table 1 and used in the following discussions.

Table 1.
Comparison between the Brucella and Ochrobactrum genus.
3.1. Pathogenicity and False Equivalence Arguments
Assuming that the Brucella and Ochrobactrum species should be part of the same genus because other investigators clustered bacteria with higher genomic divergence within the same genus follows what, in logic, is called a false equivalence structure of propositions. This case partly follows from the misapplication of the Taxonomic Principle of Balance. This taxonomy principle states that “retrieval of information is greatly facilitated if the taxa at a given categorical rank are, as far as possible, of equal size and degree of diversity” [64], which is, in any case, based on practical anthropocentric reasons, and not explicitly and accurately aimed to describe what exists in nature (the bold text shows why this principle does not support merging Ochrobactrum and Brucella because these genera show widely different internal diversity). Apparent equivalences are not uncommon since two things that are alike in some aspects are not necessarily alike in other aspects, as illustrated by many examples in taxonomy. One of the best-known is the Shigella and Escherichia dichotomy. Even though Shigella and Escherichia strains are phylogenetically entwined, they are maintained apart because of significant pangenomic differences, including chromosomal sizes, insertion sequence-mediated pseudogenization, acquisition of the pINV virulence plasmid, and above all, different typologies [32,65]. The problem is evident when the Mycobacterium cluster (the equivalence example given in [9]) is examined in detail and compared with the Ochrobactrum-Brucella case. The diagnosis, medical care, and recommended antibiotic treatment are mostly the same regardless of whether nontuberculous or tuberculous mycobacteria cause the infection. The diagnosis of lung disease caused by nontuberculous or tuberculous bacilli requires the integration of clinical, radiographic, and microbiological information and the isolation of the microorganisms from sputum by bronchoscopic lavage or biopsy with granulomatous inflammation histopathological features, and the presence of acid-fast bacilli, no matter the mycobacterial strain, and the disease overall known as tuberculosis [66]. Therefore, symptomatic patients with compatible radiographic findings should meet the microbiological criteria to diagnose mycobacterial lung disease, in all cases taking the same precautions for the appearance of antibiotic-resistant strains [67]. All these characteristics of mycobacterial infections reflect a common biological pattern that does not exist between the Brucella and Ochrobactrum organisms because the respective clinical picture, pathogenesis, virulence, infection strategies, diagnosis, treatment, lifestyle, epidemiology, and health impacts are utterly unrelated and even opposed (see below). Indeed, Mycobacterium, Ochrobactrum, and Brucella organisms have evolved in different ecological niches and under different selection forces. Thus, using the mycobacterial group (or bacteria such as Yersinia and Burkholderia) as an analogy to support joining Brucella and Ochrobactrum in a single genus is an oversimplification; as such, it has no actual taxonomical value. On the contrary, if something can be learned from these comparisons, it is that each group of living organisms must be analyzed within its own evolutionary context and from a biological, epidemiological, medical, and historical perspective.
3.2. Cladistics, Genome Comparisons, and Pangenomes
The limitations of a genus definition that considers primarily or only cladistic arguments have been underlined above, and they become evident when considering the Ochrobactrum and Brucella case.
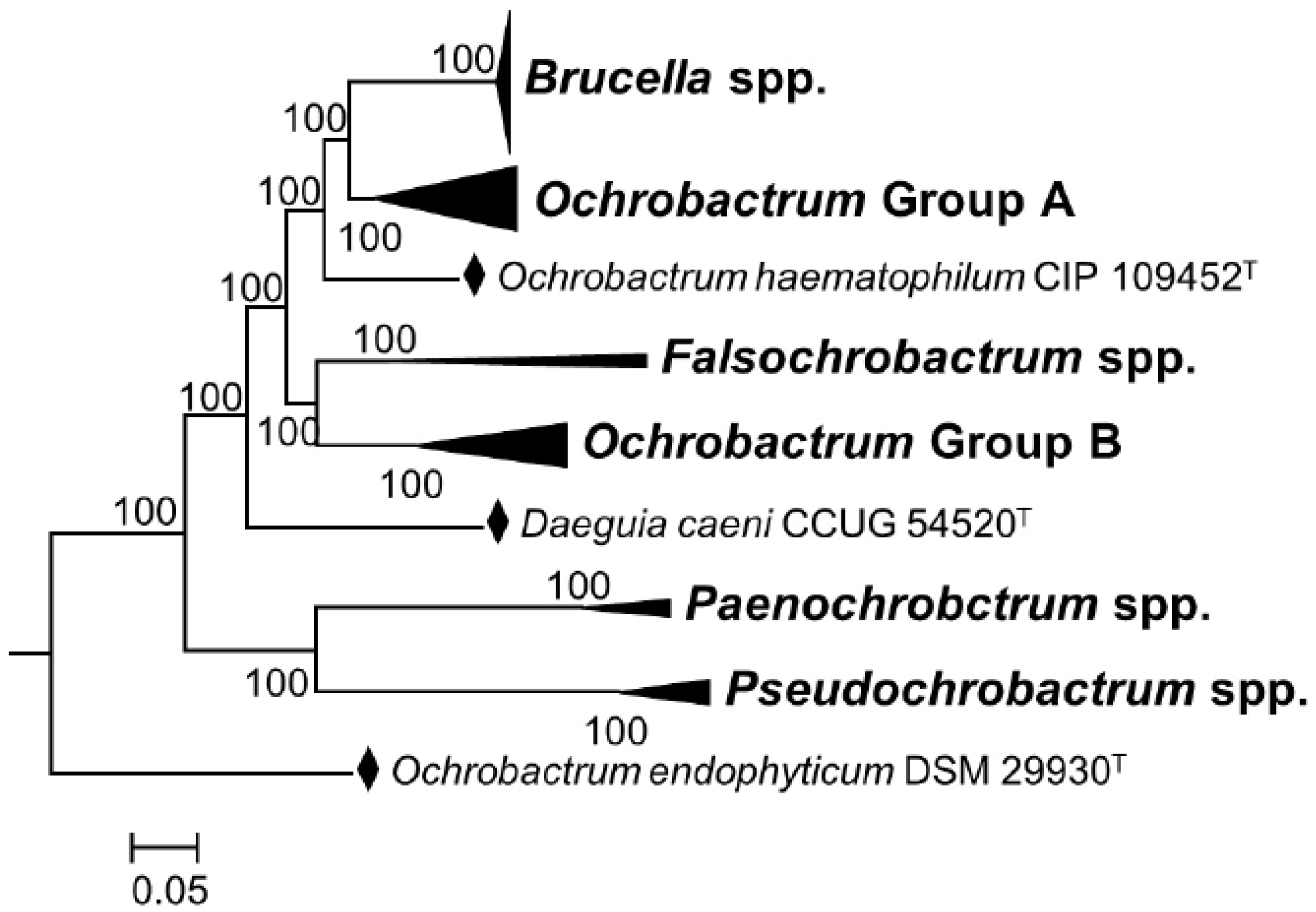
It is true that through cladistics, the members of the genus Ochrobactrum (particularly O. anthropi and O. intermedium) are the closest known relatives of Brucella organisms. However, the merit of these studies is that they are helpful to understand the widely diverging (and thus, taxonomically relevant) adaptive evolution of these bacteria, including taxonomically significant examples of exaptation (see below). However, even from a cladistic perspective, the classical and non-classical Brucella species constitute a monophyletic group that branches in a distinct and conspicuous clade from other members of the Brucellaceae, including the Ochrobactrum species [8,30]. When the analyses are focused on core genes identified in the complete genomes using bioinformatic tools, the distinctive clustering of the genus Brucella becomes evident, in contrast to the polyphyletic nature of the genus Ochrobactrum [8,30] (Figure 1). These differences are overlooked in the analyses allegedly supporting the inclusion of the latter genus in the former one [9]. They are also mainly quantitative and, thus, do not represent the profound biological implications of the genomic differences and clear-cut autapomorphies.
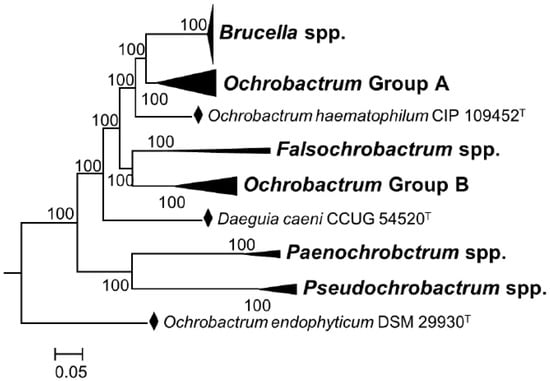
Figure 1.
