Simple Summary
Our study addresses the growing number of reports of Varroa mite resistance to amitraz, which is vital for beekeepers in managing this destructive pest. Recently, beekee-pers in France and the United States have reported the diminishing effectiveness of amitraz treatments, threatening their ability to control Varroa infestations. We collected data from French apiaries in 2020 and 2021, examining the field effectiveness of amitraz treatments and conducting laboratory sensitivity tests to the pesticide. Additionally, we genotyped 240 Varroa mites from different regions in France, focusing on a specific genetic marker previously linked to amitraz resistance. Our findings revealed regional occurrences of resistance, but the decrease in treatment efficacy was less severe compared to resistance against other pesticides. Notably, our genetic analysis suggested that the previously identified genetic marker may not directly cause amitraz resistance. These findings underscore the urgent need for ongoing monitoring and the development of new strategies to manage Varroa mites, ensuring the sustainability of beekeeping. By understanding and addressing resistance, we can maintain amitraz as a reliable tool to protect bee colonies, which are crucial due to the essential pollination services they provide.
Abstract
Resistance against amitraz in Varroa mite populations has become a subject of interest in recent years due to the increasing reports of the reduced field efficacy of amitraz treatments, especially from some beekeepers in France and the United States. The loss of amitraz as a reliable tool to effectively reduce Varroa mite infestation in the field could severely worsen the position of beekeepers in the fight to keep Varroa infestation rates in their colonies at low levels. In this publication, we present data from French apiaries, collected in the years 2020 and 2021. These data include the field efficacy of an authorized amitraz-based Varroa treatment (Apivar® ,Véto-pharma, France) and the results of laboratory sensitivity assays of Varroa mites exposed to the reference LC90 concentration of amitraz. In addition, a total of 240 Varroa mites from Eastern, Central, and Southern regions in France that were previously classified as either “sensitive” or “resistant” to amitraz in a laboratory sensitivity assay were genotyped. The genetic analyses of mite samples are focused on the β-adrenergic-like octopamine receptor, which is considered as the main target site for amitraz in Varroa mites. Special attention was paid to a single-nucleotide polymorphism (SNP) at position 260 of the ORβ-2R-L gene that was previously associated to amitraz resistance in French Varroa mites, Varroa. Our findings confirm that amitraz resistance occurs in patches or “islands of resistance”, with a less severe reduction in treatment efficacy compared to pyrethroid resistance or coumaphos resistance in Varroa mites. The results of our genetic analyses of Varroa mites call into question the hypothesis of the SNP at position 260 of the ORβ-2R-L gene being directly responsible for amitraz resistance development.
1. Introduction
The efficient and sustainable control of Varroa mites (Varroa destructor) in honey bee colonies (Apis mellifera) is characterized by ongoing challenges for beekeepers at a global scale [1]. Although several Varroa treatments with different active ingredients have been available for several decades, high colony losses are periodically reported by beekeepers from different global regions [2,3,4]. Multiple factors, such as environmental changes (climate and human land use changes) [5], inter-seasonal variations in weather, the available nectar and pollen sources, as well as honey bee and Varroa population growth [6,7], and the development of acaricide resistance in Varroa mites [8], can contribute to high colony losses due to varroosis. In addition, the complete lack of control options for bee viruses, which are transmitted by Varroa mites, allows for ever-increasing viral loads in honey bee colonies that build up continuously over time [9].
In beekeeping, one of the most prominent examples of acaricide resistance development in Varroa mites is pyrethroid resistance [10]. These molecules have been primarily formulated and applied in honey bee colonies as long-term treatments (over a period of several weeks) in the form of plastic strips. Tau-fluvalinate resistance appeared in Lombardy, Italy, soon after its 1989 authorization [11], and in the United States after its 1990 approval [12], leading to severely decreased treatment efficacy by the late 1990s [13]. Varroa resistance against flumethrin, another pyrethroid that has been authorized for Varroa control in countries all over the world, has been detected repeatedly over the last few decades—sometimes as cross-resistance against both tau-fluvalinate and flumethrin [14,15,16]. Once it occurred, the resistance of Varroa mites to pyrethroids spread quickly and led to severe reductions in treatment efficacy [10,11].
The underlying genetic mechanisms leading to pyrethroid resistance in Varroa mites have been investigated extensively over the years [10,17,18,19]. Mite populations from di-fferent countries were genotyped to identify polymorphisms that could provide more information on possible resistance mechanisms at the phenotypical level. In several research studies, single-nucleotide polymorphisms (SNPs) were identified as the source of tau-fluvalinate and/or flumethrin resistance in Varroa mites. Although the location of SNPs in the mites’ genome varied geographically, the main target mechanism of the detected variations appeared to be the same: an alteration in the voltage-gated sodium channels, the main target site for pyrethroids [20,21,22].
For many years, amitraz (a formamidine) withstood any signs of resistance development in most regions where it was used for Varroa control. The majority of colonies (62%) in Europe receive treatment with amitraz, either through strips or fumigation. In eight Western European countries (cluster I), amitraz strips are predominantly utilized, whereas in seven Eastern European countries (cluster III), amitraz fumigation is the do- minant method [23]. In Poland, amitraz use in beekeeping dates to the beginning of the 1980s. Despite a now 40+ year treatment history of amitraz in the country’s bee hives, it is still applied at present, resulting in successful treatment outcomes [24]. Similarly, in Spain, amitraz-based Varroa control products date back to the late 1990s. To date, the molecule remains an effective component of Varroa control in Spanish beekeeping operations [25].
In recent years, data from the United States and France have raised concerns that amitraz resistance among Varroa mite populations in these countries appears to be deve- loping in more and more apiaries [26,27]. In the United States, many amitraz resistance reports are based on field bioassay data (also referred to as “Pettis’s test” in beekeeping, after Dr. Jeff Pettis) [27,28,29]. In France, laboratory sensitivity assays, detecting the susceptibility of Varroa mite populations to amitraz, have been utilized as the main tool to identify amitraz resistance in French Varroa mite populations [26,28,30]. Interestingly, the occurrence of amitraz resistance in apiaries across both countries has been characterized very differently to pyrethroid resistance. Instead of a resistant Varroa mite phenotype that spreads quickly within and across apiaries and leads to severe reductions in field efficacy, amitraz resistance appears to manifest in “patches” or “islands of resistance” with—at least initially—a less severe reduction in field efficacy within the affected apiaries [27].
Concerning the genetic basis of Varroa mite resistance to amitraz, similarly to pyrethroid resistance, single-nucleotide polymorphisms have been proposed as a potential cause of amitraz resistance in French and American Varroa mite populations. By comparing the sequences of octopamine and tyramine receptor (the known targets of amitraz) genes obtained from mites collected in different apiaries with different treatment regimens in France and the United States, Hernández Rodríguez et al. indeed identified two variations in the β-adrenergic-like octopamine receptor gene Orβ-2R-L potentially associated with amitraz treatment failure events [28]. The first variation, located at position 260 in the Orβ-2R-L gene (substitution of A with G at nucleotide 260 within the ORF), was exclusively detected in French mites. The resulting amino acid substitution within the β-adrenergic-like octopamine receptor, N87S (asparagine (AAT) to serine (AGT) substitution at position 87), is positioned at the end of helix II near the residues thought to constitute the binding site for octopamine. This variation is likely to disturb the correct folding of the protein and, therefore, the efficient binding of the octopamine and of its antagonist, the amitraz. The second variation identified (substitution of T with C at position 643 of the ORF) was detected only in the mites collected in the U.S. It resulted in a tyrosine to histidine substitution at position 215 (Y215H) within the octopamine receptor due to the transition in the corresponding code (TAT to CAT). Located in the fifth transmembrane segment of the protein, this substitution is predicted to strongly reduce the stability of the receptor and to affect the interaction with its ligand.
The topic of the development of resistance against active ingredients of acaricides in Varroa mite populations has received increasing attention in recent years [20,21,22,25]. There are increasing concerns among beekeepers, veterinarians, bee inspectors, and honey bee health researchers due to an increasing number of reports of amitraz resistance in Varroa mite populations from the field. Most of these reports stem from beekeeping operations in the United States and France, with ongoing investigations in Canadian beekeeping operations underway [26,27,31].
The objective of the present study was twofold:
Gain information about the spread and magnitude of amitraz resistance in the field in French apiaries.
Investigate genetic mechanisms underlying amitraz resistance in French Varroa mite populations, focusing on the single-nucleotide polymorphism at position 260 in the Orβ-2R-L gene described by Hernández Rodríguez et al. (2021) given their identification in French Varroa populations [28].
To achieve this, the following activities were carried out during the years 2020 and 2021: (1) assessment of the treatment efficacy of an authorized Varroa control product (Apivar®) in the field; (2) assessment of the laboratory sensitivity of Varroa mites to amitraz; (3) collection and analysis of genotyping data from French Varroa mite populations.
Our study focuses on variation rather than mutation, using the term “variant” to describe alterations that can be benign, pathogenic, or of unknown significance. “Variant” is increasingly replacing “mutation” in scientific discourse.
2. Materials and Methods
2.1. Organization of the Study
In 2020 and 2021, Véto-pharma conducted laboratory assays to detect Varroa mite susceptibility to amitraz, which were carried out by the Laboratoire Départemental d’Analyses du Jura (LDA39). Varroa mites were sampled from worker and drone brood frames collected from different commercial apiaries across France and sent to the LDA39. Varroa mites were then exposed to a predetermined concentration of amitraz (LC90: Lethal Concentration for 90% of the study population) in laboratory tests. The sensitivity to amitraz of Varroa populations from different regions of France was assessed by measuring the mortality rate of the mites during the assay as an indicator of susceptibility. The 2021 assays aimed to consolidate the initial results obtained by both refining the LC90 determination and conducting additional sensitivity tests with Varroa samples from 10 different beekeepers.
Varroa mites, selected from both the dead and surviving mites tested in the laboratory assays, were later genotyped by Biomnigene laboratory. In 2021, laboratory susceptibility tests were paired with field efficacy tests using the authorized Varroa treatment, Apivar®. These field tests were carried out by the French commercial beekeepers’ Association pour le Développement de l’Apiculture provençale (ADAPI) in Provence-Alpes-Côte d’Azur (PACA) and Association pour le Développement de l’Apiculture in Occitanie (ADA Occitanie).
2.2. Apiary Locations and Samples’ Origin
To determine the reference LC90, Varroa populations were gathered in May 2020 from two “organic” apiaries from Bretagne and Occitanie, and in May 2021 from two additional “organic” apiaries in Occitanie and PACA, where amitraz treatments had not been used. Samples were collected from five to six colonies within each apiary. For this preliminary investigation, Varroa mites were pooled by apiary, regardless of colony origin.
For the main susceptibility assays, Varroa mite samples were collected in June and July 2020 from 12 apiaries across 6 French regions: Nouvelle-Aquitaine, Occitanie, PACA, Auvergne-Rhône-Alpes, Centre-Val de Loire, and Grand Est. Acknowledging the inherent variability in mite sensitivity among hives within the same apiary, sensitivity evaluations were conducted at the colony level. A sample consisted of Varroa mites from one or several frames of capped brood from a single colony. Two or three colonies were randomly sampled in each apiary.
