The Complete Mitochondrial Genome of the Chinese White Wax Scale Insect, Ericerus pela Chavannes (Hemiptera: Coccidae), with Novel Gene Arrangement and Truncated tRNA Genes
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Sample Preparation and DNA Extraction
2.2. Library Construction and Sequencing
2.3. Genome Assembly
2.4. Mitochondrial Genome Annotation
2.5. Gene Component and Structural Analysis
2.6. Phylogenetic Analysis
2.7. Gene Rearrangement Analysis
2.8. Synteny Analysis
3. Results
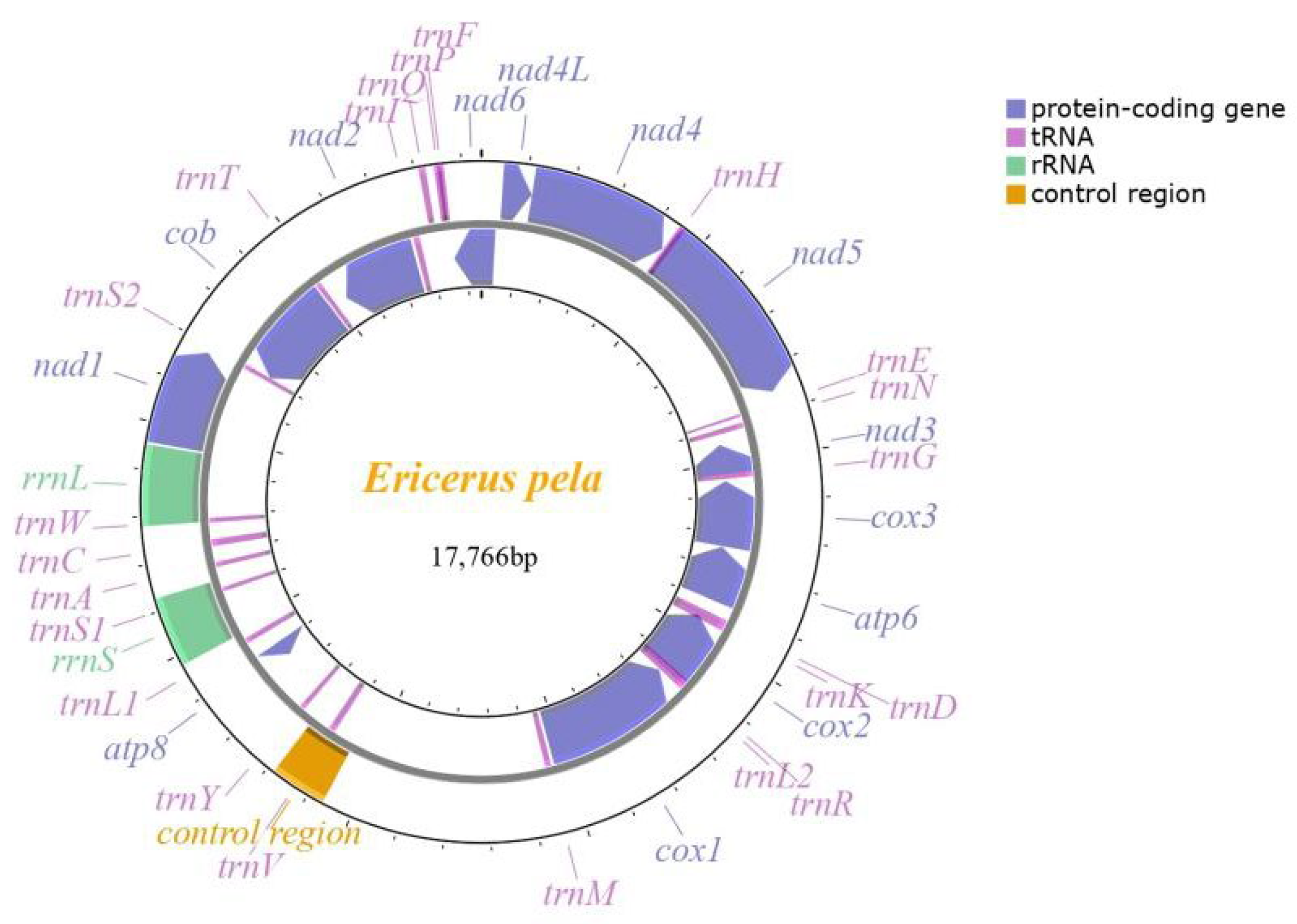
