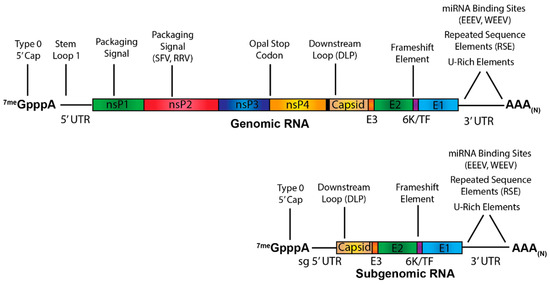
Abstract
Alphaviruses are positive-sense RNA arboviruses that are capable of causing severe disease in otherwise healthy individuals. There are many aspects of viral infection that determine pathogenesis and major efforts regarding the identification and characterization of virulence determinants have largely focused on the roles of the nonstructural and structural proteins. Nonetheless, the viral RNAs of the alphaviruses themselves play important roles in regard to virulence and pathogenesis. In particular, many sequences and secondary structures within the viral RNAs play an important part in the development of disease and may be considered important determinants of virulence. In this review article, we summarize the known RNA-based virulence traits and host:RNA interactions that influence alphaviral pathogenesis for each of the viral RNA species produced during infection. Overall, the viral RNAs produced during infection are important contributors to alphaviral pathogenesis and more research is needed to fully understand how each RNA species impacts the host response to infection as well as the development of disease.
1. Background
Alphaviruses are single-stranded, positive-sense RNA viruses that are naturally transmitted between a mosquito vector and a vertebrate host. Epizootic spillover events result in the infection of humans and horses, potentially resulting in severe disease. Alphaviruses are largely classified as either arthritogenic or encephalitic based on the symptoms of infection. Arthritogenic alphaviral infection causes disease varying from mild to severe multi-joint arthritis and can persist for several months to years past the acute phase of infection [1]. This includes Chikungunya (CHIKV) and Ross River Virus (RRV) which are capable of causing debilitating polyarthritis as well as the model alphavirus Sindbis virus (SINV), which is the causative agent of rash-arthritic diseases like Pogosta disease, Ockelbo disease, and Karelian fever [2,3,4,5]. While not associated with high rates of mortality, the high morbidity of arthritogenic alphaviral disease results in a high economic burden that is particularly damaging in regions where labor-intensive work is prevalent [6,7]. The encephalitic alphaviruses include Venezuelan, Eastern, and Western Equine Encephalitis viruses (VEEV, EEEV, and WEEV) and are capable of causing severe meningitis and encephalitis, as well as long-lasting sequelae such as seizures, paralysis, and cognitive deficits in survivors [8,9,10,11]. These viruses, while comparatively rare in regards to their incidence, typically have high mortality rates, especially in comparison to the arthritogenic alphaviruses, with viruses like EEEV having mortality rates as high as 70% in symptomatic individuals [12]. Despite the threat that alphaviruses pose to public health, there are no antiviral strategies or vaccines for preventing alphaviral infection or treating alphaviral disease. This deficit of viable therapeutics highlights the need to better understand the mechanisms behind alphaviral infection and pathogenesis in order to develop novel antiviral strategies for the mitigation of alphaviral disease.
6. Conclusions and Future Perspectives
As summarized above, the alphaviral RNAs directly contribute to virulence and pathogenesis to a significant extent. These contributions are in addition to the obvious linkage between viral replication/RNA synthetic fitness and pathogenesis, and typically involve multiple aspects of the host/pathogen interface. The mechanisms by which the viral RNAs contribute to virulence are both direct, as in acting to directly evade or resist aspects of the host innate immune response, and indirect via the modulation of the production of viral proteins during infection. The alphaviral RNA virulence determinants are often, but not always, associated with secondary structures and may be found throughout the entire length of the viral RNA. Furthermore, virulence determinants that lack defined secondary structures often act as interaction sites for host and viral RNA-binding proteins. In these instances, the virulence determinant is directly linked to the primary sequences of the viral RNAs themselves.