Phylogenetic relationships among genera of the family Brucellaceae based on whole sequence genome analysis (a black diamond denotes type strains). Node labels give percentage bootstrap support. Rhizobium etli (CFN42T) was used to root the phylogenetic tree (not shown). The tree was constructed using the maximum likelihood method, based on the general time-reversible model, as described by Ashford et al. [30] (adapted from Figure 4 of this reference).
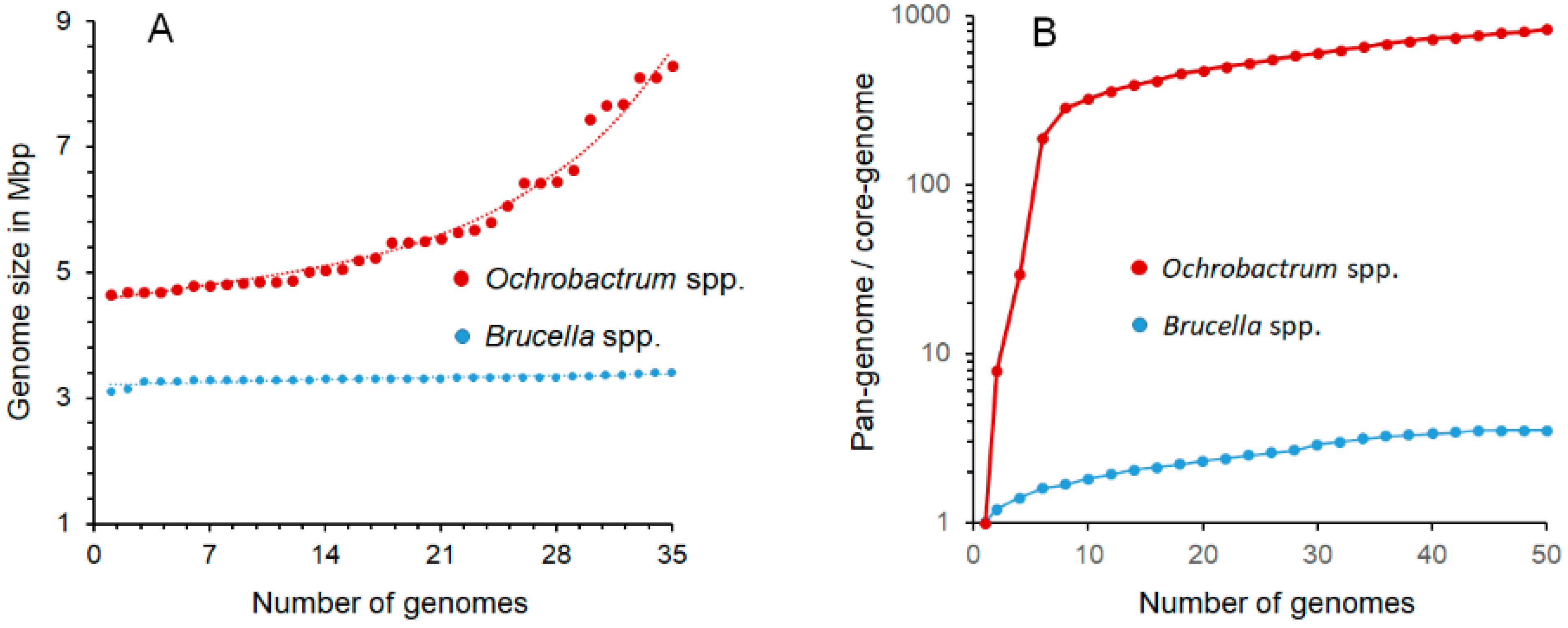
One of the most significant distinctive features of Brucella organisms is the smaller genome sizes (3.1–3.4 Mb) as compared with their closest Ochrobactrum relatives (4.7–8.3 Mb), a phenomenon linked to their different lifestyles. Despite being acknowledged [9], Hördt et al. did not consider the qualitative and quantitative significance of the ~1.6–4.9 Mb genome size variation (Table 1 and Figure 2), corresponding to the ~950–3000 gene differences between O. anthropi or O. intermedium (the closest Brucella relatives) and the Brucella species. Some of these differences are associated with the presence of variable numbers of plasmids or chromides in the Ochrobactrum species [20], while in Brucella, the genome size and absence of plasmids is a constant trait (Table 1 and Figure 2), all characteristics of great relevance (see below). Similarly unnoticed is the biological meaning of the ~170 Brucella proteins whose genes are not found in the Ochrobactrum genomes, 40% clustering into 15 genomic sections, and the ~249 genes in 13 genomic regions unique to Brucella [22,23]; also critical are the idiosyncratic insertion sequences (e.g., IS711) and genomic islands with a different evolutionary history, including those encoding one Type IV secretion system and the genes involved in synthesizing the lipopolysaccharide (LPS) O-chain. It is worth noting that both are involved in virulence and the intracellular lifestyle of Brucella organisms [23,35,46,49,50], while the two Type IV secretion systems found in Ochrobactrum seem devoted to plasmid conjugation with a different evolutionary history [68].
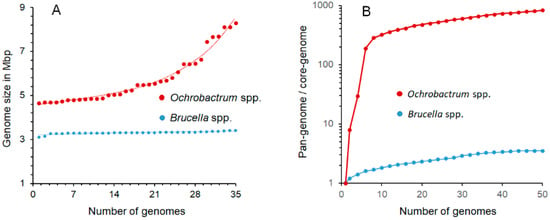
Figure 2.
Comparisons of the genome and pangenome sizes of Ochrobactrum and Brucella organisms. (A) While the smaller Brucella genomes display a narrow range of sizes, the larger Ochrobactrum genomes show significant size variations across the genus; (B) the pronounced slope of the curve (shown in logarithmic scale) with a positive trend of Ochrobactrum pangenome/core genome ratio indicates a widely open pangenome, in which the complete gene repertoire cannot be predicted with confidence, since the diversity of strains with additional genes keeps increasing due to the continuous shuffling of genes across the species. In contrast, the shallow slope of the pangenome/core genome ratio curve strongly suggests a close pangenome with no further horizontal gene exchange in the extant Brucella species. The prediction is that as more Brucella genomes are included, the numbers of conserved and accessory genes would remain, with just a limited number of additions and a proportion of ~3.6 unique-accessory genes per core conserved gene. Accordingly, the overall Brucella coding genome repertoire of the Brucella pangenome can be predicted, with some accuracy, in ~11,000 genes. Data to construct the graphs were retrieved from [20,22,24,25,27].
As expected, differences in metabolic pathways and physiological processes are also vast. For example, based on genomic predictions (see BIOCYC Summary.docx in Table S1), B. abortus 2308 and O. anthropi ATCC49188 display 254 and 313 metabolic pathways, respectively, of which 35 pathways are unique to the former, and 94 pathways are unique to the latter. The differences are even more conspicuous when the reactions involved are considered (214 versus 442 are unique to each of those two strains, respectively). The differences in numbers of transport reactions, 47 for B. abortus 2308 and 111 for O. anthropi ATCC49188, are also significant and, consistent with this, in cell envelope properties, a structure critical to understanding the ecology of any bacterium. Whereas the Ochrobactrum species keep the selective permeability barrier necessary to thrive in an open environment, the Brucella species have become sensitive to hydrophobic permeants [36], an easily observable and very meaningful phenotype. All these differences show that phenotypic differences are not restricted to autapomorphies [9] and, as shown below, there are many others.
In organisms that display large long-term effective population sizes, pangenomes arise by frequently acquiring and exchanging genes for adaptation to new niches. In general, soil environments promote the expansion of bacterial pangenome size, while host-associated habitats lead to its reduction, mainly in intracellular organisms [32]. Those bacteria with large pangenomes commonly display a relatively larger number of rRNA genes and genes for broader and diverse metabolic alternatives and the molecular machinery for exchanging exogenous genes through accessory genetic elements [32]. In contrast, bacteria with smaller pangenomes have fewer rRNA genes, fewer genes coding for metabolic routes, and a reduced capacity to exchange genes. Consequently, these divergent pangenomes have been depicted as open and closed pangenomes, respectively. Pangenome sizes influence the phylogenetic history in a given bacterial group [69], and the pangenome/core genome ratio at the genus level is more pronounced than at the species level [32].