In 2021, our focus was on two key honey-producing regions in the South of France: Occitanie and PACA. Varroa mite samples were collected from 10 apiaries in these regions. In each selected apiary, mite samples were systematically gathered from six brood frames across six individual colonies. Sampling occurred over a period of five weeks, from 7 June to 6 July, with two apiaries examined per week. Given the unexpectedly low Varroa infestation observed during the initial phase, supplementary sampling sessions were conducted between 19 July and 13 September.
The field efficacy trial with Apivar® was carried out on two apiaries with 25 colonies each, located in Auzeville-Tolosane (Department 31, Occitanie) and Avignon (Department 84, PACA region). The hives of the five beekeepers from each region were moved to these two apiaries at the end of August 2021. Apivar® treatment was inserted on 31 August in Auzeville and on 14 September in Avignon.
3. Method for Laboratory Susceptibility Test
3.1. Principle of the Method
As amitraz exhibits lipophilic properties and the amitraz-based treatments used in France are based on contact action with impregnated strips [23], the methodology employed in this study to set up laboratory susceptibility tests of Varroa mites originates from a standardized procedure outlined in the Coloss Beebook, specifically tailored for fat-soluble substances acting through contact (§3.6.3.2 of the Coloss Beebook [32]). This approach draws upon the work laid by Milani [33]. The acaricide under investigation was integrated into paraffin capsules.
3.2. Preparation of Paraffin Capsules Coated with Amitraz
These paraffin capsules were prepared as described in Milani (1995) [33] and in La Santé de l’Abeille [34]. They were formed using two Na-Ca glass discs (62 mm in diameter) and one stainless steel ring (56 mm inside diameter; 2 mm thick; 5 mm high). The interior of these capsules is entirely covered by a thin layer of paraffin (Merck ref. 1.07151.1000—melting point 46–48 °C) containing a known concentration of amitraz (Amitraz Pestanal® Sigma-Aldrich, MA, USA—ref. 45323-250MG). For each preparation, 10 g of paraffin was melted in a crystallizer placed in a water bath at 60 °C, to which the required quantity of amitraz was added and dissolved in 2 mL hexane (n-Hexane; VWR; ref. 24577.323). For negative controls, hexane alone was added.
3.3. Varroa Sampling and Laboratory Exposure of Varroa Mites to Amitraz
Varroa mites were harvested within 24 h of receiving the frames or brood fragments by individually uncapping each cell. Only mature Varroa females (founder females or young mature females) extracted from capped cells were utilized for the assay. These were brought into contact with the treated paraffin. The mites’ contact time with the miticide was shortened to 1 h instead of the 4 h described in the Beebook method. Previous LDA39 trials, not described here, showed that LC90 for amitraz and for this method was at much lower levels compared to the research on pyrethroids by Milani [33]. To avoid possible biases related to the preparation of capsules at very low concentrations, the contact time of 1 h was considered more appropriate to achieve LC90 levels between 10 and 30 ppm, similar to those obtained for pyrethroids.
Following the incubation period, mites were transferred to a Petri dish containing two or three new larvae/nymphs of worker bees. Capsules and Petri dishes were incubated at 32.5 °C with a relative humidity (RH) of approximately 70%. Mortality rates were then counted 24 h later.
During observation under a binocular magnifying glass, the mites were classified into three distinct categories:
- -
- Mobile mites: mites characterized by spontaneous movement or movement in response to external stimuli, such as brushing.
- -
- Paralyzed mites: mites exhibiting the ability to move one or more appendages but an inability to relocate.
- -
- Dead mites: mites showing no observable response following three consecutive stimuli.
Assessment was conducted upon completion of exposure to the acaricide, during the transfer of mites from the capsules to Petri dishes. Mites that were inadvertently lost or fatally injured, for instance, those entrapped between the metal ring and a glass disc, were excluded from the analysis. This assessment was reiterated 24 h post-introduction into the capsules, denoted as T0 + 24. The action of amitraz, which targets the mites’ octopamine receptors at the level of synaptic transmission and which leads to paralysis of the mites, is sub-lethal [35,36]. The death of Varroa mites occurs secondarily due to starvation related to paralysis. Therefore, mites that were paralyzed after 24 h, unable to move, were counted in the same category as dead mites.
3.4. Determination of Reference LC90 Concentration of Amitraz
As this bioassay method for substances’ contact with impregned paraffin capsules had never been used on amitraz, to our knowledge, prior to conducting laboratory sensitivity tests, it was imperative to establish the reference LC90) for amitraz in susceptible Varroa mite populations. To achieve this, Varroa populations from “organic” apiaries were selected for sampling. The selected apiaries for determining the reference LC90 were chosen based on the following criteria:
- -
- A minimum interval of 5 years devoid of amitraz application was required, in a-ccordance with observations indicating a reversion of Varroa resistance in populations unexposed to pyrethroids for at least 4 years [37,38].
- -
- Recent introduction of new queens or merging with colonies exposed to amitraz within the previous 5 years was to be avoided.
- -
- Absence of prevalent diseases such as American or European foulbrood, or nosemosis, was obligatory.
- -
- Satisfactory nutritional provisions and the inclusion of renewed frames were prere-quisites.
A series of twelve concentrations, including a negative control group, underwent testing according to the following scheme:
- -
- Three replicates consisting of 10 Varroa mites per apiary were subjected to concentrations of 0, 0.5, 1, 2, 3, 5, 7.5, and 10 ppm.
- -
- Two replicates, each comprising 10 Varroa mites per apiary, were exposed to a concentration of 12.5 ppm.
- -
- A single replicate containing 10 Varroa mites per apiary was administered to concentrations of 15, 20, 50, and 100 ppm.
To enhance the resolution of data surrounding the dose eliciting a 90% mortality rate in 2020, adjustments were made to the dilution range in 2021, as follows:
- -
- Three replicates were conducted at the following concentrations: 0, 1, 5, 7.5, 10, 12.5, 15, 20, 25, and 30 ppm.
- -
- A single replicate was administered to concentrations of 40, 50, and 100 ppm.
When there were not enough Varroa mites in the brood, the tests were adapted and carried out with 8 Varroa mites instead of 10 per capsule replicate.
3.5. Laboratory Sensitivity of Varroa Mites to the LC90
For each sample, five replicates with 8 or 10 mites per replicate were exposed to the baseline LC90 for 1 h, resulting from the preliminary work. In addition, three replicates containing 8 or 10 Varroa mites served as negative controls.
3.6. Data Analysis
The percentage of miticide effect (mortality/paralysis) was calculated as follows:
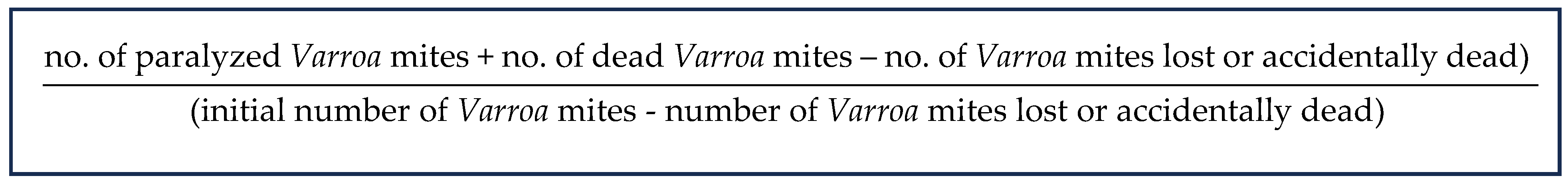
The percentage of effect obtained for the different concentrations was corrected to subtract the “natural mortality” according to the Schneider–Orelli formula: (b − k)/(1 − k) with b denoting the percentage of effect in the test and k denoting the percentage of effect in the negative control [34].
LC90 calculations were obtained using the probit transformation method. The confidence interval of the LC90 was established with a 5% risk of error. The coefficient of determination (R2) indicates the accuracy of the linear regression predictions. An R2 value close to 1 signifies a strong prediction, while a value near 0 indicates a weak prediction. The correlation coefficient (r) measures the strength of the correlation between the acaricide concentration and the percentage of effect variables. A correlation is considered strong when (r) is between 0.5 and 1, and weak when it is between 0 and 0.5.
The trials for which the percentage of effect in the negative controls was greater than 30% were invalidated according to the Beebook method (cf.§3.6.3.2 in Beebook II [32]).
Varroa mite populations were categorized into three groups according to the percentage of effect obtained at the LC90:
- -
- Sensitive population group: Varroa mites that achieved a percentage of effect at a LC90 between 75 and 100%.
- -
- Intermediate population group: Varroa mites that achieved a percentage of effect at a LC90 between 60 and 74%.
- -
- Resistant population group: Varroa mites that achieved a percentage of effect at a LC90 under 60%.
This classification was based on results from similar studies [1,4,6,8,13].
4. Genotyping of the ORβ-2R-L Gene and Potential Links to Amitraz Resistance
4.1. Genomic DNA Extraction
Genomic DNA extraction from the mites to be genotyped was carried out as described by González-Cabrera et al. (2013) [17]. Briefly, individualized mites transferred in 1.5 Eppendorf tubes were first washed with 1 mL of sterile water to remove the traces of ethanol in which they were kept until analysis. After water was removed, they were incubated at 99 °C for 10 min in 20 µL of 0.25 M NaOH. They were subsequently crushed using a plastic homogenizer and the solution was neutralized adding 10 µL of 0.25 M HCl, 5 µL of 0.5 M Tris-HCl, and 5 µL of 2% Triton X-100 before a last incubation at 99 °C for 10 min. Genomic DNA suspensions were stored at −20 °C.
4.2. Genotyping by Sanger Sequencing
Amplification of the ORβ-2R-L gene by PCR was performed in a reaction mixture of 25 µL containing 1.5 µL of genomic DNA, 12.5 µL of AccuStart II PCR SuperMix 2X (Quantabio, MA, USA) and 0.2 M of each primer. The PCR program consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 20 s, 55 °C for 20 s, and 70 °C for 1 min, and a final elongation step at 70 °C for 1 min. The primers used were ORβ-2R-L_Seq For (5′-GCTGATCTCGATCATATTGAC-3′) and ORβ-2R-L_Seq Rev (5′-CTCGAGTGGCTTGATGATCGC-3′), which were designed with the program Primer 3 u-sing a sequence of the ORβ-2R-L gene available in the NCBI database (accession number: XM_022808962.1) as a reference. They allow the amplification of a short fragment of the gene (304 bp) covering the targeted variation at position 260. After checking their length with capillary electrophoresis, PCR products were purified using 2 µL of ExoSAP ITTM (Applied Biosystems) for 5 µL of gDNA. PCR Sanger reactions were performed using 3 µL of purified fragments and the primers that were used for the amplification step. Sanger PCR products were run using SeqStudio (Applied Biosystems) and the obtained sequences were aligned with the reference sequence of the gene (accession number: XM_022808962.1).
5. Treatment Efficacy of Apivar® in the Field
Treatment Efficacy of Apivar®—Protocol 2021 (PACA and Occitanie Region)
The installation of the Apivar® treatment was carried out on August 31 in Auzeville (Occitanie region) and on 14 September in Avignon (PACA region).