3.1. General Genomic Features
3.2. Gene Rearrangement
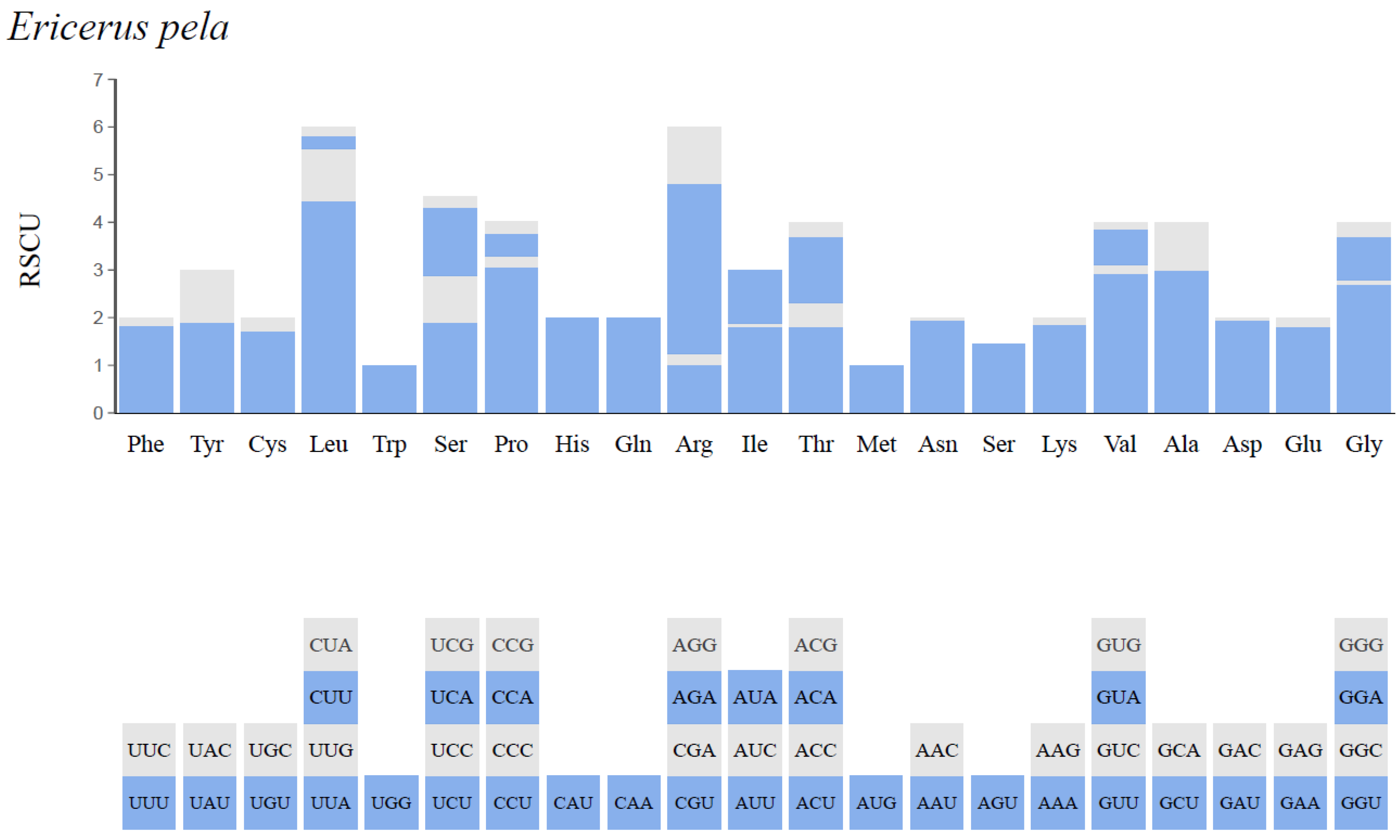
3.3. Protein-Coding Genes and Codon Usage
3.4. rRNAs and tRNAs
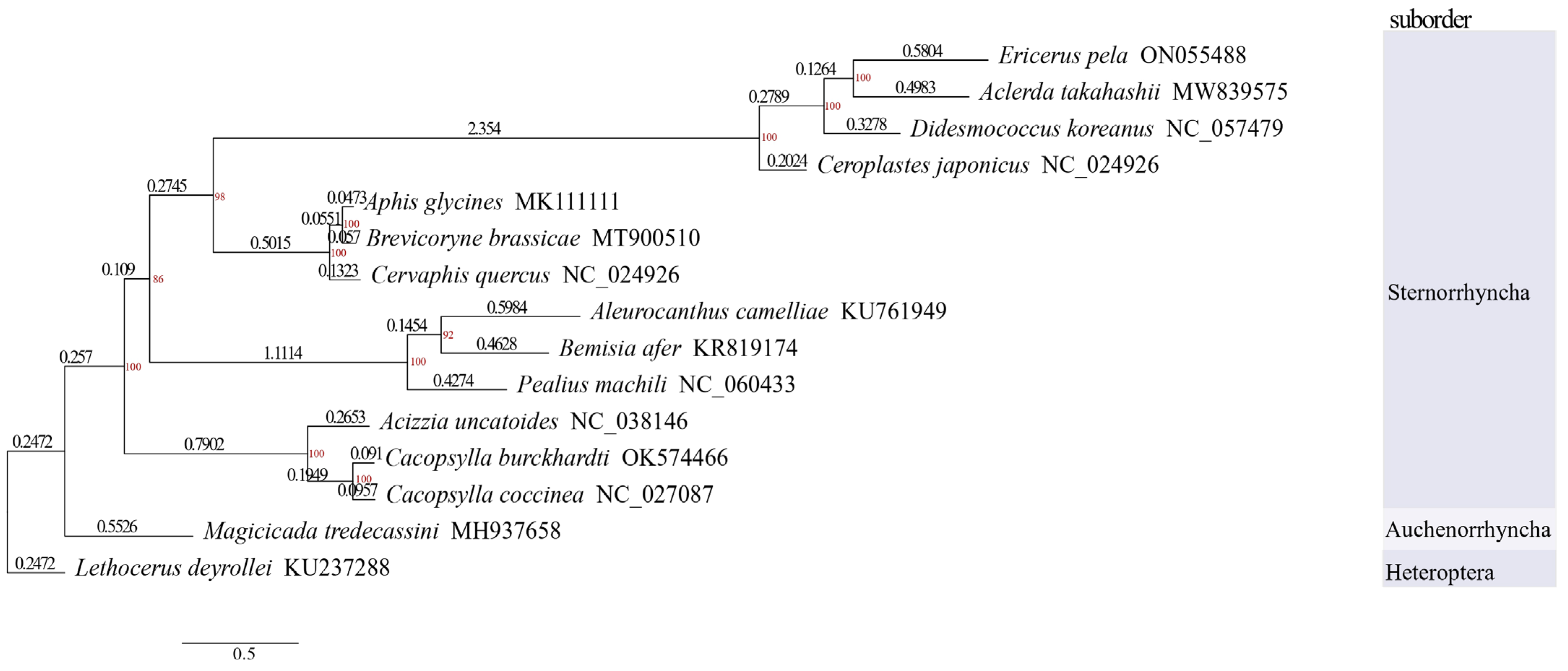
3.5. Phylogenetic Analysis
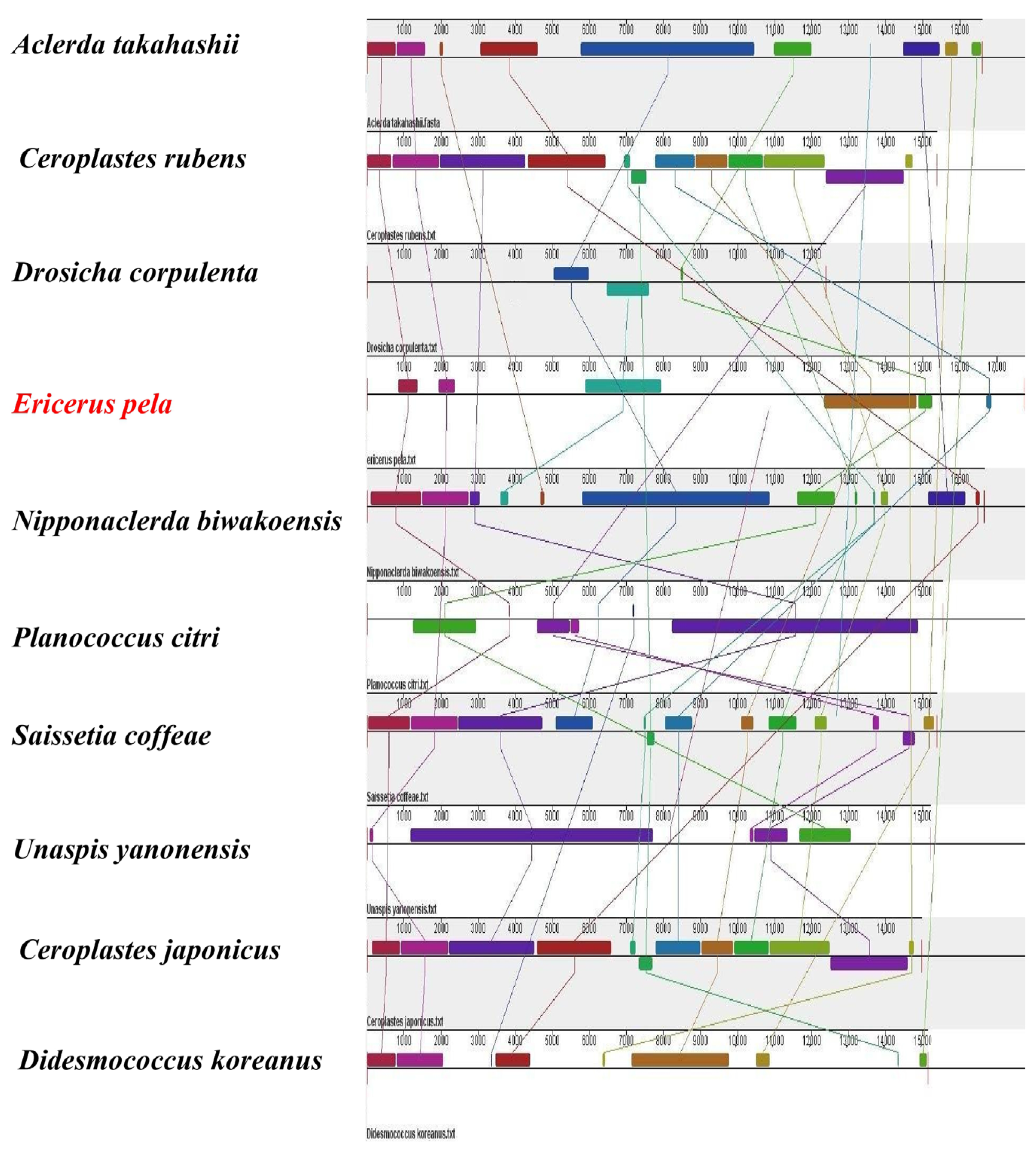
3.6. Synteny Analysis
4. Discussion
4.1. High A + T Bias
4.2. tRNA
4.3. Gene Rearrangement and Synteny Analysis
4.4. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Normark, B.B.; Okusu, A.; Morse, G.E.; Peterson, D.A.; Itioka, T.; Schneider, S.A. Phylogeny and classification of