During the evolutionary history of Brucella, plasmids and prophages were present, and cryptic sequences of these elements have remained in the genome. An investigation of 600 strains of all classical Brucella species and biovars found no free plasmids [28], and the prophage sequences detected in the hundreds of genomes of strains of Brucella species in data banks have shown widely different origins. In at least the classical Brucella, most of the prophages are defective remnants and show an uneven distribution among species [31]. Upon artificial induction, only the BiPBO1 temperate phage has been recovered from Brucella, but, so far, it is unique to Brucella inopinata BO1, and it is inactive in Ochrobactrum [31]. As expected, the absence of recombinational events and the isolated lifestyle of Brucella organisms have generated a genome that, from the evolutionary perspective, acts as a close pangenome. The gentle slope of the pangenome/core genome ratio curve for the Brucella members on a logarithmic scale shows that the genus’s increase in genetic diversity per sequenced genome added is low (Figure 2). Accordingly, the estimated core genome size for the genus is ~1000 genes with a pangenome size close to ~11,000 genes, a figure that agrees with several studies [20,22,23,24,25,26,27]. Therefore, the dispersion of the Brucella species in trees constructed using different phylogenetic strategies displays good correspondence. All this evidence suggests that at some point, the Brucella ancestor, with a larger genome than the extant species, lost its capacity for gene exchange and dispersed in different (hosts) groups that diversified by losing and degrading non-essential genes at a different rate [23,24]. At the same time, translocation of genes within the closed genomes through the concourse of insertion sequences and mutations favored the genetic drift and Brucella speciation [23,24].
In contrast with Brucella, the Ochrobactrum species possess a variable number of plasmids and lysogenic phages that promote frequent conjugation and transformation events. Some of these plasmids are very large (~1.35 Mb) and have different functions for survival in open environments [23,25]. The temperate phages are easily induced, and some strains release phage particles even under non-induced conditions. None of the Ochrobactrum phages described have activity against Brucella species [29]. In addition, the Ochrobactrum organisms contain from 4 to 12 rRNA operons and a broad spectrum of metabolic alternatives [22,37]. Consequently, Ochrobactrum organisms display the characteristic wide-open pangenome of soil bacteria subjected to continuous scattering and genomic diversification [22,23]. In the Ochrobactrum genus, the genetic repertoire is much more extensive than the gene content of the individual strains and species, and a significant number of genes per additional sequenced genome are continuously added, as shown by the pronounced pangenome/core genome curve slope (Figure 2). As expected, the estimated core genome for the Ochrobactrum genus corresponds just to ~75 genes, while the complete pangenome is, up to now, close to 74,000 genes, that is, ~1000 times larger than the core genome [22]. Therefore, predicting the actual size and content of the Ochrobactrum pangenome is, in practical terms, unfeasible because the number of genes is constantly climbing due to numerous recombination and shuffling events. As a result, correspondence between trees is not straightforward in Ochrobactrum since, as discussed in Section 2, the phylogenetic dispersion and branching of the species depend on the broad gene pool used to construct the trees. These features set a profound difference with Brucella and make cladistic comparisons with the close pangenomes of the latter subjective choices and any subsequent conclusion on the homogeneity of these bacteria untenable. Significantly, these vast multidimensional differences were already reflected in the original definition of the Ochrobactrum genus. In addition to the broad and noticeable phenotypic differences, Holmes et al. [6] noted a low degree of hybridization between the Brucella and the closest Ochrobactrum species total DNAs (~20–30%), meaningfully higher between the latter and DNA of genera of soil bacteria such as Phyllobacterium and Agrobacterium.
3.3. The Significant Differences between Intracellular Pathogens and Opportunistic Free-Living Soil Bacteria in Evolutionary Paths and Population Structures
Soil and the intracellular milieu of animal cells are utterly different environments. As a rule, free-living Alphaproteobacteria display high genetic flexibility through large flexible open genomes and accessory genetic elements that promote genetic exchange, allowing a rapid adaptation to the sudden local variations in highly diverse ecological niches of soil. In contrast, intracellular Alphaproteobacteria usually possess close smaller genomes, cryptic plasmids, or no plasmids [21,70,71]. Accordingly, while soil bacteria arrange in reticulate evolutionary units following a sympatric evolutionary strategy, the eukaryotic cell-associated bacteria assemble as clonal evolutionary units following an allopatric strategy [32,33,34].
Ochrobactrum organisms follow a sympatric speciation type, with active gene flow commensurate with their open pangenome. This phenomenon reflects their natural niche, soil, and plants’ roots [44], where the selective forces include an extensive collection of bactericidal substances and organic molecules [71]. Accordingly, Ochrobactrum organisms are commonly highly resistant to antibiotics [47,56], capable of degrading phenolic compounds [72], petroleum wastes, and an extensive collection of xenobiotics [73,74,75,76], among many other substances, and produce toxic metal-adsorbing exopolysaccharides [42], abilities complemented by their effective outer membrane permeability barrier and an array of ancillary pumps, both essential for living in open environments [25,36].
It has been long established that cell-associated bacteria share a common ancestry with free-living bacteria [77,78], and Ochrobactrum and Brucella organisms exemplify this [5,7]. However, the genomic analysis of the Brucella lineage reveals an evolutionary jump in a quick burst (punctuated evolution [79]) which probably occurred during the adaptation to an intracellular lifestyle. Bacteria that originated from this event rapidly split from the Ochrobactrum/Brucella common ancestor following an allopatric type of speciation in an isolated environment that precluded significant horizontal gene flow and led to a close pangenome. The natural niche of all well-studied Brucella organisms is the intracellular milieu of animal cells and, although they can be temporarily isolated from contaminated materials, have never been found to multiply in open environments, even in organic substrates, and are seldom in contact with other bacteria when they are metabolically active [48,50,52,80]. Consequently, the selective forces exerted over Brucella organisms are determined by the relatively stable host environments, including inimical natural host defenses, adaptive immunity effectors, and availability of nutrients within cells [35,48,52,81,82]. Consistent with these properties, they do not have the machinery to achieve the complex metabolic processes performed by Ochrobactrum organisms [38,39,40,41] and are predicted to have marked metabolic pathway reductions and acquired idiosyncratic ones (see Section 3.2). Significantly, the cell envelope of Brucella has evolved in the opposite direction of the Ochrobacrum cell envelope (see below). The minor modifications in core LPS sugars cause a profound change in selective permeability and innate immunity recognition [35,46,49,83,84] (see the previous paragraph and below). Similar to the different roles accomplished by the Type IV secretion systems in the Brucella and Ochrobactrum organisms, these are good examples of exaptation and divergent evolution and not of similarities.
As discussed in Section 2, employing ecological considerations is a critical taxonomical principle, and when pathogenicity is correctly analyzed, the ability to invade cells and thrive within an intracellular niche is a refined and multidimensional ecological adaptation. Following this, it is clear why sympatric Ochrobactrum organisms and allopatric Brucella organisms represent two markedly different groups corresponding to two separate genera.
3.4. Pathogenicity, “Risk Groups”, and Taxonomy
Like many other saprophytes and commensals living in soil environments [85], some Ochrobactrum strains can fortuitously infect and cause disease in medically compromised human hosts through catheters or other medical devices [6,22,25,44,47,51,56]. In contrast, Brucella organisms are primary intracellular pathogens that are capable of infecting a variety of healthy vertebrates through mucosae, always cause disease, are strictly linked to their hosts for perpetuation in nature, and can multiply extensively (up to 1011 bacteria per g) in some tissues and spread rapidly among individuals of the same or different host species and also from mother to offspring [46,48,52,53]. Indeed, this explains the exceedingly high contagiousness of Brucella. Therefore, claiming that “the difference between Brucella and Ochrobactrum regarding their risk-group assignment could hardly be used as an argument against their inclusion in the same genus” [9] is mistaken since there is a marked conceptual difference between “risk group” and pathogenesis. The above-summarized differences manifest different infection dynamics and, hence, profound biological differences that need to be reflected in a truthful taxonomy, as explained below.