Each hive was equipped with a screened bottom-board, allowing for Varroa mite counts to easily be obtained on white sheets of sticky paper. Counts were performed twice a week right after the application of treatments and once a week later on. Initial counts were conducted from 11 to 14 days before the application of the Apivar® treatment. Mite counts were then continued until three weeks after the application of the second control treatment (see below: control treatments).
On the day of application of the Apivar® strips (Day 0 or D0) and on the day of their removal (Day 70 or D70), the colonies were evaluated with the ColEval method [39], weighed, and sampled to assess the rate of phoretic Varroa mites per 100 bees (PVM100bees). On day 35 of the treatment, the strips were cleaned with a hive tool and repositioned in the center of the brood nest (Figure 1).
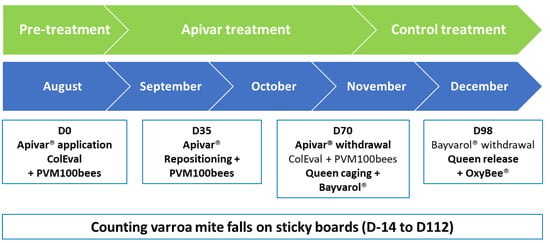
Figure 1.
Time schedule of Apivar® application in the field in the late summer of 2021.
Two control treatments were applied. When the Apivar® (D70) strips were removed, the queens were caged and four Bayvarol® (flumethrin) strips were inserted between the brood frames. After 28 days, these strips were removed, the queens released, and an Oxybee® (oxalic acid) treatment was applied at a rate of 5 mL per occupied interframe.
The average treatment efficacies, calculated using the formula below, correspond to the total number of Varroa falls observed over the 10 weeks of Apivar® treatment (Days 0 to 70 or D0–70) compared to the total number of Varroa mites fallen during the entire period (Apivar® + Bayvarol® and Oxybee® control treatments; Days 0 to 112 or D0–112).

6. Results
6.1. Laboratory Sensitivity of Varroa Mites Exposed to Amitraz LC90 and Genotyping of Varroa Mites in 2020
6.1.1. Laboratory Sensitivity of Varroa Mites to the LC90 of Amitraz (2020)
Preliminary tests for the reference LC90 of amitraz in Varroa mites conducted in 2020 resulted in the determination of an LC90 of 28 ppm [CI95 (confidence interval 95): 22–36]. The coefficient of determination (R2) obtained was 0.8, indicating a good prediction of the linear regression. The correlation coefficient (r) was 0.9, indicating a high strength of the correlation between the variables “acaricide concentration” and “percentage of mite mortality”.
Out of the 18 colonies sampled for the main sensitivity test, valid amitraz sensitivity assay results were obtained for 751 Varroa mites (excluding negative controls) from 17 co-lonies across 9 different apiaries (Figure 2). The results from one colony were excluded because the mortality in the negative control group of mites, not exposed to amitraz, was too high (>30%, see Table 1). Of these 17 colonies, mite samples from 15 colonies were considered susceptible to amitraz, as per the pre-defined mortality rate of at least 75% (Table 1). Varroa mites from 2/3 of these colonies (n = 10) showed a mortality rate of 95% or higher during and after the exposure to amitraz LC90 in the laboratory. Varroa mite samples from three colonies demonstrated a mortality rate around the expected 90% mortality, between 89% and 94%. The remaining two mite samples that were classified as susceptible to amitraz demonstrated mortality rates of 77% and 78% (Table 1).
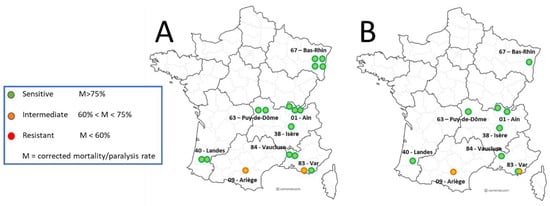
Figure 2.
Laboratory sensitivity of Varroa mites exposed to amitraz LC90 and genotyping of Varroa mites; 2020 distribution. (A) Distribution map of Varroa population phenotyping—by colony (A) and by apiary (B).

Table 1.
Overview of the mortality rate of Varroa mites after exposure to the LC90 of amitraz in the laboratory (2020). The sample size (n) represents the total number of mites tested over six French regions: Nouvelle-Aquitaine, Occitanie, Provence-Alpes-Côte d’Azur, Auvergne-Rhône-Alpes, Centre-Val de Loire, and Grand Est. The mortality rates of those total samples are given in percentage (%) for LC90 test assays and negative controls.
Varroa mites from two out of the seventeen sampled colonies were classified as showing intermediate sensitivity (60–75% mortality rate) in response to amitraz exposure in the laboratory (Table 1). None of the mite samples—and, consequently, the Varroa populations of the respective colonies—was classified as resistant against amitraz (Table 1).
The overall LC90 mortality/paralysis rate for susceptible populations is 92% (n = 751), very close to the 90% that was expected, which supports the estimate of the LC90 obtained in preliminary tests.
No correlation was found in this study between the infestation rate and the level of sensitivity of Varroa mites. The four populations with the lowest mite sensitivity results (between 73% and 78% mortality rate) come from hives with Varroa brood infestation rate estimates between 1.9% and 5.4%. The 13 most sensitive mite populations (between 89% and 100% mortality rate) corresponded to the highly variable infestation rates in the respective honey bee colonies, with values between a minimum of 1% and a maximum value of 22.8% Varroa mite infestation.
6.1.2. Genotyping of Varroa Mites Previously Tested in the 2020 Laboratory Assay
A total of 105 Varroa mites from all mites that were tested in the 2020 laboratory assays (53 and 52 mites, selected randomly among the dead and the surviving mites, respectively; see Table A1 and Table A2 for more details) were later genotyped and grouped into one of three genotypes, depending on the base pair detected at location 260 of their ORβ-2R-L gene: (1) A/A (wild-type) (n = 25), (2) A/G (heterozygous) (n = 15), and (3) G/G (homozygous variation) (n = 65). The genotyped mites originated from fifteen different colonies: twelve that were classified as sensitive and two that were classified as showing intermediate sensitivity to amitraz at the end of the laboratory sensitivity assay. The test did not provide valid results for the remaining colony (Table A1 and Table A2).
Thus, a total of 80 of the Varroa mites previously tested in the laboratory assay carried the variation at position 260 of the ORβ-2R-L gene, with 65 of them presenting a homozygous genotype.
Looking at the potential link between Varroa mite sensitivity to amitraz and the genotypes determined for 105 of the assay-tested mites, we were unable to identify a direct relationship between the variation at position 260 and a resistant phenotype in mites that had survived the assay.
Table 2 indicates that, in both groups, mites that perished during or shortly after exposure to amitraz in the laboratory and mites that survived the assay, a majority of mites carried the previously identified variation. The percentage of mites carrying the variation was very similar in both groups (75% in mites that had died after amitraz exposure and 77.3% in mites that had survived amitraz exposure), not showing a significant difference (Chi2 = 0.0029765, df = 1, p = 0.9565) (Table 2). The only significant difference (Chi2 = 7.606, df = 2, p = 0.005817) between both groups (perished and surviving mites) is the distribution of homozygous and heterozygous individuals: 24.5% of surviving mites carried the A/G genotype, whereas the same was true for only 3.8% of perished mites (Table 2). Likewise, 52.8% of mites carried the homozygous G/G genotype in the group of surviving mites, compared to 71.2% among the perished mites.

Table 2.
Genotyping results for Varroa mites, sampled after LC90 exposure to amitraz. Results show the base at position 260 of the Varroa mite ORβ-2R-L gene (2020).
6.2. Laboratory Sensitivity of Varroa Mites Exposed to Amitraz LC90, Treatment Efficacy of Apivar® in the Field, and Genotyping of Varroa Mites in 2021
6.2.1. Laboratory Sensitivity of Varroa Mites to the LC90 of Amitraz (2021)
To determine LC90, the results obtained for the two additional apiaries in 2021 were compiled with the results obtained for two other apiaries sampled in 2020, resulting in an estimate of the LC90 of 25 ppm [CI95: 21–29]. The coefficient of determination (R2) obtained was 0.7, which indicates a good prediction of linear regression, and the correlation coefficient (r) was 0.8, indicating a high correlation strength between the variables of acaricide concentration and percentage of mortality effect.
Although a total of 84 brood frames were sampled to test the laboratory sensitivity of Varroa mites to amitraz, only 25 in vitro assays resulted in valid sensitivity rates (8 samples from Occitanie and 17 samples from PACA). The other samples showed too-low infestation rates, and problems related to the excess mortality of Varroa mites in the brood during transport or in negative controls during the tests.
The average level of susceptibility of Varroa populations to LC90 (25 ppm amitraz) exposure is 76 ± 17%. This sensitivity varies from 58 ± 13% to 96 ± 6% depending on the beekeeping operation, with contrasting profiles depending on the region (87% ± 10% in Occitanie vs. 70 ± 17% in PACA) (Table 3). In total, of the 25 colonies sampled, 15 Varroa populations were classified as sensitive to amitraz (75–100% mortality), 6 were classified as intermediate (60–75% mortality), and 4 were classified as resistant (<60% mortality).

Table 3.
Overview of the mortality rate of Varroa mites after exposure to the LC90 of amitraz in the laboratory (2021). The sample size (n) represents the total number of mites tested. Mortality rates of those total samples are given in percentage (%) for LC90 test assays and negative controls. (Géographic maps.)
6.2.2. Genotyping of Varroa Mites Previously Tested in the 2021 Laboratory Assay
A total of 135 Varroa mites (59 surviving mites and 76 dead mites at the end of the laboratory sensitive assay) from eight different colonies sampled for the laboratory sensitivity test and included in the field efficacy test with Apivar® were genotyped in 2021. Two of the colonies had previously been classified as resistant in the laboratory sensitivity assay, five colonies were classified as showing intermediate sensitivity to amitraz, and the remaining one was classified as sensitive to amitraz (Table 4).

Table 4.
Overview of genotyped Varroa mites from laboratory assay samples, presenting the mites´ origin, laboratory sensitivity results of the colony, and the status of the mites after amitraz exposure in the laboratory (survivors vs. dead Varroa mites).
Similar to the genotyping results from the year 2020, the 2021 analyses revealed a higher percentage of mites carrying the homozygous G/G genotype compared to the homozygous wild-type A/A. This trend was observed in both groups of Varroa mites (those that survived and those that perished during amitraz exposure in the laboratory) (Table 5). Additionally, as in 2020, the heterozygous mites constituted the smallest portion of the three genotypes: 8.9% in 2021 vs. 14.3% in 2020 (Table 2 and Table 5).

Table 5.
Genotyping results for Varroa mites, sampled after LC90 exposure to amitraz. Results show the base at position 260 of the Varroa mite ORβ-2R-L gene (2021).
Between the Varroa mites that survived amitraz exposure in the laboratory and Varroa mites which did not survive, there was only a slight difference (Chi2 = 2.5128, df = 1, p = 0.1129) in the percentage of mites carrying the variation (G/G) with a homozygous genotype (Table 5). When referring to the heterozygous genotype, as in 2020, surviving mites were more likely to carry this genotype, at 15.3% compared to 3.9% of heterozygous mites within the pool of mites that perished during the laboratory assay exposure to amitraz.