Overall, the critical contributions of the alphaviral RNAs to pathogenesis have been established, but much work remains to identify the full extent to which the alphaviral RNAs are intertwined with pathogenesis and the precise mechanisms by which the viral RNA influence disease. While the knowledge compiled above represents the current state of understanding in this regard, it also highlights the presence of significant critical gaps in the understanding of the role of viral RNAs in pathogenesis.
Author Contributions
Conceptualization: A.T.L. and K.J.S.; writing original draft preparation: A.T.L. and K.J.S.; review and editing: A.T.L. and K.J.S.; supervision, K.J.S. All authors have read and agreed to the published version of the manuscript. Authorship is limited to those who have contributed substantially to the work reported.
Funding
This work was funded by grants from the National Institute of Allergy and Infectious Diseases (NIH-NIAID), specifically R01 AI153275 to K.J.S., and by a COBRE program grant from the National Institute of General Medical Sciences (NIGMS), P20 GM125504 to K.J.S. and R. Lamont. A.T.L. was supported by an NIH-NIAID-funded predoctoral fellowship, T32 AI132146. Additional support was received from the Integrated Programs in Biomedical Sciences (IPIBS) to A.T.L. and a generous startup package from the University of Louisville to K.J.S.
Acknowledgments
We thank the members of the K. J. Sokoloski laboratory for their valuable input and discussions of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kurkela, S.; Helve, T.; Vaheri, A.; Vapalahti, O. Arthritis and arthralgia three years after Sindbis virus infection: Clinical follow-up of a cohort of 49 patients. Scand. J. Infect. Dis. 2008, 40, 167–173. [Google Scholar] [CrossRef]
- Kurkela, S.; Manni, T.; Myllynen, J.; Vaheri, A.; Vapalahti, O. Clinical and Laboratory Manifestations of Sindbis Virus Infection: Prospective Study, Finland, 2002–2003. J. Infect. Dis. 2005, 191, 1820–1829. [Google Scholar] [CrossRef]
- Kurkela, S.; Rätti, O.; Huhtamo, E.; Uzcátegui, N.Y.; Nuorti, J.P.; Laakkonen, J.; Manni, T.; Helle, P.; Vaheri, A.; Vapalahti, O. Sindbis Virus Infection in Resident Birds, Migratory Birds, and Humans, Finland. Emerg. Infect. Dis. 2008, 14, 41–47. [Google Scholar] [CrossRef]
- Sissoko, D.; Malvy, D.; Ezzedine, K.; Renault, P.; Moscetti, F.; Ledrans, M.; Pierre, V. Post-Epidemic Chikungunya Disease on Reunion Island: Course of Rheumatic Manifestations and Associated Factors over a 15-Month Period. PLoS Negl. Trop. Dis. 2009, 3, e389. [Google Scholar] [CrossRef]
- Farnon, E.C.; Sejvar, J.J.; Staples, J.E. Severe disease manifestations associated with acute chikungunya virus infection*. Crit. Care Med. 2008, 36, 2682–2683. [Google Scholar] [CrossRef]
- Cardona-Ospina, J.A.; Villamil-Gómez, W.E.; Jimenez-Canizales, C.E.; Castañeda, D.M.; Rodríguez-Morales, A.J. Estimating the burden of disease and the economic cost attributable to chikungunya, Colombia, 2014. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 793–802. [Google Scholar] [CrossRef]
- Seyler, T.; Hutin, Y.; Ramanchandran, V.; Ramakrishnan, R.; Manickam, P.; Murhekar, M. Estimating the burden of disease and the economic cost attributable to chikungunya, Andhra Pradesh, India, 2005–2006. Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H. Medically important arboviruses of the United States and Canada. Clin. Microbiol. Rev. 1994, 7, 89–116. [Google Scholar] [CrossRef]