The opportunistic Ochrobactrum species induce an acute proinflammatory pyogenic response triggered by multiple innate immune mechanisms [35,51,86], and the “virulence” factors proposed [22,56] are, in specific terms, merely accidental. Ochrobactrum “virulence” has been assigned to antibiotic efflux pumps, the type IV secretion systems, and the presence of LPS genes [22,56]. However, antibiotic efflux pumps are not, sensu stricto, virulence factors. Moreover, the Ochrobactrum type IV secretion systems seem devoted to conjugation and not virulence (see above). Likewise, the LPS (which displays endotoxicity, in contrast to the LPS of the Brucella) is characteristic of all Gram-negative bacteria and not a specific trait of pathogens. No genes for any of the known widespread true bacterial virulence-associated factors are present in the 130 genomes of Ochrobactrum species analyzed [56]. Clearly, as with any other opportunistic microorganism, Ochrobactrum human infections do not depend on the bacterium’s intrinsic “virulence” properties but on the host’s immune status and iatrogenic causes [47]. Indeed, human infections described as occurring in immunocompetent hosts [47] have corresponded to a case of septic shock that followed parental administration of a solution heavily contaminated with O. anthropi [87] and to a patient that had undergone multiple invasive procedures, including urethral catheterization [88]. After a few days or even hours in animal and cell models, their rapid elimination highlights this absence of real pathogenicity [35].
In contrast to Ochrobactrum, the Brucella organisms display furtive behavior manifested in low proinflammatory responses, prolonged incubation times, and long-lasting infections [35,46,48,81,89]. This furtive behavior is related to many significant modifications in the so-called pathogen-associated molecular patterns of surface structures that, in contrast, remain unmodified in Ochrobactrum [35]. The list of differences is extensive and includes specific interactions of these two bacterial groups with phagocytes and cell receptors, complement, and other innate and adaptive immunity factors [81]. For example, the subtle structural differences of Ochrobactrum and Brucella LPS core oligosaccharide explain why the latter displays a comparatively reduced interaction with MD2, the TLR4 LPS co-receptor critical in the immune response [35,49,81,83]. An essential set of meaningful differences includes the sophisticated machinery for intracellular trafficking and replication inside cells carried by Brucella organisms, totally absent in Ochrobactrum [35,46]. These and other molecular differences eventually display multidimensional emergent heuristic attributes that reflect the deep understanding of living organisms’ essence, shaping them in space and time.
In summary, the argument that Brucella and Ochrobactrum are similar because “Ochrobactrum species are also known from clinical specimens, including its type species” [9] is incomplete because it does not include the complexities of the pathogenicity mechanisms (“virulence factors”). Similarly, it does not consider the fact that hosts display a wide variety of environments, rather than being a global environment, and that the barriers preventing infections are affected by the host immune status and medical manipulations. As stressed above, ecological considerations play a key role in genus definitions. From an ecological and microbiological perspective, bacterial pathogenicity is a complex phenomenon that cannot be treated in taxonomy either generally (“clinical specimens”) or as a unidimensional low-to-high quantitative property (“risk groups”) [9].
4. The Practical Arguments Derived from Pathogenicity and Virulence
Since the description of the genus Brucella by Meyer and Shaw 100 years ago [1], scientists have known that members of this genus were dangerous intracellular pathogens of animals and humans, causing severe human suffering and economic losses in the livestock industry worldwide. As said, minimal standards for inclusion in the genus stressed pathogenicity [2], and new members, keeping their zoonotic pathogen potential, have been incorporated into the genus in the last two decades [1]. Medical, microbiology, and bacteriology textbooks and manuals of bacterial classification such as the Prokaryotes and Bergey’s consistently define, as a chief characteristic, the solid pathogenic nature of Brucella organisms, describing how to recognize species, diagnose, prevent, and treat an infection [52,90]; however, by no means in the case of Ochrobactrum species. From a practical perspective, including soil environmental bacteria in the Brucella genus is nontrivial; it causes difficulties by confusing veterinarians, medical doctors, epidemiologists, scientists, and official authorities devoted to the treatment, surveillance, and control of brucellosis. These conundrums are particularly relevant in low- and middle-income countries where Brucella species remain endemic and sophisticated diagnostic tools are scarce.
4.1. Animal Brucellosis and Ochrobactrum
In addition to its zoonotic character, brucellosis is an economically relevant disease of important domestic animals, including bovines, caprines, ovines, swine, canines, yacks, water buffaloes, camels, reindeer, and others [52,91]. Therefore, the correct identification of Brucella is essential for monitoring the disease in livestock. Countries expend millions of dollars on brucellosis control programs to reach brucellosis-free status, significantly impacting international trade. Ochrobactrum species are ubiquitous in soil and water, and they are common contaminants in bacteriological cultures, bovine embryos, bovine semen, soil-cattle farms, food animal products, as well as bovine, ovine, and swine tissues [54,92,93,94,95,96,97]. Consequently, renaming Ochrobactrum organisms as Brucella would bring confusion to those countries that have eradicated brucellosis from livestock. For example, reporting the isolation of “Brucella intermedium” (Ochrobactrum intermedium) on Brucella-selective agar after necropsy of imported cattle in Japan [97] would immediately catch the attention of health authorities, causing unnecessary troubles. Likewise, in Bosnia and Germany, Ochrobactrum organisms have been isolated from tissues after necropsy of animals suspected to have brucellosis [54]. Undeniably, using the same genus name for ubiquitous soil bacteria and dangerous pathogenic organisms increases the chances of committing mistakes in vaccination, testing, and slaughtering during the execution of programs devoted to controlling and eradicating the disease from livestock, thus, causing economic distress and health problems.
4.2. Human Brucellosis versus Ochrobactrum Infections
Except for sporadic Brucella canis infections and the rare cases caused by non-classical atypical Brucella BO strains (only two cases reported), all other Brucella human infections (regardless of the Brucella species) can be diagnosed by straightforward, simple, fast, and inexpensive serological tests [55]. This characteristic rests in the shared surface LPS epitopes of all smooth Brucella species causing disease [55]. These serological assays can also be used to follow the course of brucellosis after antibiotic treatment and assess the prevalence and incidence of human and animal brucellosis [55,58,98]. Due to the antigenic surface differences among Ochrobactrum organisms, the serological tests are not practical and, in all respects, absent. Consequently, the diagnosis, identification, and treatment of ochrobacteriosis follow entirely different strategies [47]. Moreover, safe and effective vaccines were developed many years ago for the prophylaxis of animal brucellosis [62], limiting zoonotic infections as a key additional benefit [63]. In contrast, vaccines neither exist nor are necessary against opportunistic Ochrobactrum. Thus, including Ochrobactrum organisms in the genus Brucella, rather than helping, creates confusion in an already complicated diagnosis, epidemiological surveys, prevention, and treatment of human brucellosis.
The lack of plasmids and confined environment of Brucella species, accentuated by the absence of significant human-to-human transmission, relates to their stable susceptibility to antibiotics used in the best regimes [59]. Tetracycline and aminoglycoside sensitivity has remained constant throughout time, even in isolates recovered from relapsed patients [58], and only a low degree of resistance to rifampin has been detected in vitro in a few isolates [99,100]. The stable sensitivity of antibiotics of the classical Brucella species can be extended to all the known extant members of the genus, as revealed by whole-genome comparisons among the different species. Thus, independently of the Brucella species causing the disease, medical doctors can apply, with confidence, effective and well-defined antibiotic therapy protocols [101]. The treatment is the same even in those few infections caused by non-classical Brucella species that do not display the usual LPS structure, and the identification is performed by other means (e.g., PCR or MALDI-TOF MS) [60]. In contrast, Ochrobactrum species show broad antibiotic resistance and can develop de novo antibiotic resistance [47,56]. Consequently, testing antibiotic susceptibility in ochrobacteriosis is mandatory [47,61]. In addition, it is necessary to understand that, in ochrobacteriosis, the antibiotics generally used are a monotherapy of ciprofloxacin, trimethoprim, or sulfamethoxazole administrated for a few days or weeks [47,56], whereas the standard protocols in non-complicated brucellosis are a combined use of doxycycline plus streptomycin or rifampin for eight weeks [101].