In a more detailed portrayal of the results, Figure 3 shows the partitioning of genotypes (A/A, G/G, and A/G) in individual mite samples (survivors vs. perished mites) for each individual colony and summarized across all genotyped mites (Figure 3).
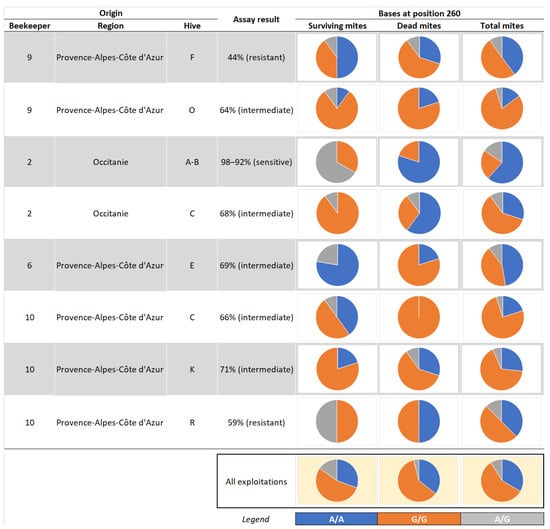
Figure 3.
Partitioning of genotypes of Varroa mite samples collected in 2021 after mites’ exposure to the LC90 of amitraz in the laboratory.
Furthermore, Table A3 presents the individual results for each genotyped Varroa mite, detailing the origin, classification after the laboratory assay (sensitive, intermediate, or resistant), and the outcome of the assay (survived vs. perished).
Additional variations were found in the ORβ-2R-L gene (nature and positions of the variations described in the table reported in Table A4). We noted that most of them were detected in single surviving mites, making it difficult to establish an association with amitraz resistance in Varroa mites. The variated bases AT at positions 344–345 (found in 19 out of the 59 surviving mites), were present in surviving mites on the same allele as base A at position 260.
6.2.3. Treatment Efficacy of Apivar® (2021) in the Occitanie and PACA Regions
Nine colonies were excluded from the final evaluation of the field efficacy: five in Occitanie and four in PACA. The reasons for exclusion included the need to replace the queen at the beginning of the test (n = 3), initial Varroa infestation being too low (<300 Varroa mites; n = 2), and significant weakening of the bee population at the end of treatment (“non-value” < 3000 bees; n = 4).
The mean efficacy in all tested colonies in both regions at the end of the 10-week treatment was 93% (SD ± 6.4) (n = 41 colonies). Twenty-one of these colonies demonstrated an efficacy of 95% or higher, seven of these in PACA and fourteen in Occitanie. Of the 20 colonies below this threshold, 9 did not achieve 90% efficiency (Figure 4). The mean field efficacy after 10 weeks in the two different regions was 95.5% (SD ± 2.7) in Occitanie (n = 14/20 > 95% Eff.; n = 2 < 90% Eff.) and 90.5% (SD ± 7.8) in PACA (n = 7/21 > 95% Eff.; n = 7 < 90% Eff.) (Table 6, Figure 5)
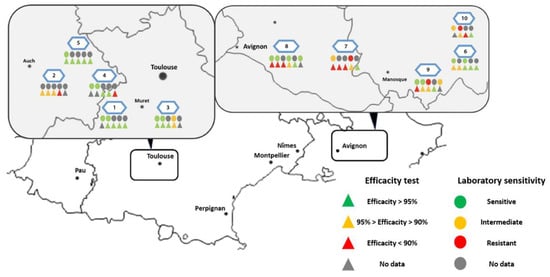
Figure 4.
Map presenting field efficacy and sensitivity results assessed per colony in each apiary.

Table 6.
Summary table of amitraz field efficacy results (SD averages, both globally and segmented by region).
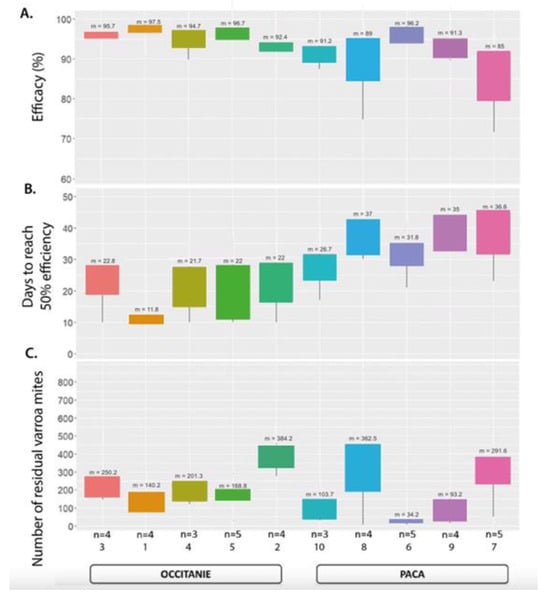
Figure 5.
Boxplots of calculated efficacy for the Apivar treatment (A), days to achieve 50% efficacy (B), and numbers of residual Varroa mites after treatment (C). Numbers (n) of hive per apiary and means (m) are indicated.
A comprehensive comparison of Varroa populations concerning amitraz sensitivity in the laboratory and Apivar® treatment efficacy in the field across 50 colonies was not fully achieved. Despite sampling 84 brood frames, only 25 in vitro assays yielded valid Varroa mite mortality rates (8 samples from Occitanie and 17 from PACA). The remaining samples were compromised by low mite infestation, excess Varroa mortality in the brood during transport, or high mortality in negative controls during the tests.
The amitraz sensitivity–Apivar® efficacy relationship could only be studied on 16 colonies (9 colonies were excluded from the field trial between Varroa mite sampling and the end of the trial). Despite the small amount of data, the results show a tendency towards better treatment efficacy in colonies with more sensitive Varroa populations (Spearman test, p < 0.05, rho = 0.5). This correlation is no longer significant when only the 11 colonies monitored in PACA are considered, revealing a regional effect.
7. Discussion
7.1. Geographic Distribution and Spread and Magnitude of Amitraz Resistance Detected in the Field in French Apiaries
Over the course of two years (2020–2021), the laboratory sensitivity of Varroa mites to amitraz and the field efficacy of an amitraz-based Varroa treatment were investigated on mite populations of 75 honey bee colonies in 19 apiaries across France. The sampled apiaries were predominantly located in the Eastern, Central, and Southern regions of France. The main reasons for the focus on these geographic regions were as follows: (1) amitraz resistance in French Varroa mite populations was first reported from the Auvergne-Rhône-Alpes (AURA) region in the east of the country [26]; (2) the southern regions of France offer a high density of honey bee colonies and commercial beekeeping operations.
The results of the laboratory assay in 2020, including Varroa mites from 17 colonies of 9 different apiaries, do not indicate widespread amitraz resistance in the sampled regions of AURA, Bretagne, Grand Est, and Occitanie. Varroa mite samples from 15 out of 17 sampled colonies were classified as “sensitive” to amitraz. However, it is important to note that, due to shipment constraints during the COVID-19 pandemic in 2020 and the resulting difficulties in testing Varroa mites for the laboratory assays when they were still alive, the total sample size of 17 colonies remains relatively low. On the other hand, the mites tested in the assay were sourced from nine apiaries across the country, including the AURA region in Eastern France and some apiaries in which resistance had previously been reported. For the investigation of the genetic basis of amitraz resistance in Varroa destructor (see below), the number of mites available for genotyping was more than adequate.
In 2021, we decided to focus on two regions in the South of France that are of high importance for the national honey production [40]: Occitanie and Provence-Alpes-Côte d’Azur (short: PACA). For this project, a field efficacy trial, consisting of a 10-week application with the authorized amitraz-based product Apivar®, was added to the labo-ratory assays. We found that, in both tests (laboratory assay and field efficacy trial), mites from the Occitanie region were largely sensitive to amitraz and the test results (mortality rate in the assay; treatment efficacy in percentage) were superior compared with the PACA region. With a treatment efficacy of 95.5% in the field, the result for the Occitanie region was compliant with the required minimum Varroa treatment efficacy of 95% for synthetic acaricides defined by the European Medicines Agency (EMA) [41]. The result of 90.5% efficacy in the PACA region was below the required minimum efficacy for synthetic treatments—the standard for organic Varroa treatments is slightly lower, with a required minimum efficacy of 90% [41].
The difference in field efficacy between the PACA and Occitanie regions may be explained by the regional differences in treatment history. Participating beekeepers from the Occitanie region reported that they had treated the honey bee colonies included in this trial with Apivar® strips for 10–14 weeks in previous years (maximum treatment period of Apivar® as per product label: 10 weeks). On the other hand, beekeepers from the PACA region reported that they had treated the colonies included in the present trial with Apivar® for up to 24 weeks in previous years [42]. The prolonged and possibly repeated exposure of Varroa mite populations to an amitraz-based treatment which, in France, is authorized for a maximum treatment period of 10 weeks, could have contributed to the desensitization of Varroa mites in the PACA region to amitraz. For a more detailed analysis of the correlation between the off-label use of authorized and unauthorized amitraz-based Varroa treatments, we suggest further investigations including a higher sample size of honey bee colonies with a known treatment history. It is crucial for honey bee health, as well as the global beekeeping industry, to determine the potential long-term outcomes of the off-label use of Varroa treatments and raise awareness of the consequences of risking desensitization of Varroa mites against present and future treatment options.
Overall, the results of the present field efficacy trial support the hypotheses presented by Rinkevich (2020) [27]: 1. amitraz resistance occurs in “patches” or “islands” of resistance, rather than affecting all honey bee colonies located in a single apiary or even broader geographic regions within a short period of time; 2. the reduction in amitraz efficacy in the field clearly develops more slowly than pyrethroid or coumaphos resistance in Varroa mites [11,13,31].
In addition, this trial demonstrated that, across regions, the measured mortality rate of mites in the laboratory assay was lower compared to the corresponding field efficacy result (in percentage) after treatment with Apivar®. This indicates that mite sensitivity is likely not the only relevant factor for treatment efficacy in the field. This finding confirms similar results obtained in previous studies where field efficacy was compared to the mortality rate of Varroa mites in sensitivity assays [14]. A global result of 93% field efficacy (n = 41) corresponded to a laboratory mortality rate of 77.3% (n = 16). Even when we consider that the standard for treatment efficacy in the field is 95% mortality, and treatments such as Apivar® have been formulated accordingly, when we exposed mites in the laboratory to the LC90, the mean difference between field efficacy and laboratory mortality was slightly higher than expected. This could be due to different factors, such as a higher natural mite mortality rate in honey bee colonies in the field [14] or a lack of long-term exposure of Varroa mites to amitraz in the laboratory, which does not correspond to the weeks-long treatment exposure in the field and therefore does not result in comparable mite mortality. While conditions in a live honey bee colony are clearly more variable than the standardized conditions in the laboratory, the mites have much more opportunity for avoidance behavior inside a honey bee colony compared to a spatially limited paraffin capsule in the laboratory. This contradicts the observed reduced laboratory mortality compared to the exposure in a honey bee colony. Future studies on Varroa mite resistance should consider the difference between laboratory mortality and field efficacy—which has been observed before—carefully.