- De La Monte, S.M.; Bonilla, N.J.; De Urdaneta, A.G.; Hutchins, G.M.; Castro, F. The Systemic Pathology of Venezuelan Equine Encephalitis Virus Infection in Humans. Am. J. Trop. Med. Hyg. 1985, 34, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Ronca, S.E.; Dineley, K.T.; Paessler, S. Neurological Sequelae Resulting from Encephalitic Alphavirus Infection. Front. Microbiol. 2016, 7, 959. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.E. Emergence and re-emergence of viral diseases of the central nervous system. Prog. Neurobiol. 2010, 91, 95–101. [Google Scholar] [CrossRef]
- Steele, K.R.D.; Glass, P.; Hart, M.; Ludwig, G.; Pratt, W.; Parker, M.; Smith, J. Chapter 12: Alphavirus Encephalitides. In Medical Aspects of Biological Warfare; Reed, W., Ed.; Department of the Army: Arlington, VI, USA, 2007; pp. 241–270. [Google Scholar]
- Hardy, W.R.; Strauss, J.H. Processing the nonstructural polyproteins of sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 1989, 63, 4653–4664. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef]
- Ghosh, A.; Lima, C.D. Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA 2010, 1, 152–172. [Google Scholar] [CrossRef]
- Akhrymuk, I.; Frolov, I.; Frolova, E. Both RIG-I and MDA5 detect alphavirus replication in concentration-dependent mode. Virology 2016, 487, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Sokoloski, K.J.; Haist, K.C.; Morrison, T.E.; Mukhopadhyay, S.; Hardy, R.W. Noncapped Alphavirus Genomic RNAs and Their Role during Infection. J. Virol. 2015, 89, 6080–6092. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Linehan, M.M.; Iwasaki, A.; Pyle, A.M. RIG-I Selectively Discriminates against 5′-Monophosphate RNA. Cell Rep. 2019, 26, 2019–2027.e4. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, A.T.; Landers, V.D.; Westcott, C.E.; Sokoloski, K.J. Production of Noncapped Genomic RNAs Is Critical to Sindbis Virus Disease and Pathogenicity. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Hyde, J.L.; Chen, R.; Trobaugh, D.; Diamond, M.S.; Weaver, S.C.; Klimstra, W.B.; Wilusz, J. The 5′ and 3′ ends of alphavirus RNAs—Non-coding is not non-functional. Virus Res. 2015, 206, 99–107. [Google Scholar] [CrossRef]
- Frolov, I.; Hardy, R.; Rice, C.M. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 2001, 7, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- White, L.J.; Wang, J.-G.; Davis, N.L.; Johnston, R.E. Role of Alpha/Beta Interferon in Venezuelan Equine Encephalitis Virus Pathogenesis: Effect of an Attenuating Mutation in the 5′ Untranslated Region. J. Virol. 2001, 75, 3706–3718. [Google Scholar] [CrossRef] [PubMed]
- Kinney, R.M.; Chang, G.J.; Tsuchiya, K.R.; Sneider, J.M.; Roehrig, J.; Woodward, T.M.; Trent, D.W. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J. Virol. 1993, 67, 1269–1277. [Google Scholar] [CrossRef]
- Spotts, D.R.; Reich, R.M.; Kalkhan, M.A.; Kinney, R.M.; Roehrig, J. Resistance to Alpha/Beta Interferons Correlates with the Epizootic and Virulence Potential of Venezuelan Equine Encephalitis Viruses and Is Determined by the 5′ Noncoding Region and Glycoproteins. J. Virol. 1998, 72, 10286–10291. [Google Scholar] [CrossRef]
- Logue, C.H.; Sheahan, B.J.; Atkins, G.J. The 5′ untranslated region as a pathogenicity determinant of Semliki Forest virus in mice. Virus Genes 2008, 36, 313–321. [Google Scholar] [CrossRef]