In summary, the infectiveness, anamnesis, diagnosis, course of the disease, treatment, and recovery in brucellosis depart from opportunistic ochrobacteriosis in such a way that, from a practical standpoint, the nomen periculosum rule applies in all its terms [102]. The appropriateness of applying this rule is highlighted by the fact that the Brucella organisms are Class 3 pathogens and among the most typical causes of laboratory-acquired infections [103,104,105], which is, indeed, not the case of Ochrobactrum organisms. Therefore, it is necessary to keep Brucella and Ochrobactrum as two separate genera for safety and common-sense reasons.
5. Concluding Remarks
Cladograms do not denote everything within the context of taxonomy or evolution. In modern prokaryotic systematics, we have chosen to compare genomic sequences of the extant species because it is the most accessible and expedited manner to explore relatedness. However, a phylogenetic tree is a two-dimensional representation, not an entire taxonomic monograph, and an even less complete evolutionary history of the embodied living systems. Living organisms must not be categorized following the only rules of two-dimensional taxonomy since they are multidimensional dynamic systems that evolve in real-time according to complex spatial-temporal contingencies with a practically infinite number of genetic combinations. There are many examples in biology in which two genera closely related by cladistics are kept apart following different criteria. We can live with this if we understand the context in which taxonomy works. The evolution of pathogenesis (and many other life episodes) is not a bidimensional flat-looking circumstance but a multidimensional emergent process requiring complex and elaborated analysis. In this regard, the Brucella and Ochrobactrum case is paradigmatic. It illustrates how intracellular pathogens of animals diversify from soil bacteria into a totally distinct group, which, as demonstrated, comprises a distinct and clearly defined genus that should not be clustered with Ochrobactrum. As established, Brucella and Ochrobactrum genera must be maintained separately, and we urge researchers, culture collections, and databases to keep their canonical nomenclature.
Taxonomy is a valuable and practical endeavor that commonly ends with a Linnaean nomenclature. Unless we arbitrarily adopt an unnatural epistemological pan-mathematicism, any natural taxonomic scheme must incorporate analytical and typological properties (including pathogenicity). Not doing this overlooks the essence and purpose of taxonomy to serve as a valuable instrument for other disciplines. As recently underlined by scientists working with a variety of microorganisms, who have also been affected by unilateral taxonomical decisions, “Science depends on nomenclature, but nomenclature is not science,” arguing that taxonomy requires the input of a broader community of microbiologists, other scientists, and professionals working with the organisms because they are affected by such decisions [106]. Ignoring the genetic, structural, biological, medical, and practical differences between Ochrobactrum and Brucella to merge them in a single genus is not granted when the analytical and typological properties are considered and, in addition, it conveys serious risks. Moreover, one of the purposes of taxonomy is to provide names wisely linked to a set of biological characteristics uniformly shared by the living beings under a given designation.
For one hundred years, “brucellosis” has been linked to a disease caused by members of the genus Brucella because, regardless of the species, all induce the same syndrome. Before the genus was established, the disease received at least fifty different names, causing significant confusion in the anamnesis and diagnosis [1]. Therefore, brucellosis is a meaningful name representing a malady caused by a relatively homogeneous group of pathogenic bacteria. From the practical and historical perspective, changing the genus of Ochrobactrum to “Brucella” would alter the mainstream of microbiology laboratories, medical doctors and veterinarians, microbial collections, genome databases, microbiological and medical books, product names, and many other practical issues, mainly targeting low- and middle-income countries where the disease is endemic and/or currently emerging.
Names of microorganisms have relevant implications in medical and veterinary practices, among many other areas. If taxonomy constitutes a system of information storage, then the relevant and factual information must be easily retrievable and not confusing. Following this, what is, for example, the information retrieved from names such as “Brucella anthropi,” “Brucella pecoris,” or “Brucella lupini” (actually Ochrobactrum anthropi, Ochrobactrum pecoris, and Ochrobactrum lupini)? Is there a “Brucella anthropi”pathogen-specific to humans, such as Brucella ovis is to sheep, Brucella canis to dogs, or Brucella ceti to dolphins? Is there a new brucellosis agent of domestic livestock as “Brucella pecoris” denotes? Do yellow beans pose a risk for transmitting brucellosis and, therefore, should they be treated as a source of pathogenic selected agents? Symptomatic of the lack of practical value of merging these bacteria in a single genus, some authors simultaneously use Brucella and Ochrobactrum for the same bacterium in the publication’s title to avoid misunderstandings [107].
A phylogenetic tree is a valuable representation of an evolutionary path that species have followed and a principle to establish hypotheses. However, taxonomical ranks are not equivalent to phylogenetic analyses, as exemplified by a proper understanding of pathogenicity. Following Jacques Monod’s (1910–1976) premise, we are dealing “with living systems not with the living matter”; accordingly, we should acknowledge their multidimensional complexity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11030377/s1, Table S1: Compartive properties between Brucella abortus 2308 and Ochrobactrum anthropi ATCC49188.
Author Contributions
Conceptualization, E.M., J.M.B., J.J.L., J.P.G. and I.M.; methodology, E.M., J.M.B. and I.M.; validation, E.M., J.M.B., J.J.L., J.P.G. and I.M.; formal analysis, E.M., J.M.B., J.J.L., J.P.G. and I.M.; investigation, E.M., J.M.B., I.M., E.M., J.P.G. and I.M.; data curation, E.M., J.J.L. and I.M.; writing—original draft, E.M., J.M.B., J.J.L., J.P.G. and I.M.; writing—review and editing, E.M., J.M.B., J.J.L., J.P.G. and I.M.; supervision, E.M. and I.M.; project administration, E.M. and J.P.G.; funding acquisition, E.M., J.M.B., J.P.G. and I.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors wish to express their gratitude for the support of the National Academy of Sciences of Costa Rica (E.M.), the Aragón Government (Grupo de Investigación A13_17R) (J.M.B.), the Centre National de la Recherche Scientifique (CNRS) and the Institut National de la Santé et de la Recherche Médicale (INSERM), and by the Fondation pour la Recherche Médicale (FRM) grant DEQ20170336745 (J.P.G.) and the Institute of Tropical University of Navarra- Health funders (Fundación la CAIXA -LCF/PR/PR13/11080005- and Fundación Caja Navarra, Fundación María Francisca de Roviralta, Ubesol and Inversiones Garcilaso de la Vega S.L) and MINECO (PID2019-107601RA-C32) (I.M.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moreno, E. The one hundred year journey of the genus Brucella (Meyer and Shaw 1920). FEMS Microbiol. Rev. 2020, 45, fuaa045. [Google Scholar] [CrossRef] [PubMed]