7.2. Genotyping of the ORβ-2R-L Gene and Potential Links to Amitraz Resistance
The genotyping analyses carried out in 2020 and 2021 included a total of 240 Varroa mites from 14 French apiaries in the eastern, central, and southern regions of France. The sequences constituted for all mites were compared to the sequence of the ORβ-2R-L gene referenced in NCBI.
The substitution of A with G at position 260 described by Hernández Rodríguez et al. [24] was identified, with a strong representativeness (70.8%) in all analyzed samples (70.8%), whatever their geographic localization and the status of their original colony, as defined by laboratory sensitivity tests (sensitive, intermediate, or resistant). Nevertheless, we were not able to establish a direct link between the presence of this variation and the development of a resistance to amitraz. Indeed, although many of the surviving mites of laboratory and field bioassays clearly carried the variation, no clear pattern in the distribution of the genotypes between perished and surviving mites was possible. Surviving mites did not carry the variation more often than mites that had died from amitraz exposure. Notably, out of the 170 mites carrying the variation at position 260, 143 (84.12%) were homozygous and 27 (15.88%) were heterozygous. As previously, it was difficult to establish a correlation between these two variated genotypes and a possible resistance to amitraz due to their similar distribution between the surviving and the dead mites.
Genotyping analyses allowed for the identification of other, previously undescribed variations in the ORβ-2R-L gene (Table A4). Two of them particularly drew our attention: the variations located at positions 344–345 that were detected in the Varroa mites analyzed in 2021 and were more frequently present in the “resistant” mites (ratio 19/3 between surviving and dead mites), suggesting a potential involvement in the acquisition of the resistance to amitraz. This hypothesis would merit further exploration. The involvement of these variations in the development of resistance would be inopportune given their low representativity (variations detected in 9.17% of the mites analyzed).
The genome of V. destructor was, for a long time, believed to be particularly stable. Genetic bottleneck events, haplodiploid sex determination, and the mite’s sibling system contribute to this low genetic diversity [43]. However, in recent years, several studies have demonstrated that the genetic variation of the Varroa mite genome is higher than previously suspected, and that mite populations can differ significantly with respect to certain traits. V. destructor has as original obligate host the Asian honey bee, A. cerana (Hymenoptera; Apidae) [44]. In 1952, it switched from A. cerana to A. mellifera colonies in eastern Russia, producing what was named the Korean haplotype of the mite [45]. Another switch event occurred around 1957, leading to the creation of a second haplotype, the Japanese haplotype [45]. Both haplotypes (with a dominance of the Korean haplotype) first colonized the west of their countries of origin [46,47,48,49] and they are now present all around the world. These host switch events did not occur without any genetic changes. Several variants of both haplotypes were identified in different areas of Asia [49]. A more recent study suggests that the genomic diversity of the populations across the world could be even higher [50,51,52,53]. Whole genome analyses of 63 V. destructor and V. jacobsoni mites collected in their native ranges from both their ancestral and novel hosts allowed the identification of previously undiscovered mitochondrial lineages on the novel host, as well as the equivalent of tens of individuals involved in the initial host switch, demonstrating that a modest gene flow remains between mites adapted to their host [50]. Analyses of the fine scale population structure of V. destructor in a managed apiary setting highlighted the hierarchical genetic variation between apiaries, between colonies within an apiary, and even within colonies [54]. The authors also reported a modest increase in the total variation over time within individuals, possibly due to the de novo generation of diversity or, more probably, (due to the short time scales of the study) to the horizontal transmission of mites between colonies [54]. It is also now well known that a few populations of A. mellifera can survive V. destructor infestation without treatments by means of natural selection. A study was designed in 2016 to investigate potential genetic changes in mite populations from Sweden occurring in response to their host adaptation [55]. A comparison of mites collected in 2009 and 2017–2018 from Varroa-resistant A. mellifera populations and from neighboring mite-susceptible colonies highlighted significant changes in the genetic structure of the mites during the time frame of the study, with more pronounced variations in the V. destructor population collected in the mite-resistant honeybee colonies. This suggests that, like other parasites such as human lice [56], the human plasmodia, or mammal Leishmania [57], V. destructor reciprocally adapts to its host’s adaptations [55], and this coevolution occurs under the direct influence of environmental factors (climatic conditions, food resources, exposure to pesticides, and pollution). The variations that appeared in the genome of V. destructor following treatments with acaricides to develop resistance are perfect examples [24]. Another source of V. destructor genome variability is the possible hybridization with V. jacobsoni [58], another Varroa species mostly found within A. cerana but which causes less damage compared to V. destructor. With 99.7% of their genomes in common, both species are very similar genetically [59]. All events detailed above are all arguments that could explain the presence and the geographic specificity of the variation at position 260 (as well as the variations at positions 344–345), in addition to the development of a resistance to amitraz for which no correlation has been clearly established in this study.
8. Conclusions
The present results call into question the hypothesis of a single-nucleotide polymorphism as the direct cause of amitraz resistance in Varroa mites, while an association between amitraz resistance and the occurrence of the variation at position 260 cannot be completely ruled out. However, we suspect that amitraz resistance development is more likely to include additional genetic variants and evolutionary steps, thus making the detection of amitraz resistance by markers more complex than detecting the presence of a SNP.
Further, the potential effects of off-label treatments (exposure to higher doses, a longer exposure period than recommended, the use of unauthorized treatments etc.) on resistance development should be the focus of future investigations.
Author Contributions
U.M. performed the study and was a major contributor in writing the manuscript, B.R. performed the genetic analysis, and wrote part of the materials and methods and part of the results. A.D. performed and analyzed the genetics. A.V. analyzed and interpreted the sensibility data (LDA39 expert) and wrote part of the materials and methods. M.A.R.R. interpretated and reviewed the manuscript. A.H. participated in reviewing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been sponsored by Veto-pharma.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Authors used R to perform the Chi-square tests. RStudio 2023.09.0+463 “Desert Sunflower” Release (b51c81cc303d4b52b010767e5b30438beb904641, 2023-09-25) for windows. Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) RStudio/2023.09.0+463 Chrome/114.0.5735.289 Electron/25.5.0 Safari/537.36.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its.
Acknowledgments
We would like to thank Biomnigene for performing genotyping assays and statistical analyses and LDA39 for performing laboratory sensitivity tests. We also appreciate the collaboration of Alexandre Douablin, Bénédicte Rognon, and Alain Viry in helping with the writing and paper review.
Conflicts of Interest
Authors U.M., M.A.R.R. and A.H. were employed by the company Véto-pharma. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix A

Table A1.
Table showing the mites´ origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa surviving mite samples collected in 2020.
Table A1.
Table showing the mites´ origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa surviving mite samples collected in 2020.
| Origin | Sensitivity Test | Genotyping Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| % of Mites Dead 24 h after Exposure at the LC90 | Population Classification | Number of Surviving Mites Analyzed | Biomnigene Identifiant | Base at Position 260 | |||||
| Apiary | Town | Hive | A/A | G/G | A/G | ||||
| 11 | SAINT-AUBIN | A | 89 | sensitive | 2 | R1 | X | ||
| R2 | X | ||||||||
| 11 | SAINT-AUBIN | B | 78 | sensitive | 6 | R3 | X | ||
| R4 | X | ||||||||
| R5 | X | ||||||||
| R6 | X | ||||||||
| R7 | X | ||||||||
| R8 | X | ||||||||
| 12 | COUSSA | C | 73 | intermediary | 10 | R9 | X | ||
| R10 | X | ||||||||
| R11 | X | ||||||||
| R12 | X | ||||||||
| R13 | X | ||||||||
| R14 | X | ||||||||
| R15 | X | ||||||||
| R16 | X | ||||||||
| R17 | X | ||||||||
| R18 | X | ||||||||
| 13 | ENTRAIGUES | D | 95 | sensitive | 2 | R19 | X | ||
| R20 | X | ||||||||
| 13 | ENTRAIGUES | E | 98 | sensitive | 1 | R21 | X | ||
| 14 | LE-REVEST-LES-EAUX | F | 95 | sensitive | 2 | R22 | X | ||
| R23 | X | ||||||||
| 14 | LE-REVEST-LES-EAUX | G | 74 | intermediary | 1 | R24 | X | ||
| 15 | ST GEORGES D'ESPERANCHE | H | 77 | sensitive | 12 | R25 | X | ||
| R26 | X | ||||||||
| R27 | X | ||||||||
| R28 | X | ||||||||
| R29 | X | ||||||||
| R30 | X | ||||||||
| R31 | X | ||||||||
| R32 | X | ||||||||
| R33 | X | ||||||||
| R34 | X | ||||||||
| R35 | X | ||||||||
| R36 | X | ||||||||
| 16 | JASSERON | L | ND | ND | 4 | R37 | X | ||
| R38 | X | ||||||||
| R39 | X | ||||||||
| R40 | X | ||||||||
| 16 | JASSERON | K | 91 | sensitive | 6 | R41 | X | ||
| R42 | X | ||||||||
| R43 | X | ||||||||
| R44 | X | ||||||||
| R45 | X | ||||||||
| R46 | X | ||||||||
| 17 | VILLEREVERSURE | n° 2 | 78 | sensitive | 2 | R47 | X | ||
| R48 | X | ||||||||
| 17 | VILLEREVERSURE | n° 3 | 94 | sensitive | 2 | R49 | X | ||
| R50 | X | ||||||||
| 18 | ORLEAT | M | 97 | sensitive | 1 | R51 | X | ||
| 18 | ORLEAT | N | 98 | sensitive | 1 | R52 | X | ||
| 19 | MAISONSGOUTTE | Green | 96 | sensitive | 1 | R53 | X | ||
| N | 12 | 28 | 13 | ||||||
| % | 22.6 | 52.8 | 24.5 | ||||||

Table A2.
Table showing the mites origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa dead mite samples collected in 2020.
Table A2.