- Kobiler, D.; Rice, C.M.; Brodie, C.; Shahar, A.; Dubuisson, J.; Halevy, M.; Lustig, S. A Single Nucleotide Change in the 5′ Noncoding Region of Sindbis Virus Confers Neurovirulence in Rats. J. Virol. 1999, 73, 10440–10446. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J.; Griffin, D.E.; Zhang, H.; Niesters, H.G.; Strauss, J.H. Attenuation of Sindbis virus neurovirulence by using defined mutations in nontranslated regions of the genome RNA. J. Virol. 1992, 66, 7121–7127. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.L.; Gardner, C.L.; Kimura, T.; White, J.P.; Liu, G.; Trobaugh, D.; Huang, C.; Tonelli, M.; Paessler, S.; Takeda, K.; et al. A Viral RNA Structural Element Alters Host Recognition of Nonself RNA. Science 2014, 343, 783–787. [Google Scholar] [CrossRef]
- Reynaud, J.M.; Kim, D.Y.; Atasheva, S.; Rasalouskaya, A.; White, J.P.; Diamond, M.S.; Weaver, S.C.; Frolova, E.; Frolov, I. IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon. PLoS Pathog. 2015, 11, e1004863. [Google Scholar] [CrossRef]
- Nickens, D.G.; Hardy, R.W. Structural and functional analyses of stem–loop 1 of the Sindbis virus genome. Virology 2008, 370, 158–172. [Google Scholar] [CrossRef]
- Frolov, I.; Schlesinger, S. Translation of Sindbis virus mRNA: Analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 1996, 70, 1182–1190. [Google Scholar] [CrossRef]
- Ventoso, I. Adaptive Changes in Alphavirus mRNA Translation Allowed Colonization of Vertebrate Hosts. J. Virol. 2012, 86, 9484–9494. [Google Scholar] [CrossRef]
- Lemaire, P.A.; Anderson, E.; Lary, J.; Cole, J.L. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008, 381, 351–360. [Google Scholar] [CrossRef]
- Toribio, R.; López, R.T.; Boskovic, J.; Ventoso, I. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res. 2016, 44, 4368–4380. [Google Scholar] [CrossRef]
- Sanz, M.A.; Almela, E.G.; Carrasco, L. Translation of Sindbis Subgenomic mRNA is Independent of eIF2, eIF2A and eIF2D. Sci. Rep. 2017, 7, srep43876. [Google Scholar] [CrossRef]
- Ventoso, I.; Sanz, M.A.; Molina, S.; Berlanga, J.J.; Carrasco, L.; Esteban, M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006, 20, 87–100. [Google Scholar] [CrossRef]
- Toribio, R.; Díaz-López, I.; Berlanga, J.J.; Molina-Jiménez, F.; Majano, P.; Ventoso, I. Naturally Occurring and Engineered Alphaviruses Sensitive to Double-Stranded-RNA-Activated Protein Kinase Show Restricted Translation in Mammalian Cells, Increased Sensitivity to Interferon, and Marked Oncotropism. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Pfeffer, M.; Kinney, R.M.; Rügerkaadena, O. The Alphavirus 3′-Nontranslated Region: Size Heterogeneity and Arrangement of Repeated Sequence Elements. Virology 1998, 240, 100–108. [Google Scholar] [CrossRef]
- Hardy, R.W.; Rice, C.M. Requirements at the 3′ End of the Sindbis Virus Genome for Efficient Synthesis of Minus-Strand RNA. J. Virol. 2005, 79, 4630–4639. [Google Scholar] [CrossRef]
- Chen, R.; Wang, E.; Tsetsarkin, K.A.; Weaver, S.C. Chikungunya Virus 3′ Untranslated Region: Adaptation to Mosquitoes and a Population Bottleneck as Major Evolutionary Forces. PLoS Pathog. 2013, 9, e1003591. [Google Scholar] [CrossRef]
- Garcia-Moreno, M.; Sanz, M.A.; Carrasco, L. A Viral mRNA Motif at the 3′-Untranslated Region that Confers Translatability in a Cell-Specific Manner. Implications for Virus Evolution. Sci. Rep. 2016, 6, srep19217. [Google Scholar] [CrossRef]
- Filomatori, C.V.; Merwaiss, F.; Bardossy, E.S.; Alvarez, D.E. Impact of alphavirus 3′UTR plasticity on mosquito transmission. Semin. Cell Dev. Biol. 2021, 111, 148–155. [Google Scholar] [CrossRef]