- Corbel, M.J.; Brinley Morgan, W.J. Proposal for minimal standards for descriptions of new species and biotypes of the genus Brucella. Int. J. Syst. Evol. Microbiol. 1975, 25, 83–89. [Google Scholar]
- De Ley, J.; Mannheim, W.; Segers, P.; Lievens, A.; Deninj, M.; Vanhoucke, M.; Gillis, M. Ribosomal ribonucleic acid cistron similarities and taxonomic neighborhood of Brucella and CDC group Vd. Int. J. Syst. Bacteriol. 1987, 37, 35–42. [Google Scholar] [CrossRef]
- Dorsch, M.; Moreno, E.; Stackebrandt, E. Nucleotide sequence of the 16S rRNA from Brucella abortus. Nucleic Acids Res. 1989, 17, 1765. [Google Scholar] [CrossRef][Green Version]
- Moreno, E.; Stackebrandt, E.; Dorsch, M.; Wolters, J.; Busch, M.; Mayer, H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 1990, 172, 3569–3576. [Google Scholar] [CrossRef]
- Holmes, B.; Popoff, M.; Kiredjian, M.; Kersters, K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as Group Vd. Int. J. Syst. Bacteriol. 1988, 38, 406–416. [Google Scholar] [CrossRef]
- Velasco, J.; Romero, C.; López-Goñi, I.; Leiva, J.; Díaz, R.; Moriyón, I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 1998, 48, 759–768. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Cloeckaert, A.; Zygmunt, M.S.; Foster, J.T. Taxonomic organization of the family Brucellaceae based on a phylogenomic approach. Front. Microbiol. 2020, 10, 3083. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Malik, V. The genus: A natural or arbitrary entity. Plant. Arch. 2017, 17, 251–257. [Google Scholar]
- Bartlett, H.H. The concept of the genus: I. History of the generic concept in botany. Bull. Torrey Bot. Club 1940, 67, 349–362. [Google Scholar] [CrossRef]
- Rosselló-Móra, R. Towards a taxonomy of Bacteria and Archaea based on interactive and cumulative data repositories. Environ. Microbiol. 2012, 14, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Puigbo, P.; Wolf, Y.I.; Koonin, E.V. The tree and net components of prokaryote evolution. Genome Biol. Evol. 2010, 2, 745–756. [Google Scholar] [CrossRef]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 2020, 11, e02475-19. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Puppe, F. Heuristic classification. In Systematic Introduction to Expert Systems; Springer: Berlin/Heidelberg, Germany, 1993; pp. 131–148. [Google Scholar]
- Mayr, E. Systematics and the Origin of Species; Columbia University Press: New York, NY, USA, 1942. [Google Scholar]
- Wood, B.; Collard, M. The human genus. Science 1999, 284, 65–71. [Google Scholar] [CrossRef]
- Teyssier, C.; Marchandin, H.; Masnou, A.; Jeannot, J.; de Buochberg, M.S.; Jumas-Bilak, E. Pulsed-field gel electrophoresis to study the diversity of whole-genome organization in the genus Ochrobactrum. Electrophoresis 2005, 26, 2898–2907. [Google Scholar] [CrossRef]
- Moreno, E. Genome evolution within the alpha Proteobacteria: Why do some bacteria not possess plasmids and others exhibit more than one different chromosome? FEMS Microbiol. Rev. 1998, 22, 255–275. [Google Scholar] [CrossRef]
- Gohil, K.; Rajput, V.; Dharne, M. Pan-genomics of Ochrobactrum species from clinical and environmental origins reveals distinct populations and possible links. Genomics 2020, 112, 3003–3012. [Google Scholar] [CrossRef]
- Wattam, A.R.; Foster, J.T.; Mane, S.P.; Beckstrom-Sternberg, S.M.; Beckstrom-Sternberg, J.M.; Dickerman, A.W.; Keim, P.; Pearson, T.; Shukla, M.; Ward, D.V.; et al. Comparative phylogenomics and evolution of the Brucellae reveal a path to virulence. J. Bacteriol. 2014, 196, 920–930. [Google Scholar] [CrossRef]
- Suárez-Esquivel, M.; Chaves-Olarte, E.; Moreno, E.; Guzmán-Verri, C. Brucella genomics: Macro and micro evolution. Int. J. Mol. Sci. 2020, 21, 7749. [Google Scholar] [CrossRef] [PubMed]
- Teyssier, C.; Marchandin, H.; Jean-Pierre, H.; Diego, I.; Darbas, H.; Jeannot, J.L.; Gouby, A.; Jumas-Bilak, E. Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J. Med. Microbiol. 2005, 54, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, Y.; Zang, J.; Li, Y.; Bie, P.; Lu, Y.; Wu, Q. Analysis of pan-genome to identify the core genes and essential genes of Brucella spp. Mol. Genet. Genom. 2016, 291, 905–912. [Google Scholar] [CrossRef]
- Sankarasubramanian, J.; Vishnu, U.S.; Gunasekaran, P.; Rajendhran, J. Identification of genetic variants of Brucella spp. through genome-wide association studies. Infect. Genet. Evol. 2017, 56, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.E. Current concepts in the Taxonomy of the genus Brucella. In Animal Brucellosis; Nielsen, K.H., Duncan, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 1990; pp. 1–17. [Google Scholar]
- Jäckel, C.; Hertwig, S.; Scholz, H.C.; Nockler, K.; Reetz, J.; Hammerl, J.A. Prevalence, host range, and comparative genomic analysis of temperate Ochrobactrum phages. Front. Microbiol. 2017, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Ashford, R.T.; Muchowski, J.; Koylass, M.; Scholz, H.C.; Whatmore, A.M. Application of whole genome sequencing and pan-family multi-locus sequence analysis to characterize relationships within the family Brucellaceae. Front. Microbiol. 2020, 11, 1329. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Göllner, C.; Al Dahouk, S.; Nöckler, K.; Reetz, J.; Hertwig, S. Analysis of the first temperate broad host range brucellaphage (BiPBO1) isolated from B. inopinata. Front. Microbiol. 2016, 7, 24. [Google Scholar] [CrossRef]
- Rouli, L.; Merhej, V.; Fournier, P.-E.; Raoult, D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015, 7, 72–85. [Google Scholar] [CrossRef]
- Moreno, E.; Cloeckaert, A.; Moriyón, I. Brucella evolution and taxonomy. Vet. Microbiol. 2002, 90, 209–227. [Google Scholar] [CrossRef]
- Moreno, E. In search of a bacterial species definition. Rev. Biol. Trop. 1997, 45, 735–771. [Google Scholar]
- Barquero-Calvo, E.; Conde-Álvarez, R.; Chacón-Díaz, C.; Quesada-Lobo, L.; Martirosyan, A.; Guzmán-Verri, C.; Iriarte, M.; Manček-Keber, M.; Jerala, R.; Gorvel, J.-P.; et al. The differential interaction of Brucella and Ochrobactrum with innate immunity reveals traits related to the evolution of stealthy pathogens. PLoS ONE 2009, 4, e5893. [Google Scholar] [CrossRef] [PubMed]
- Velasco, J.; Bengoechea, J.A.; Brandenburg, K.; Lindner, B.; Seydel, U.; González, D.; Zähringer, U.; Moreno, E.; Moriyón, I. Brucella abortus and its closest phylogenetic relative, Ochrobactrum spp., differ in outer membrane permeability and cationic peptide resistance. Infect. Immun. 2000, 68, 3210–3218. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Wegmann, U.; Akinyemi, N.; Oguntoyinbo, F.A.; Sayavedra, L.; Mayer, M.J.; Narbad, A. Complete genome sequence of Ochrobactrum haematophilum FI11154, isolated from kunu-zaki, a nigerian millet-based fermented food. Genome Announc. 2018, 6, e00428-18. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Inzana, T.J.; Williams, K.P.; Mane, S.P.; Shukla, M.; Almeida, N.F.; Dickerman, A.W.; Mason, S.; Moriyón, I.; O’Callaghan, D.; et al. Comparative genomics of early-diverging Brucella strains reveals a novel lipopolysaccharide biosynthesis pathway. mBio 2012, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Barbier, T.; Nicolas, C.; Letesson, J.-J. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 2011, 585, 2929–2934. [Google Scholar] [CrossRef]