Table showing the mites origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa dead mite samples collected in 2020.
| Origin | Sensitivity Test | Genotyping Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| % of Mites dead 24 h after Exposure at the LC90 | Population Classification | Number of Dead Mites Analyzed | Biomnigene Identifiant | Base at Position 260 | |||||
| Exploitation Name | Town | Hive | A/A | G/G | A/G | ||||
| 11 | SAINT-AUBIN | n° 1 | 89 | sensitive | 4 | S1 | X | ||
| S2 | X | ||||||||
| S3 | X | ||||||||
| S4 | X | ||||||||
| 11 | SAINT-AUBIN | n° 2 | 78 | sensitive | 6 | S5 | X | ||
| S6 | X | ||||||||
| S7 | X | ||||||||
| S8 | X | ||||||||
| S9 | X | ||||||||
| S10 | X | ||||||||
| 12 | COUSSA | n° 72 | 73 | intermediary | 10 | S11 | X | ||
| S12 | X | ||||||||
| S13 | X | ||||||||
| S14 | X | ||||||||
| S15 | X | ||||||||
| S16 | X | ||||||||
| S17 | X | ||||||||
| S18 | X | ||||||||
| S19 | X | ||||||||
| S20 | X | ||||||||
| 15 | ST GEORGES D'ESPERANCHE | n° 3 | 77 | sensitive | 12 | S21 | X | ||
| S22 | X | ||||||||
| S23 | X | ||||||||
| S24 | X | ||||||||
| S25 | X | ||||||||
| S26 | X | ||||||||
| S27 | X | ||||||||
| S28 | X | ||||||||
| S29 | X | ||||||||
| S30 | X | ||||||||
| S31 | X | ||||||||
| S32 | X | ||||||||
| 16 | JASSERON | n° 4 | ND | ND | 2 | S33 | X | ||
| S34 | X | ||||||||
| 16 | JASSERON | n° 6 | 91 | sensitive | 8 | S35 | X | ||
| S36 | X | ||||||||
| S37 | X | ||||||||
| S38 | X | ||||||||
| S39 | X | ||||||||
| S40 | X | ||||||||
| S41 | X | ||||||||
| S42 | X | ||||||||
| 17 | VILLEREVERSURE | n° 2 | 78 | sensitive | 4 | S43 | X | ||
| S44 | X | ||||||||
| S45 | X | ||||||||
| S46 | X | ||||||||
| 17 | VILLEREVERSURE | n° 3 | 94 | sensitive | 6 | S47 | X | ||
| S48 | X | ||||||||
| S49 | X | ||||||||
| S50 | X | ||||||||
| S51 | X | ||||||||
| S52 | X | ||||||||
| N | 13 | 37 | 2 | ||||||
| % | 25.0 | 71.2 | 3.8 | ||||||

Table A3.
Table showing the mites´ origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa mite samples collected in 2021.
Table A3.
Table showing the mites´ origin, classification after the laboratory assay (sensitive, intermediate, or resistant) as well as genotyping results for Varroa mite samples collected in 2021.
| Origin | Assay Results | Number of Mites Analyzed | Genotyping Result | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Surviving Mites | Dead Mites | Identifiant | Bases at Position 260 | ||||||
| Beekeeper | Town | Hive | A/A | G/G | A/G | ||||
| 9 | MONTAGNAC-MONTPEZAT | F | 44% (resistant) | 10 | V3.1 | X | |||
| V3.2 | X | ||||||||
| V3.3 | X | ||||||||
| V3.4 | X | ||||||||
| V3.5 | X | ||||||||
| V3.6 | X | ||||||||
| V3.7 | X | ||||||||
| V3.8 | X | ||||||||
| V3.9 | X | ||||||||
| V3.10 | X | ||||||||
| 9 | MONTAGNAC-MONTPEZAT | F | 44% (resistant) | 10 | V3.11 | X | |||
| V3.12 | X | ||||||||
| V3.13 | X | ||||||||
| V3.14 | X | ||||||||
| V3.15 | X | ||||||||
| V3.16 | X | ||||||||
| V3.17 | X | ||||||||
| V3.18 | X | ||||||||
| V3.19 | X | ||||||||
| V3.20 | X | ||||||||
| 9 | MONTAGNAC-MONTPEZAT | O | 64% (intermediate) | 10 | V3.21 | X | |||
| V3.22 | X | ||||||||
| V3.23 | X | ||||||||
| V3.24 | X | ||||||||
| V3.25 | X | ||||||||
| V3.26 | X | ||||||||
| V3.27 | X | ||||||||
| V3.28 | X | ||||||||
| V3.29 | X | ||||||||
| V3.30 | X | ||||||||
| 9 | MONTAGNAC-MONTPEZAT | O | 64% (intermediate) | 10 | V3.31 | X | |||
| V3.32 | X | ||||||||
| V3.33 | X | ||||||||
| V3.34 | X | ||||||||
| V3.35 | X | ||||||||
| V3.36 | X | ||||||||
| V3.37 | X | ||||||||
| V3.38 | X | ||||||||
| V3.39 | X | ||||||||
| V3.40 | X | ||||||||
| 2 | CLERMONT-LE-FORT | A | 98-92% (sensitive) | 3 | V3.41 | X | |||
| V3.42 | X | ||||||||
| V3.43 | X | ||||||||
| 2 | CLERMONT-LE-FORT | A | 98-92% (sensitive) | 10 | V3.44 | X | |||
| V3.45 | X | ||||||||
| V3.46 | X | ||||||||
| V3.47 | X | ||||||||
| V3.48 | X | ||||||||
| V3.49 | X | ||||||||
| V3.50 | X | ||||||||
| V3.51 | X | ||||||||
| V3.52 | X | ||||||||
| V3.53 | X | ||||||||
| 2 | CLERMONT-LE-FORT | B | 68% (intermediate) | 10 | V3.54 | X | |||
| V3.55 | X | ||||||||
| V3.56 | X | ||||||||
| V3.57 | X | ||||||||
| V3.58 | X | ||||||||
| V3.59 | X | ||||||||
| V3.60 | X | ||||||||
| V3.61 | X | ||||||||
| V3.62 | X | ||||||||
| V3.63 | X | ||||||||
| 2 | CLERMONT-LE-FORT | B | 68% (intermediate) | 10 | V3.64 | X | |||
| V3.65 | X | ||||||||
| V3.66 | X | ||||||||
| V3.67 | X | ||||||||
| V3.68 | X | ||||||||
| V3.69 | X | ||||||||
| V3.70 | X | ||||||||
| V3.71 | X | ||||||||
| V3.72 | X | ||||||||
| V3.73 | X | ||||||||
| 6 | ENTRESSEN | E | 69% (intermediate) | 9 | V3.74 | X | |||
| V3.75 | X | ||||||||
| V3.76 | X | ||||||||
| V3.77 | X | ||||||||
| V3.78 | X | ||||||||
| V3.79 | X | ||||||||
| V3.80 | X | ||||||||
| V3.81 | X | ||||||||
| V3.82 | X | ||||||||
| 6 | ENTRESSEN | E | 69% (intermediate) | 10 | V3.83 | X | |||
| V3.84 | X | ||||||||
| V3.85 | X | ||||||||
| V3.86 | X | ||||||||
| V3.87 | X | ||||||||
| V3.88 | X | ||||||||
| V3.89 | X | ||||||||
| V3.90 | X | ||||||||
| V3.91 | X | ||||||||
| V3.92 | X | ||||||||
| 10 | VACHERE | C | 66% (intermediate) | 10 | V3.93 | X | |||
| V3.94 | X | ||||||||
| V3.95 | X | ||||||||
| V3.96 | X | ||||||||
| V3.97 | X | ||||||||
| V3.98 | X | ||||||||
| V3.99 | X | ||||||||
| V3.100 | X | ||||||||
| V3.101 | X | ||||||||
| V3.102 | X | ||||||||
| 10 | VACHERE | C | 66% (intermediate) | 10 | V3.103 | X | |||
| V3.104 | X | ||||||||
| V3.105 | X | ||||||||
| V3.106 | X | ||||||||
| V3.107 | X | ||||||||
| V3.108 | X | ||||||||
| V3.109 | X | ||||||||
| V3.110 | X | ||||||||
| V3.111 | X | ||||||||
| V3.112 | X | ||||||||
| 10 | VACHERE | K | 71% (intermediate) | 5 | V3.113 | X | |||
| V3.114 | X | ||||||||
| V3.115 | X | ||||||||
| V3.116 | X | ||||||||
| V3.117 | X | ||||||||
| 10 | VACHERE | K | 71% (intermediate) | 10 | V3.118 | X | |||
| V3.119 | X | ||||||||
| V3.120 | X | ||||||||
| V3.121 | X | ||||||||
| V3.122 | X | ||||||||
| V3.123 | X | ||||||||
| V3.124 | X | ||||||||
| V3.125 | X | ||||||||
| V3.126 | X | ||||||||
| V3.127 | X | ||||||||
| 10 | VACHERE | R | 59% (resistant) | 2 | V3.128 | X | |||
| V3.129 | X | ||||||||
| 10 | VACHERE | R | 59% (resistant) | 6 | V3.130 | X | |||
| V3.131 | X | ||||||||
| V3.132 | X | ||||||||
| V3.133 | X | ||||||||
| V3.134 | X | ||||||||
| V3.135 | X | ||||||||
| Survivors | 18 | 32 | 9 | ||||||
| Dead mites | 27 | 46 | 3 | ||||||
| Total mites | 45 | 78 | 12 | ||||||

Table A4.
Table reporting the nature and positions of the variations found in the 135 sequenced samples in 2021.
Table A4.
Table reporting the nature and positions of the variations found in the 135 sequenced samples in 2021.