- Morley, V.J.; Noval, M.G.; Chen, R.; Weaver, S.C.; Vignuzzi, M.; Stapleford, K.A.; Turner, P.E. Chikungunya virus evolution following a large 3′UTR deletion results in host-specific molecular changes in protein-coding regions. Virus Evol. 2018, 4, vey012. [Google Scholar] [CrossRef]
- Hawman, D.W.; Carpentier, K.S.; Fox, J.; May, N.A.; Sanders, W.; Montgomery, S.A.; Moorman, N.J.; Diamond, M.S.; Morrison, T.E. Mutations in the E2 Glycoprotein and the 3′ Untranslated Region Enhance Chikungunya Virus Virulence in Mice. J. Virol. 2017, 91, e00816-17. [Google Scholar] [CrossRef]
- Garneau, N.L.; Sokoloski, K.J.; Opyrchal, M.; Neff, C.P.; Wilusz, C.J.; Wilusz, J. The 3′ Untranslated Region of Sindbis Virus Represses Deadenylation of Viral Transcripts in Mosquito and Mammalian Cells. J. Virol. 2008, 82, 880–892. [Google Scholar] [CrossRef]
- Sokoloski, K.J.; Dickson, A.M.; Chaskey, E.L.; Garneau, N.L.; Wilusz, C.J.; Wilusz, J. Sindbis Virus Usurps the Cellular HuR Protein to Stabilize Its Transcripts and Promote Productive Infections in Mammalian and Mosquito Cells. Cell Host Microbe 2010, 8, 196–207. [Google Scholar] [CrossRef]
- Dickson, A.M.; Anderson, J.R.; Barnhart, M.D.; Sokoloski, K.J.; Oko, L.; Opyrchal, M.; Galanis, E.; Wilusz, C.J.; Morrison, T.E.; Wilusz, J. Dephosphorylation of HuR Protein during Alphavirus Infection Is Associated with HuR Relocalization to the Cytoplasm*. J. Biol. Chem. 2012, 287, 36229–36238. [Google Scholar] [CrossRef]
- Trobaugh, D.; Gardner, C.L.; Sun, C.; Haddow, A.D.; Wang, E.; Chapnik, E.; Mildner, A.; Weaver, S.C.; Ryman, K.D.; Klimstra, W.B. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 2014, 506, 245–248. [Google Scholar] [CrossRef]
- Jopling, C.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Shimakami, T.; Yamane, D.; Jangra, R.K.; Kempf, B.J.; Spaniel, C.; Barton, D.J.; Lemon, S.M. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 941–946. [Google Scholar] [CrossRef]
- Trobaugh, D.W.; Sun, C.; Bhalla, N.; Gardner, C.L.; Dunn, M.D.; Klimstra, W.B. Cooperativity between the 3′ untranslated region microRNA binding sites is critical for the virulence of eastern equine encephalitis virus. PLoS Pathog. 2019, 15, e1007867. [Google Scholar] [CrossRef]
- Gardner, C.L.; Burke, C.W.; Tesfay, M.Z.; Glass, P.J.; Klimstra, W.B.; Ryman, K.D. Eastern and Venezuelan Equine Encephalitis Viruses Differ in Their Ability To Infect Dendritic Cells and Macrophages: Impact of Altered Cell Tropism on Pathogenesis. J. Virol. 2008, 82, 10634–10646. [Google Scholar] [CrossRef]
- López, P.; Girardi, E.; Mounce, B.C.; Weiss, A.; Chane-Woon-Ming, B.; Messmer, M.; Kaukinen, P.; Kopp, A.; Bortolamiol-Becet, D.; Fendri, A.; et al. High-Throughput Fluorescence-Based Screen Identifies the Neuronal MicroRNA miR-124 as a Positive Regulator of Alphavirus Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Kunec, D.; Osterrieder, N. Codon Pair Bias Is a Direct Consequence of Dinucleotide Bias. Cell Rep. 2016, 14, 55–67. [Google Scholar] [CrossRef]
- Butt, A.M.; Nasrullah, I.; Tong, Y. Genome-Wide Analysis of Codon Usage and Influencing Factors in Chikungunya Viruses. PLoS ONE 2014, 9, e90905. [Google Scholar] [CrossRef] [PubMed]
- Di Giallonardo, F.; Schlub, T.E.; Shi, M.; Holmes, E.C. Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species. J. Virol. 2017, 91, e02381-16. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Odon, V.; Fros, J.J.; Goonawardane, N.; Dietrich, I.; Ibrahim, A.; Alshaikhahmed, K.; Nguyen, D.; Simmonds, P. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res. 2019, 47, 8061–8083. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and Characterization of Interferon-Induced Proteins That Inhibit Alphavirus Replication. J. Virol. 2007, 81, 11246–11255. [Google Scholar] [CrossRef] [PubMed]