- Barbier, T.; Zúñiga-Ripa, A.; Moussa, S.; Plovier, H.; Sternon, J.F.; Lázaro-Antón, L.; Conde-Álvarez, R.; De Bolle, X.; Iriarte, M.; Moriyón, I.; et al. Brucella central carbon metabolism: An update. Crit. Rev. Microbiol. 2017, 44, 182–211. [Google Scholar] [CrossRef]
- Ronneau, S.; Moussa, S.; Barbier, T.; Conde-Álvarez, R.; Zúñiga-Ripa, A.; Moriyón, I.; Letesson, J.-J. Brucella, nitrogen and virulence. Crit Rev. Microbiol. 2014, 7828, 507–525. [Google Scholar] [CrossRef]
- Ozdemir, G.; Ozturk, T.; Ceyhan, N.; Isler, R.; Cosar, T. Heavy metal biosorption by biomass of Ochrobactrum anthropi producing exopolysaccharide in activated sludge. Bioresour. Technol. 2003, 90, 71–74. [Google Scholar] [CrossRef]
- Morais, P.V.; Branco, R.; Francisco, R. Chromium resistance strategies and toxicity: What makes Ochrobactrum tritici 5bvl1 a strain highly resistant. Biometals 2011, 24, 401–410. [Google Scholar] [CrossRef]
- Bathe, S.; Achouak, W.; Hartmann, A.; Heulin, T.; Schloter, M.; Lebuhn, M. Genetic and phenotypic microdiversity of Ochrobactrum spp. FEMS Microbiol. Ecol. 2006, 56, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.E.; Willems, A.; Abril, A.; Planchuelo, A.M.; Rivas, R.; Ludena, D.; Mateos, P.F.; Martinez-Molina, E.; Velazquez, E. Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl. Environ. Microbiol. 2005, 71, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Roop, R.M.; Barton, I.S.; Hopersberger, D.; Martin, D.W. Uncovering the hidden credentials of Brucella virulence. Microbiol. Mol. Biol. Rev. 2021, 85, e00021-19. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.P.; Tony Pembroke, J. The genus Ochrobactrum as major opportunistic pathogens. Microorganisms 2020, 8, 1797. [Google Scholar] [CrossRef]
- González-Espinoza, G.; Arce-Gorvel, V.; Mémet, S.; Gorvel, J.P. Brucella: Reservoirs and niches in animals and humans. Pathogens 2021, 10, 186. [Google Scholar] [CrossRef]
- Conde-Álvarez, R.; Arce-Gorvel, V.; Iriarte, M.; Manček-Keber, M.; Barquero-Calvo, E.; Palacios-Chaves, L.; Chacón-Díaz, C.; Chaves-Olarte, E.; Martirosyan, A.; von Bargen, K.; et al. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog. 2012, 8, e1002675. [Google Scholar] [CrossRef]
- Celli, J.; De Chastellier, C.; Franchini, D.M.; Pizarro-Cerda, J.; Moreno, E.; Gorvel, J.-P. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 2003, 198, 545–556. [Google Scholar] [CrossRef]
- Zhu, M.; Zhao, X.; Zhu, Q.; Zhang, Z.; Dai, Y.; Chen, L.; Liang, Z. Clinical characteristics of patients with Ochrobactrum anthropi bloodstream infection in a Chinese tertiary-care hospital: A 7-year study. J. Infect. Public Health 2018, 11, 873–877. [Google Scholar] [CrossRef]
- Moreno, E.; Moriyón, I. The genus Brucella. In The Prokaryotes. Volume 5. Proteobacteria; Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Dworkin, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 3, pp. 315–455. [Google Scholar]
- Corner, L.A.A. Three aspects of bovine brucellosis: Epidemiology, the role of bulls and vaccines. N. S. W. Vet. Proc. 1983, 19, 47–48. [Google Scholar]
- Kampfer, P.; Huber, B.; Busse, H.J.; Scholz, H.C.; Tomaso, H.; Hotzel, H.; Melzer, F. Ochrobactrum pecoris sp. nov., isolated from farm animals. Int. J. Syst. Evol. Microbiol. 2011, 61, 2278–2283. [Google Scholar] [CrossRef]
- Yagupsky, P.; Morata, P.; Colmenero, J.D. Laboratory diagnosis of human brucellosis. Clin. Microbiol. Rev. 2019, 33, e00073-19. [Google Scholar] [CrossRef]
- Yagel, Y.; Sestito, S.; Motro, Y.; Shnaiderman-Torban, A.; Khalfin, B.; Sagi, O.; Navon-Venezia, S.; Steinman, A.; Moran-Gilad, J. Genomic characterization of antimicrobial resistance, virulence, and phylogeny of the genus Ochrobactrum. Antibiotics 2020, 9, 177. [Google Scholar] [CrossRef]
- Corbel, M.J.; Alton, G.G.; Banai, M.; Díaz, R.; Dranovskaia, B.A.; Elberg, S.S.; Garin-Bastuji, B.; Kolar, J.; Mantovani, A.; Mousa, A.M.; et al. Brucellosis in Humans and Animals; WHO Press: Geneva, Switzerland, 2006; Volume 52, ISBN 9241547138. [Google Scholar]
- Ariza, J. Brucellosis: An update. The perspective from the Mediterranean basin. Rev. Med. Microbiol. 1999, 10, 125–135. [Google Scholar]
- García-Lobo, J.M.; Agüero, J.; Solera, J. La duradera y notoria sensibilidad de Brucella a las tetraciclinas. Enferm. Infecc. Microbiol. Clin. 1999, 17, 366–367. [Google Scholar]
- Rouzic, N.; Desmier, L.; Cariou, M.-E.; Gay, E.; Foster, J.T.; Williamson, C.H.D.; Schmitt, F.; Le Henaff, M.; Le Coz, A.; Lorléac’h, A. First case of brucellosis caused by an amphibian-type Brucella. Clin. Infect. Dis. 2021, 72, e404–e407. [Google Scholar] [CrossRef]
- Thoma, B.; Straube, E.; Scholz, H.C.; Al-Dahouk, S.; Zoller, L.; Pfeffer, M.; Neubauer, H.; Tomaso, H. Identification and antimicrobial susceptibilities of Ochrobactrum spp. Int. J. Med. Microbiol. 2009, 299, 209–220. [Google Scholar] [CrossRef]
- Blasco, J.M.; Moreno, E.; Moriyón, I. Brucellosis vaccines and vaccine candidates. In Veterinary Vaccines. Principles and Applications, 1st ed.; Metwally, S., Viljoen, G.J., El Idrissi, A., Eds.; FAO: Rome, Italy; Wiley & Sons: Hobeken, NJ, USA, 2021; pp. 295–316. [Google Scholar]
- Nicoletti, P. Relationship between animal and human disease. In Brucellosis: Clinical and Laboratory Aspects; Young, E.D., Corbel, M.J., Eds.; CRC Press: Boca Raton, FL, USA, 1989; pp. 41–52. ISBN 0-8493-6661-5. [Google Scholar]
- Mayr, E. Two empires or three? Proc. Natl. Acad. Sci. USA 1998, 95, 9720–9723. [Google Scholar] [CrossRef]
- Seferbekova, Z.; Zabelkin, A.; Yakovleva, Y.; Afasizhev, R.; Dranenko, N.O.; Alexeev, N.; Gelfand, M.S.; Bochkareva, O.O. High rates of genome rearrangements and pathogenicity of Shigella spp. Front. Microbiol. 2021, 12, 12. [Google Scholar] [CrossRef]
- Ryu, Y.J.; Koh, W.-J.; Daley, C.L. Diagnosis and treatment of nontuberculous mycobacterial lung disease: Clinicians’ perspectives. Tuberc. Respir. Dis. 2016, 79, 74–84. [Google Scholar] [CrossRef]
- Johnson, M.M.; Odell, J.A. Nontuberculous mycobacterial pulmonary infections. J. Thorac. Dis. 2014, 6, 210. [Google Scholar]
- de los Ángeles Giusti, M.; Pistorio, M.; Lozano, M.J.; Torres Tejerizo, G.A.; Salas, M.E.; Martini, M.C.; López, J.L.; Draghi, W.O.; Del Papa, M.F.; Pérez-Mendoza, D.; et al. Genetic and functional characterization of a yet-unclassified rhizobial Dtr (DNA-transfer-and-replication) region from a ubiquitous plasmid conjugal system present in Sinorhizobium meliloti, in Sinorhizobium medicae, and in other nonrhizobial Gram-negatives. Plasmid 2012, 67, 199–210. [Google Scholar] [CrossRef]
- Doolittle, W.F.; Zhaxybayeva, O. On the origin of prokaryotic species. Genome Res. 2009, 19, 744–756. [Google Scholar] [CrossRef]
- Dicenzo, G.C.; Finan, T.M. The divided bacterial genome: Structure, function, and evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Eberhard, W.G. Evolution in bacterial plasmids and levels of selection. Q. Rev. Biol. 1990, 65, 3–22. [Google Scholar] [CrossRef]
- El-Sayed, W.S.; Ibrahim, M.K.; Abu-Shady, M.; El-Beih, F.; Ohmura, N.; Saiki, H.; Ando, A. Isolation and identification of a novel strain of the genus Ochrobactrum with phenol-degrading activity. J. Biosci. Bioeng. 2003, 96, 310–312. [Google Scholar] [CrossRef]