| Origin | Assay Results | Number of Mites Analyzed | Genotyping Results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Surviving Mites | Dead Mites | Identifiant | Bases of Interest at 260 | Other Mutations Found (Mutated base(s)^wt base(s) (position)) | ||||||||||||||
| Beekeeper | Town | Hive | A/A | G/G | A/G | W^T (146) | R^G (235) | T^C (248) | R^G (262) | R^A (320) | AT^CA (344/345) | MW^CA (344-345) | Y^C (366) | Y^T (375) | ||||
| 9 | MONTAGNAC-MONTPEZAT | F | 44% (resistant) | 10 | V3.1 | X | X | |||||||||||
| V3.2 | X | |||||||||||||||||
| V3.3 | X | X | ||||||||||||||||
| V3.4 | X | X | ||||||||||||||||
| V3.5 | X | X | ||||||||||||||||
| V3.6 | X | |||||||||||||||||
| V3.7 | X | X | ||||||||||||||||
| V3.8 | X | X | X | |||||||||||||||
| V3.9 | X | |||||||||||||||||
| V3.10 | X | |||||||||||||||||
| 9 | MONTAGNAC-MONTPEZAT | F | 44% (resistant) | 10 | V3.11 | X | ||||||||||||
| V3.12 | X | |||||||||||||||||
| V3.13 | X | |||||||||||||||||
| V3.14 | X | |||||||||||||||||
| V3.15 | X | |||||||||||||||||
| V3.16 | X | |||||||||||||||||
| V3.17 | X | |||||||||||||||||
| V3.18 | X | |||||||||||||||||
| V3.19 | X | X | ||||||||||||||||
| V3.20 | X | |||||||||||||||||
| 9 | MONTAGNAC-MONTPEZAT | O | 64% (intermediate) | 10 | V3.21 | X | ||||||||||||
| V3.22 | X | |||||||||||||||||
| V3.23 | X | |||||||||||||||||
| V3.24 | X | |||||||||||||||||
| V3.25 | X | |||||||||||||||||
| V3.26 | X | |||||||||||||||||
| V3.27 | X | X | X | |||||||||||||||
| V3.28 | X | |||||||||||||||||
| V3.29 | X | |||||||||||||||||
| V3.30 | X | |||||||||||||||||
| 9 | MONTAGNAC-MONTPEZAT | O | 64% (intermediate) | 10 | V3.31 | X | ||||||||||||
| V3.32 | X | |||||||||||||||||
| V3.33 | X | |||||||||||||||||
| V3.34 | X | |||||||||||||||||
| V3.35 | X | |||||||||||||||||
| V3.36 | X | |||||||||||||||||
| V3.37 | X | |||||||||||||||||
| V3.38 | X | |||||||||||||||||
| V3.39 | X | |||||||||||||||||
| V3.40 | X | |||||||||||||||||
| Genotyping Results | ||||||||||||||||||
| Identifiant | Bases of interest at 260 | Other mutations found (mutated base(s)^wt base(s) (position)) | ||||||||||||||||
| A/A | G/G | A/G | W^T (146) | R^G (235) | T^C (248) | R^G (262) | R^A (320) | AT^CA (344/345) | MW^CA (344-345) | Y^C (366) | Y^T (375) | |||||||
| 2 | CLERMONT-LE-FORT | A | 98-92% (sensitive) | 3 | V3.41 | X | ||||||||||||
| V3.42 | X | X | X | |||||||||||||||
| V3.43 | X | |||||||||||||||||
| 2 | CLERMONT-LE-FORT | A | 98-92% (sensitive) | 10 | V3.44 | X | ||||||||||||
| V3.45 | X | |||||||||||||||||
| V3.46 | X | |||||||||||||||||
| V3.47 | X | |||||||||||||||||
| V3.48 | X | |||||||||||||||||
| V3.49 | X | |||||||||||||||||
| V3.50 | X | |||||||||||||||||
| V3.51 | X | |||||||||||||||||
| V3.52 | X | |||||||||||||||||
| V3.53 | X | |||||||||||||||||
| 2 | CLERMONT-LE-FORT | B | 68% (intermediate) | 10 | V3.54 | X | ||||||||||||
| V3.55 | X | |||||||||||||||||
| V3.56 | X | X | X | |||||||||||||||
| V3.57 | X | |||||||||||||||||
| V3.58 | X | |||||||||||||||||
| V3.59 | X | |||||||||||||||||
| V3.60 | X | |||||||||||||||||
| V3.61 | X | |||||||||||||||||
| V3.62 | X | |||||||||||||||||
| V3.63 | X | |||||||||||||||||
| 2 | CLERMONT-LE-FORT | B | 68% (intermediate) | 10 | V3.64 | X | ||||||||||||
| V3.65 | X | |||||||||||||||||
| V3.66 | X | |||||||||||||||||
| V3.67 | X | |||||||||||||||||
| V3.68 | X | |||||||||||||||||
| V3.69 | X | |||||||||||||||||
| V3.70 | X | |||||||||||||||||
| V3.71 | X | |||||||||||||||||
| V3.72 | X | |||||||||||||||||
| V3.73 | X | |||||||||||||||||
| 6 | ENTRESSEN | E | 69% (intermediate) | 9 | V3.74 | X | X | |||||||||||
| V3.75 | X | X | ||||||||||||||||
| V3.76 | X | X | ||||||||||||||||
| V3.77 | X | X | ||||||||||||||||
| V3.78 | X | X | ||||||||||||||||
| V3.79 | X | X | ||||||||||||||||
| V3.80 | X | X | ||||||||||||||||
| V3.81 | X | X | ||||||||||||||||
| V3.82 | X | X | ||||||||||||||||
| 6 | ENTRESSEN | E | 69% (intermediate) | 10 | V3.83 | X | ||||||||||||
| V3.84 | X | |||||||||||||||||
| V3.85 | X | |||||||||||||||||
| V3.86 | X | X | ||||||||||||||||
| V3.87 | X | |||||||||||||||||
| V3.88 | X | X | ||||||||||||||||
| V3.89 | X | |||||||||||||||||
| V3.90 | X | |||||||||||||||||
| V3.91 | X | |||||||||||||||||
| V3.92 | X | |||||||||||||||||
| Genotyping results | ||||||||||||||||||
| Identifiant | Bases of interest at 260 | Other mutations found (mutated base(s)^wt base(s) (position)) | ||||||||||||||||
| A/A | G/G | A/G | W^T (146) | R^G (235) | T^C (248) | R^G (262) | R^A (320) | AT^CA (344/345) | MW^CA (344-345) | Y^C (366) | Y^T (375) | |||||||
| 10 | VACHERE | C | 66% (intermediate) | 10 | V3.93 | X | X | |||||||||||
| V3.94 | X | |||||||||||||||||
| V3.95 | X | X | ||||||||||||||||
| V3.96 | X | |||||||||||||||||
| V3.97 | X | |||||||||||||||||
| V3.98 | X | X | ||||||||||||||||
| V3.99 | X | |||||||||||||||||
| V3.100 | X | |||||||||||||||||
| V3.101 | X | |||||||||||||||||
| V3.102 | X | |||||||||||||||||
| 10 | VACHERE | C | 66% (intermediate) | 10 | V3.103 | X | ||||||||||||
| V3.104 | X | |||||||||||||||||
| V3.105 | X | |||||||||||||||||
| V3.106 | X | |||||||||||||||||
| V3.107 | X | |||||||||||||||||
| V3.108 | X | |||||||||||||||||
| V3.109 | X | |||||||||||||||||
| V3.110 | X | |||||||||||||||||
| V3.111 | X | |||||||||||||||||
| V3.112 | X | |||||||||||||||||
| 10 | VACHERE | K | 71% (intermediate) | 5 | V3.113 | X | ||||||||||||
| V3.114 | X | |||||||||||||||||
| V3.115 | X | |||||||||||||||||
| V3.116 | X | |||||||||||||||||
| V3.117 | X | X | ||||||||||||||||
| 10 | VACHERE | K | 71% (intermediate) | 10 | V3.118 | X | ||||||||||||
| V3.119 | X | |||||||||||||||||
| V3.120 | X | |||||||||||||||||
| V3.121 | X | |||||||||||||||||
| V3.122 | X | |||||||||||||||||
| V3.123 | X | |||||||||||||||||
| V3.124 | X | |||||||||||||||||
| V3.125 | X | |||||||||||||||||
| V3.126 | X | |||||||||||||||||
| V3.127 | X | |||||||||||||||||
| 10 | VACHERE | R | 59% (resistant) | 2 | V3.128 | X | ||||||||||||
| V3.129 | X | X | ||||||||||||||||
| 10 | VACHERE | R | 59% (resistant) | 6 | V3.130 | X | ||||||||||||
| V3.131 | X | |||||||||||||||||
| V3.132 | X | |||||||||||||||||
| V3.133 | X | |||||||||||||||||
| V3.134 | X | |||||||||||||||||
| V3.135 | X | |||||||||||||||||
References
- Mondet, F.; Parejo, M.; Meixner, M.D.; Costa, C.; Kryger, P.; Andonov, S.; Servin, B.; Basso, B.; Bieńkowska, M.; Bigio, G.; et al. Evaluation of suppressed mite reproduction (SMR) reveals potential for Varroa resistance in European honey bees (Apis mellifera L.). Insects 2020, 11, 595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bruckner, S.; Wilson, M.; Aurell, D.; Rennich, K.; Vanengelsdorp, D.; Steinhauer, N.; Williams, G.R. A national survey of managed honey bee colony losses in the USA: Results from the Bee Informed Partnership for 2017–2018, 2018–2019, and 2019–2020. J. Apic. Res. 2023, 62, 429–443. [Google Scholar] [CrossRef]
- Stahlmann-Brown, P.; Hall, R.J.; Pragert, H.; Robertson, T. Varroa appears to drive persistent increases in New Zealand colony losses. Insects 2022, 13, 589. [Google Scholar] [CrossRef]
- Mutinelli, F.; Pinto, A.; Barzon, L.; Toson, M. Some considerations about winter colony losses in Italy according to the Coloss questionnaire. Insects 2022, 13, 1059. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodney, T.R.; Ida, C.; Renata, S.L.; Shelley, H.; Rob, C.; Pierre, G.; Marta, G.M.; Stephen, P.; Leonard, F.; Amro, Z. Land use changes associated with declining honey bee health across temperate North America. Environ. Res. Lett. 2023, 18, 064042. [Google Scholar] [CrossRef]
- Smoliński, S.; Langowska, A.; Glazaczow, A. Raised seasonal temperatures reinforce autumn Varroa destructor infestation in honey bee colonies. Sci. Rep. 2021, 11, 22256. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jack, C.J.; de Bem Oliveira, I.; Kimmel, C.B.; Ellis, J.D. Seasonal differences in Varroa destructor population growth in western honey bee (Apis mellifera) colonies. Front. Ecol. Evol. 2023, 11, 1102457. [Google Scholar] [CrossRef]
- Stahlmann-Brown, P.; Robertson, T. Report on the 2023 New Zealand Colony Loss Survey; MPI Technical Paper No: 2024/01; Ministry for Primary Industries: Wellington, New Zealand, March 2021; ISBN 978-1-991087-53-9. [Google Scholar]
- Posada-Florez, F.; Lamas, Z.S.; Hawthorne, D.J.; Ryabov, E.V. Pupal cannibalism by worker honey bees contributes to the spread of deformed wing virus. Sci. Rep. 2021, 11, 8989. [Google Scholar] [CrossRef]
- Mitton, G.A.; Meroi Arcerito, F.; Cooley, H.; Fernández de Landa, G.; Eguaras, M.J.; Ruffinengo, S.R.; Maggi, M.D. More than sixty years living with Varroa destructor: A review of acaricide resistance. Int. J. Pest Manag. 2022, 1–18. [Google Scholar] [CrossRef]
- Marco, L.; Colombo, M.; Spreafico, M. Ineffectiveness of Apistan® treatment against the mite Varroa jacobsoni Oud in several districts of Lombardy (Italy). Apidologie 1995, 26, 67–72. [Google Scholar] [CrossRef]
- Johnson, R.; Henry, P.; May, R.B. Synergistic interactions between in-hive miticides in Apis mellifera. J. Econ. Entomol. 2009, 102, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Patti, E.; Frank, E.; James, B.; Gary, E.; William, W. Detection of resistance in US Varroa jacobsoni Oud. (Mesostigmata: Varroidae) to the acaricide fluvalinate. Apidologie 1999, 30, 13–17. [Google Scholar] [CrossRef]
- Jérôme, T. Monitoring Varroa jacobsoni resistance to pyrethroids in western Europe. Apidologie 1998, 29, 537–546. [Google Scholar]
- Thompson, H.M.; Brown, M.A.; Ball, R.F.; Bew, M.H. First report of Varroa destructor resistance to pyrethroids in the UK. Apidologie 2002, 33, 357–366. [Google Scholar] [CrossRef]
- Mitton, G.A.; Quintana, S.; Giménez Martínez, P.; Mendoza, Y.; Ramallo, G.; Brasesco, C.; Villalba, A.; Eguaras, M.J.; Maggi, M.D.; Ruffinengo, S.R. First record of resistance to flumethrin in a Varroa population from Uruguay. J. Apic. Res. 2016, 55, 422–427. [Google Scholar] [CrossRef]