- Bick, M.J.; Carroll, J.-W.N.; Gao, G.; Goff, S.P.; Rice, C.M.; MacDonald, M.R. Expression of the Zinc-Finger Antiviral Protein Inhibits Alphavirus Replication. J. Virol. 2003, 77, 11555–11562. [Google Scholar] [CrossRef]
- Ryman, K.D.; White, L.J.; Johnston, R.E.; Klimstra, W.B. Effects of PKR/RNase L-Dependent and Alternative Antiviral Pathways on Alphavirus Replication and Pathogenesis. Viral Immunol. 2002, 15, 53–76. [Google Scholar] [CrossRef]
- Bréhin, A.-C.; Casadémont, I.; Frenkiel, M.-P.; Julier, C.; Sakuntabhai, A.; Desprès, P. The large form of human 2′,5′-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology 2009, 384, 216–222. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.M.H.; Zhao, J.; Li, S.; MacDonald, M.R.; Rice, C.M.; Gao, X.; Gao, G. Sindbis Virus Can Exploit a Host Antiviral Protein To Evade Immune Surveillance. J. Virol. 2016, 90, 10247–10258. [Google Scholar] [CrossRef]
- Jones, J.E.; Long, K.M.; Whitmore, A.C.; Sanders, W.; Thurlow, L.R.; Brown, J.A.; Morrison, C.R.; Vincent, H.; Peck, K.M.; Browning, C.; et al. Disruption of the Opal Stop Codon Attenuates Chikungunya Virus-Induced Arthritis and Pathology. mBio 2017, 8, e01456-17. [Google Scholar] [CrossRef]
- Tuittila, M.T.; Santagati, M.G.; Roytta, M.; Maatta, J.A.; Hinkkanen, A.E. Replicase Complex Genes of Semliki Forest Virus Confer Lethal Neurovirulence. J. Virol. 2000, 74, 4579–4589. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Kelly, C.L.H.; Ledermann, J.P.; Powers, A.M. Effects of an Opal Termination Codon Preceding the nsP4 Gene Sequence in the O’Nyong-Nyong Virus Genome on Anopheles gambiae Infectivity. J. Virol. 2006, 80, 4992–4997. [Google Scholar] [CrossRef]
- Li, G.P.; Rice, C.M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: Studies of translational readthrough and its effect on virus replication. J. Virol. 1989, 63, 1326–1337. [Google Scholar] [CrossRef]
- Firth, A.E.; Wills, N.M.; Gesteland, R.F.; Atkins, J.F. Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011, 39, 6679–6691. [Google Scholar] [CrossRef]
- Kendra, J.; Advani, V.M.; Chen, B.; Briggs, J.W.; Zhu, J.; Bress, H.J.; Pathy, S.M.; Dinman, J.D. Functional and structural characterization of the chikungunya virus translational recoding signals. J. Biol. Chem. 2018, 293, 17536–17545. [Google Scholar] [CrossRef]
- Kutchko, K.M.; Madden, E.; Morrison, C.; Plante, K.S.; Sanders, W.; Vincent, H.A.; Cisneros, M.C.C.; Long, K.M.; Moorman, N.J.; Heise, M.T.; et al. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res. 2018, 46, 3657–3670. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.-P.; Rehwinkel, J.; Kato, H.; Takeuchi, O.; Akira, S.; Way, M.; Schiavo, G.; Sousa, C.R.E. Activation of MDA5 Requires Higher-Order RNA Structures Generated during Virus Infection. J. Virol. 2009, 83, 10761–10769. [Google Scholar] [CrossRef]
- Rudd, P.A.; Wilson, J.; Gardner, J.; Larcher, T.; Babarit, C.; Le, T.T.; Anraku, I.; Kumagai, Y.; Loo, Y.-M.; Gale, M.; et al. Interferon Response Factors 3 and 7 Protect against Chikungunya Virus Hemorrhagic Fever and Shock. J. Virol. 2012, 86, 9888–9898. [Google Scholar] [CrossRef]