- Veeranagouda, Y.; Paul, P.V.E.; Gorla, P.; Siddavattam, D.; Karegoudar, T.B. Complete mineralisation of dimethylformamide by Ochrobactrum sp. DGVK1 isolated from the soil samples collected from the coalmine leftovers. Appl. Microbiol. Biotechnol. 2006, 71, 369–375. [Google Scholar] [CrossRef]
- Zhang, R.; Cui, Z.; Jiang, J.; He, J.; Gu, X.; Li, S. Diversity of organophosphorus pesticide-degrading bacteria in a polluted soil and conservation of their organophosphorus hydrolase genes. Can. J. Microbiol. 2005, 51, 337–343. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Park, K.; Joo, G.-J.; Jeong, E.-M.; Kim, J.-E.; Rhee, I.-K. Glutathione-dependent biotransformation of the fungicide chlorothalonil. J. Agric. Food Chem. 2004, 52, 4192–4196. [Google Scholar] [CrossRef]
- Ermakova, I.T.; Shushkova, T.V.; Sviridov, A.V.; Zelenkova, N.F.; Vinokurova, N.G.; Baskunov, B.P.; Leontievsky, A.A. Organophosphonates utilization by soil strains of Ochrobactrum anthropi and Achromobacter sp. Arch. Microbiol. 2017, 199, 665–675. [Google Scholar] [CrossRef]
- Margulis, L. Symbiosis in Cell Evolution: Life and Its Environment on the Early Earth; Freeman, W.H., Ed.; W H Freeman & Co.: San Francisco, CA, USA, 1981; ISBN 0716712555. [Google Scholar]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M. Microbial trait evolution is dominated by frequent and rare pulsed evolution. bioRxiv 2021. [Google Scholar] [CrossRef]
- de Bagües, M.P.J.; Iturralde, M.; Arias, M.A.; Pardo, J.; Cloeckaert, A.; Zygmunt, M.S. The new strains Brucella inopinata BO1 and Brucella species 83-210 behave biologically like classic infectious Brucella species and cause death in murine models of infection. J. Infect. Dis. 2014, 210, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Barquero-Calvo, E.; Chaves-Olarte, E.; Weiss, D.S.; Guzmán-Verri, C.; Chacón-Díaz, C.; Rucavado, A.; Moriyón, I.; Moreno, E. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE 2007, 2, e631. [Google Scholar] [CrossRef] [PubMed]
- Martirosyan, A.; Moreno, E.; Gorvel, J.-P.P. An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol. Rev. 2011, 240, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Conde-Álvarez, R.; Ståhle, J.; Holst, O.; Iriarte, M.; Zhao, Y.; Arce-Gorvel, V.; Hanniffy, S.S.; Gorvel, J.-P.P.; Moriyón, I.; et al. Structural studies of lipopolysaccharide-defective mutants from Brucella melitensis identify a core oligosaccharide critical in virulence. J. Biol. Chem. 2016, 291, 7727–7741. [Google Scholar] [CrossRef]
- Martirosyan, A.; Gorvel, J.-P. Brucella evasion of adaptive immunity. Future Microbiol. 2013, 8, 147–154. [Google Scholar] [CrossRef]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar] [CrossRef]
- Cieslak, T.J.; Drabick, C.J.; Robb, M.L. Pyogenic infections due to Ochrobactrum anthropi. Clin. Infect. Dis. 1996, 22, 845–847. [Google Scholar] [CrossRef]
- Kettaneh, A.; Weill, F.X.; Poilane, I.; Fain, O.; Thomas, M.; Herrmann, J.L.; Hocqueloux, L. Septic shock caused by Ochrobactrum anthropi in an otherwise healthy host. J. Clin. Microbiol. 2003, 41, 1339–1341. [Google Scholar] [CrossRef]
- Ozdemir, D.; Soypacaci, Z.; Sahin, I.; Bicik, Z.; Sencan, I. Ochrobactrum anthropi endocarditis and septic shock in a patient with no prosthetic valve or rheumatic heart disease: Case report and review of the literature. Jpn. J. Infect. Dis. 2006, 59, 264–265. [Google Scholar]
- Spink, W.W. The natural course of brucellosis. In The Nature of Brucellosis; The University of Minnesota Press: Minneapolis, MN, USA, 1956; pp. 145–170. [Google Scholar]
- Scholz, H.C.; Banai, M.; Cloeckaert, A.; Kämpfer, P.; Whatmore, A.M. Brucella. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2018; pp. 1–38. ISBN 9781118960608. [Google Scholar]
- Moreno, E. Retrospective and prospective perspectives on zoonotic brucellosis. Front. Microbiol. 2014, 5, 213. [Google Scholar] [CrossRef]
- Alonso, C.A.; Kwabugge, Y.A.; Anyanwu, M.U.; Torres, C.; Chah, K.F. Diversity of Ochrobactrum species in food animals, antibiotic resistance phenotypes and polymorphisms in the blaOCH gene. FEMS Microbiol. Lett. 2017, 364, 364. [Google Scholar] [CrossRef]
- Alvarado-Capó, Y.; González, N.P.; García-Aguila, L.; Freire-Seijo, M.; Martínez, Y.; Kosky, R.G. Ochrobactrum anthropi, contaminants of in vitro culture of sugarcane cells and tissues. Biotecnol. Veg. 2007, 7, 211–214. [Google Scholar]
- Bielanski, A.; Bergeron, H.; Lau, P.C.K.; Devenish, J. Microbial contamination of embryos and semen during long term banking in liquid nitrogen. Cryobiology 2003, 46, 146–152. [Google Scholar] [CrossRef]
- Choi, G.-M.; Kim, K.M.; Yun, C.-S.; Lee, S.Y.; Kim, S.Y.; Wee, J.-H.; Im, W.-T. Ochrobactrum soli sp. nov., Isolated from a Korean cattle farm. Curr. Microbiol. 2020, 77, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Cobo, F.; Concha, A. Environmental microbial contamination in a stem cell bank. Lett. Appl. Microbiol. 2007, 44, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Tamura, T.; Okamura, Y.; Kabamoto, A.; Moriwaki, S.; Eto, M. Isolation and characterization of Ochrobactrum intermedium from imported cattle serologically diagnosed with bovine brucellosis. J. Jpn. Vet. Med. Assoc. 2010, 63, 615–619. [Google Scholar] [CrossRef]
- Ducrotoy, M.J.; Bertu, W.J.; Matope, G.; Cadmus, S.; Conde-Álvarez, R.; Gusi, A.M.; Welburn, S.; Ocholi, R.; Blasco, J.M.; Moriyón, I. Brucellosis in Sub-Saharan Africa: Current challenges for management, diagnosis and control. Acta Trop. 2017, 165, 179–193. [Google Scholar] [CrossRef]
- Deshmukh, A.; Hagen, F.; Al Sharabasi, O.; Abraham, M.; Wilson, G.; Doiphode, S.; Al Maslamani, M.; Meis, J.F. In vitro antimicrobial susceptibility testing of human Brucella melitensis isolates from Qatar between 2014–2015. BMC Microbiol. 2015, 15, 121. [Google Scholar] [CrossRef]
- Rubinstein, E.; Lang, R.; Shasha, B.; Hagar, B.; Diamanstein, L.; Joseph, G.; Anderson, M.; Harrison, K. In vitro susceptibility of Brucella melitensis to antibiotics. Antimicrob. Agents Chemother. 1991, 35, 1925–1927. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ariza, J.; Bosilkovski, M.; Cascio, A.; Colmenero, J.D.; Corbel, M.J.; Falagas, M.E.; Memish, Z.A.; Roushan, M.R.H.; Rubinstein, E.; Sipsas, N.V.; et al. Perspectives for the treatment of brucellosis in the 21st century: The Ioannina recommendations. PLoS Med. 2007, 4, e317. [Google Scholar] [CrossRef] [PubMed]
- Judicial Commission Proposal to emend the International Code of Nomenclature of Bacteria. Int. J Syst. Bacteriol. 1985, 35, 123. [CrossRef]
- Bouza, E.; Sánchez-Carrillo, C.; Hernan-Gómez, S.; González, M.J. Laboratory-acquired brucellosis: A Spanish national survey. J. Hosp. Infect. 2005, 61, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Baron, E.J.; Miller, J.M. Bacterial and fungal infections among diagnostic laboratory workers: Evaluating the risks. Diagn. Microbiol. Infect. Dis. 2008, 60, 241–246. [Google Scholar] [CrossRef]
- Singh, K. Laboratory-acquired infections. Clin. Infect. Dis. 2009, 49, 142–147. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Tahon, G. Science depends on nomenclature, but nomenclature is not science. Nat. Rev. Microbiol. 2022, 20, 123–124. [Google Scholar] [CrossRef]
- Li, S.Y.; Huang, Y.E.; Chen, J.Y.; Lai, C.H.; Mao, Y.C.; Huang, Y.T.; Liu, P.Y. Genomics of Ochrobactrum pseudogrignonense (newly named Brucella pseudogrignonensis) reveals a new blaoxa subgroup. Microb. Genom. 2021, 7, 000626. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).