- González-Cabrera, J.; Davies, T.G.; Field, L.M.; Kennedy, P.J.; Williamson, M.S. An amino acid substitution (L925V) associated with resistance to pyrethroids in Varroa destructor. PLoS ONE 2013, 8, e82941. [Google Scholar] [CrossRef] [PubMed]
- González-Cabrera, J.; Rodríguez-Vargas, S.; Davies, T.G.; Field, L.M.; Schmehl, D.; Ellis, J.D.; Krieger, K.; Williamson, M.S. Novel mutations in the voltage-gated sodium channel of pyrethroid-resistant Varroa destructor populations from the southeastern USA. PLoS ONE 2016, 11, e0155332. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Millán-Leiva, A.; Marín, Ó.; Christmon, K.; vanEngelsdorp, D.; González-Cabrera, J. Mutations associated with pyrethroid resistance in Varroa mite, a parasite of honey bees, are widespread across the United States. Pest. Manag. Sci. 2021, 77, 3241–3249. [Google Scholar] [CrossRef] [PubMed]
- Alissandrakis, E.; Ilias, A.; Tsagkarakou, A. Pyrethroid target site resistance in Greek populations of the honey bee parasite Varroa destructor (Acari: Varroidae). J. Apic. Res. 2017, 56, 625–630. [Google Scholar] [CrossRef]
- Esengül, E.; Nafiye, K.; Mustafa, R.; Emre, I. Geographical distribution of pyrethroid resistance mutations in Varroa destructor across Türkiye and a European overview. Exp. Appl. Acarol. 2024, 92, 1–13. [Google Scholar] [CrossRef]
- Yarsan, E.; Yilmaz, F.; Sevin, S.; Akdeniz, G.; Celebi, B.; Hasan Ozturk, S.; Nurhan Ayikol, S.; Karatas, U.; Hasan, E.; Fidan, N.; et al. Investigation of resistance against to flumethrin using against Varroa destructor in Türkiye. Vet. Res. Commun. 2024. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Brodschneider, R.; Schlagbauer, J.; Arakelyan, I.; Ballis, A.; Brus, J.; Brusbardis, V.; Cadahía, L.; Charrière, J.D.; Chlebo, R.; Coffey, M.F.; et al. Spatial clusters of Varroa destructor control strategies in Europe. J. Pest. Sci. 2023, 96, 759–783. [Google Scholar] [CrossRef]
- Piotr, S.; Piotr, S.; Krystyna, P. The amitraz strips efficacy in control of Varroa destructor after many years application of amitraz in apiaries. J. Apic. Sci. 2013, 57, 107–121. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Hernández-Rodríguez, C.S.; González-Cabrera, J. Assessing the resistance to acaricides in Varroa destructor from several Spanish locations. Parasitol Res. 2020, 119, 3595–3601. [Google Scholar] [CrossRef] [PubMed]
- Gabrielle, A.; Benjamin, P.; Précillia, C.; Christelle, S. Inventory of Varroa destructor susceptibility to amitraz and tau-fluvalinate in France. Exp. Appl. Acarol. 2020, 82, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, F. Detection of amitraz resistance and reduced treatment efficacy in the Varroa Mite, Varroa destructor, within commercial beekeeping operations. PLoS ONE 2020, 15, e0227264. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rodríguez, C.S.; Moreno-Martí, S.; Almecija, G.; Christmon, K.; Johnson, J.D.; Ventelon, M.; vanEngelsdorp, D.; Cook, S.C.; González-Cabrera, J. Resistance to amitraz in the parasitic honey bee mite Varroa destructor is associated with mutations in the β-adrenergic-like octopamine receptor. J. Pest. Sci. 2022, 95, 1179–1195. [Google Scholar] [CrossRef]
- Pettis, J.S.; Shimanuki, H.; Feldlaufer, M.F. Detection of Varroa resistant to fluvalinate. Am. Bee J. 1998, 138, 538–541. [Google Scholar]
- Almecija, G.; Poirot, B. Résistances de Varroa destructor aux Acaricides: Conséquences sur L’efficacité des Traitements. Application au Tau-Fluvalinate et à L’amitraze. Doctoral Thesis, University of Tours, Tours, France, 12 October 2021. [Google Scholar]
- Morfin, N.; Rawn, D.; Petukhova, T.; Kozak, P.; Eccles, L.; Chaput, J.; Pasma, T.; Guzman-Novoa, E. Surveillance of synthetic acaricide efficacy against Varroa destructor in Ontario, Canada. Can. Entomol. 2022, 154, 1. [Google Scholar] [CrossRef]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D. Standard methods for Varroa research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Milani, N. The resistance of Varroa jacobsoni Oud. To pyrethroids—A laboratory assay. Apidologie 1995, 26, 415–429. [Google Scholar] [CrossRef]
- Sandon, Y.; Viry, A. Lutte contre le Varroa—Etude de sensibilité/résistance à l’amitraze chez Varroa destructor. La Santé de l’Abeille 2017, 277, 47–56. [Google Scholar]
- Giles, D.P.; Rothwell, D.N. The sub-lethal activity of amidines on insects and acarids. Pestic. Sci. 1983, 14, 303–312. [Google Scholar] [CrossRef]
- Evans, P.D.; Gee, J.D. Action of formamidine pesticides on octopamine receptors. Nature 1980, 287, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Milani, N.; Vedova, G. Decline in the proportion of mites resistant to fluvalinate in a population of Varroa destructor not treated with pyrethroids. Apidologie 2002, 33, 417–422. [Google Scholar] [CrossRef]
- Elzen, P.; Westervelt, D. A scientific note on reversion of fluvalinate resistance to a degree of susceptibility in Varroa destructor. Apidologie 2004, 35, 519–520. [Google Scholar] [CrossRef]
- Maisonnasse, A.; Hernandez, J.; Le Quintec, C.; Cousin, M.; Beri, C.; Kretzschamat, A. Evaluation de la structure des colonies d’abeilles, création et utilisation de la méthode ColEval (Colony Evaluation). Innov. Agron. 2016, 53, 27–37. [Google Scholar] [CrossRef]
- Avelin, C. Observatoire de la production de miel et de gelée royale. Programme Apicole Européen. FranceAgriMer. Edition Juillet. 2022. Available online: https://www.apipro-ffap.fr/Observatoire-de-la-production-de-miel-et-gelee-royale-2022 (accessed on 20 May 2024).
- Guideline on Veterinary Medicinal Products Controlling Varroa Destructor Parasitosis in Bees EMA/CVMP/EWP/459883/2008-Rev.1. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-veterinary-medicinal-products-controlling-varroa-destructor-parasitosis-bees-revision-1_en.pdf (accessed on 20 May 2024).
- Rapport D’étude Evaluation de L’efficacité du Traitement Apivar® en Lien Avec la Sensibilité des Populations de Varroas à L’amitraze. Microsoft Word—Rapport_EfficacitéApivar2021_v04082022 2021. Available online: https://www.adaoccitanie.org (accessed on 20 May 2024).
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010, 103, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Nurit, E.; Alexander, M. Varroa mite evolution: A neglected aspect of worldwide bee collapses? (2020). Curr. Opin. Insect Sci. 2020, 39, 21–26. [Google Scholar]
- Oldroyd, B.P. Coevolution while you wait: Varroa jacobsoni, a new parasite of western honeybees. Trends Ecol. Evol. 1999, 14, 312–315. [Google Scholar] [CrossRef]
- Ayan, A.; Aldemır, O.S.; Selamoglu, Z. Analysis of COI gene region of Varroa destructor in honey bees (Apis mellifera) in province of Siirt. Turkish J. Vet. Res. 2017, 1, 20–23. [Google Scholar]
- Hajializadeh, Z.; Asadi, M.; Kavousi, H. First report of Varroa genotype in western Asia based on genotype identification of Iranian Varroa destructor populations (Mesostigmata: Varroidae) using RAPD marker. Syst. Appl. Acarol. 2018, 23, 199–205. [Google Scholar] [CrossRef]
- Ogihara, M.H.; Yoshiyama, M.; Morimoto, N.; Kimura, K. Dominant honeybee colony infestation by Varroa destructor (Acari: Varroidae) K haplotype in Japan. Appl. Entomol. Zool. 2020, 55, 189–197. [Google Scholar] [CrossRef]
- Navajas, M.; Anderson, D.L.; De Guzman, L.I.; Huang, Z.Y.; Clement, J.; Zhou, T.; Le Conte, Y. New Asian types of Varroa destructor: A potential new threat for world apiculture. Apidologie 2010, 41, 181–193. [Google Scholar] [CrossRef]
- Techer, M.A.; Roberts, J.M.K.; Cartwright, R.A.; Mikheyev, A.S. The first steps toward a global pandemic: Reconstructing the demographic history of parasite host switches in its native range. Mol. Ecol. 2021, 31, 1358–1374. [Google Scholar] [CrossRef]
- Gajic, B.; Radulovic, Z.; Stevanovic, J.; Kulisic, Z.; Vucicevic, M.; Simeunovic, P.; Stanimirovic, Z. Variability of the honey bee mite Varroa destructor in Serbia, based on mtDNA analysis. Exp. Appl. Acarol. 2013, 61, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Gajić, B.; Stevanović, J.; Radulović, Ž.; Kulišić, Z.; Vejnović, B.; Glavinić, U.; Stanimirović, Z. Haplotype identification and detection of mitochondrial DNA heteroplasmy in Varroa destructor mites using ARMS and PCR-RFLP methods. Exp. Appl. Acarol. 2016, 70, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Gajić, B.; Muñoz, I.; De la Rúa, P.; Stevanović, J.; Lakić, N.; Kulišić, Z.; Stanimirović, Z. Coexistence of genetically different Varroa destructor in Apis mellifera colonies. Exp. Appl. Acarol. 2019, 78, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Dynes, T.L.; De Roode, J.C.; Lyons, J.I.; Berry, J.A.; Delaplane, K.S.; Brosi, B.J. Fine scale population genetic structure of Varroa destructor, an ectoparasitic mite of the honey bee (Apis mellifera). Apidologie 2017, 48, 93–101. [Google Scholar] [CrossRef]
- Beaurepaire, A.L.; Moro, A.; Mondet, F.; Le Conte, Y.; Neumann, P.; Locke, B. Population genetics of ectoparasitic mites suggest arms race with honeybee hosts. Sci. Rep. 2019, 9, 11355. [Google Scholar] [CrossRef]
- Ascunce, M.S.; Toups, M.A.; Kassu, G.; Fane, J.; Scholl, K.; Reed, D.L. Nuclear genetic diversity in human lice (Pediculus humanus) reveals continental differences and high inbreeding among worldwide populations. PLoS ONE 2013, 8, e57619. [Google Scholar] [CrossRef] [PubMed]
- Nozais, J.P. The origin and dispersion of human parasitic diseases in the old world (Africa, Europe and Madagascar). Mem. Inst. Oswaldo Cruz. 2003, 98 (Suppl. I), 13–19. [Google Scholar] [CrossRef] [PubMed]
- Dietemann, V.; Beaurepaire, A.; Page, P.; Yañez, O.; Buawangpong, N.; Chantawannakul, P.; Neumann, P. Population genetics of ectoparasitic mites Varroa spp. in Eastern and Western honey bees. Parasitology 2019, 146, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Techer, M.A.; Rane, R.V.; Grau, M.L.; Roberts, J.M.K.; Sullivan, S.T.; Liachko, I.; Childers, A.K.; Evans, J.D.; Mikheyev, A.S. Divergent evolutionary trajectories following speciation in two ectoparasitic honey bee mites. Commun. Biol. 2019, 2, 357. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).