- Frolova, E.; Frolov, I.; Schlesinger, S. Packaging signals in alphaviruses. J. Virol. 1997, 71, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Firth, A.E.; Atasheva, S.; Frolova, E.I.; Frolov, I. Conservation of a Packaging Signal and the Viral Genome RNA Packaging Mechanism in Alphavirus Evolution. J. Virol. 2011, 85, 8022–8036. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E.; Chung, B.Y.; Fleeton, M.N.; Atkins, J.F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.E.; Kulcsar, K.A.; Schultz, K.L.W.; Riley, C.P.; Neary, J.T.; Marr, S.; Jose, J.; Griffin, D.E.; Kuhn, R.J. Functional Characterization of the Alphavirus TF Protein. J. Virol. 2013, 87, 8511–8523. [Google Scholar] [CrossRef] [PubMed]
- Kendra, J.A.; de la Fuente, C.; Brahms, A.; Woodson, C.; Bell, T.M.; Chen, B.; Khan, Y.A.; Jacobs, J.L.; Kehn-Hall, K.; Dinman, J.D. Ablation of Programmed −1 Ribosomal Frameshifting in Venezuelan Equine Encephalitis Virus Results in Attenuated Neuropathogenicity. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.-W.; Firth, A.E.; Atkins, J. Frameshifting in Alphaviruses: A Diversity of 3′ Stimulatory Structures. J. Mol. Biol. 2010, 397, 448–456. [Google Scholar] [CrossRef]
- Frolova, E.I.; Gorchakov, R.; Pereboeva, L.; Atasheva, S.; Frolov, I. Functional Sindbis Virus Replicative Complexes Are Formed at the Plasma Membrane. J. Virol. 2010, 84, 11679–11695. [Google Scholar] [CrossRef]
- Wielgosz, M.M.; Huang, H.V. A novel viral RNA species in Sindbis virus-infected cells. J. Virol. 1997, 71, 9108–9117. [Google Scholar] [CrossRef]
- Levin, J.G.; Friedman, R.M. Analysis of Arbovirus Ribonucleic Acid Forms by Polyacrylamide Gel Electrophoresis. J. Virol. 1971, 7, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Bruton, C.J.; Kennedy, S.I.T. Semliki Forest Virus Intracellular RNA: Properties of the Multi-stranded RNA Species and Kinetics of Positive and Negative Strand Synthesis. J. Gen. Virol. 1975, 28, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Albulescu, I.C.; Tas, A.; Scholte, F.; Snijder, E.; van Hemert, M. An in vitro assay to study chikungunya virus RNA synthesis and the mode of action of inhibitors. J. Gen. Virol. 2014, 95, 2683–2692. [Google Scholar] [CrossRef]
- Weiss, B.; Goran, D.; Cancedda, R.; Schlesinger, S. Defective Interfering Passages of Sindbis Virus: Nature of the Intracellular Defective Viral RNA. J. Virol. 1974, 14, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, S.; Weiss, B.; Dohner, D. Defective particles in alphavirus infections. Med. Biol. 1975, 53, 372–379. [Google Scholar]
- Poirier, E.; Goic, B.; Tomé-Poderti, L.; Frangeul, L.; Boussier, J.; Gausson, V.; Blanc, H.; Vallet, T.; Loyd, H.; Levi, L.I.; et al. Dicer-2-Dependent Generation of Viral DNA from Defective Genomes of RNA Viruses Modulates Antiviral Immunity in Insects. Cell Host Microbe 2018, 23, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.; Schlesinger, S. Defective interfering particles of Sindbis virus do not interfere with the homologous virus obtained from persistently infected BHK cells but do interfere with Semliki Forest virus. J. Virol. 1981, 37, 840–844. [Google Scholar] [CrossRef]
- Bruton, C.J.; Porter, A.; Kennedy, S.I.T. Defective-Interfering Particles of Semliki Forest Virus: Intracellular Events During Interference. J. Gen. Virol. 1976, 31, 397–416. [Google Scholar] [CrossRef] [PubMed]
- Levi, L.I.; Rezelj, V.V.; Henrion-Lacritick, A.; Erazo, D.; Boussier, J.; Vallet, T.; Bernhauerová, V.; Suzuki, Y.; Carrau, L.; Weger-Lucarelli, J.; et al. Defective viral genomes from chikungunya virus are broad-spectrum antivirals and prevent virus dissemination in mosquitoes. PLoS Pathog. 2021, 17, e1009110. [Google Scholar] [CrossRef]
- Langsjoen, R.M.; Muruato, A.E.; Kunkel, S.R.; Jaworski, E.; Routh, A. Differential Alphavirus Defective RNA Diversity between Intracellular and Extracellular Compartments Is Driven by Subgenomic Recombination Events. mBio 2020, 11. [Google Scholar] [CrossRef]
- Forrester, N.L.; Guerbois, M.; Adams, A.P.; Liang, X.; Weaver, S.C. Analysis of Intrahost Variation in Venezuelan Equine Encephalitis Virus Reveals Repeated Deletions in the 6-Kilodalton Protein Gene. J. Virol. 2011, 85, 8709–8717. [Google Scholar] [CrossRef] [PubMed]
- Petterson, E.; Stormoen, M.; Evensen, Ø.; Mikalsen, A.B.; Haugland, Ø. Natural infection of Atlantic salmon (Salmo salar L.) with salmonid alphavirus 3 generates numerous viral deletion mutants. J. Gen. Virol. 2013, 94, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- Portner, A.; Kingsbury, D.W. Homologous Interference by Incomplete Sendai Virus Particles: Changes in Virus-Specific Ribonucleic Acid Synthesis. J. Virol. 1971, 8, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Dimmock, N.J.; Kennedy, S.I.T. Prevention of Death in Semliki Forest Virus-Infected Mice by Administration of Defective-Interfering Semliki Forest Virus. J. Gen. Virol. 1978, 39, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D.T.; Dimmock, N.J. Modulation of Semliki Forest Virus-Induced Infection of Mice by Defective-Interfering Virus. J. Infect. Dis. 1984, 150, 98–104. [Google Scholar] [CrossRef]
- Thomson, M.; Dimmock, N. Common Sequence Elements in Structurally Unrelated Genomes of Defective Interfering Semliki Forest Virus. Virology 1994, 199, 366–375. [Google Scholar] [CrossRef]
- Thomson, M.; White, C.L.; Dimmock, N.J. The Genomic Sequence of Defective Interfering Semliki Forest Virus (SFV) Determines Its Ability to Be Replicated in Mouse Brain and to Protect against a Lethal SFV Infectionin Vivo. Virology 1998, 241, 215–223. [Google Scholar] [CrossRef][Green Version]
- Tapia, K.; Kim, W.-K.; Sun, Y.; Mercado-López, X.; Dunay, E.; Wise, M.; Adu, M.; López, C.B. Defective Viral Genomes Arising In Vivo Provide Critical Danger Signals for the Triggering of Lung Antiviral Immunity. PLoS Pathog. 2013, 9, e1003703. [Google Scholar] [CrossRef] [PubMed]
- Roux, L.; Holland, J.J. Role of defective interfering particles of sendai virus in persistent infections. Virology 1979, 93, 91–103. [Google Scholar] [CrossRef]
- Calain, P.; Monroe, M.C.; Nichol, S.T. Ebola Virus Defective Interfering Particles and Persistent Infection. Virology 1999, 262, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A. Characterization of West Nile virus persistent infections in genetically resistant and susceptible mouse cells I. Generation of defective nonplaquing virus particles. Virology 1982, 116, 84–98. [Google Scholar] [CrossRef]
- Ogura, T.; Tanaka, J.; Kamiya, S.; Sato, H.; Ogura, H.; Hatano, M. Human Cytomegalovirus Persistent Infection in a Human Central Nervous System Cell Line: Production of a Variant Virus with Different Growth Characteristics. J. Gen. Virol. 1986, 67, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Sekellick, M.J.; Marcus, P.I. Persistent infection I. Interferon-inducing defective-interfering particles as mediators of cell sparing: Possible role in persistent infection by vesicular stomatitis virus. Virology 1978, 85, 175–186. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).