armored scale insects (Hemiptera: Coccomorpha: Diaspididae). Zootaxa 2019, 4616, zootaxa.4616.1.1. [Google Scholar] [CrossRef]
- Ross, L.; Shuker, D.M. Scale insects. Curr. Biol. 2009, 19, R184–R186. [Google Scholar] [CrossRef] [PubMed]
- Szklarzewicz, T. The ovaries of scale insects (Hemiptera, Coccinea). Morphology and phylogenetic conclusions. Folia Histochem. Cytobiol. 1998, 36, 157–165. [Google Scholar] [PubMed]
- Cook, L.G.; Gullan, P.J.; Trueman, H.E. A preliminary phylogeny of the scale insects (Hemiptera: Sternorrhyncha: Coccoidea) based on nuclear small-subunit ribosomal DNA. Mol. Phylogenet. Evol. 2002, 25, 43–52. [Google Scholar] [CrossRef]
- Gruwell, M.E.; Morse, G.E.; Normark, B.B. Phylogenetic congruence of armored scale insects (Hemiptera: Diaspididae) and their primary endosymbionts from the phylum Bacteroidetes. Mol. Phylogenet. Evol. 2007, 44, 267–280. [Google Scholar] [CrossRef]
- Ellenrieder, N.V.; Stocks, I.C. A new species of mealybug in the genus Paracoccus Ezzat & McConnell from North America (Insecta: Coccoidea: Pseudococcidae). Zootaxa 2014, 3873, 25–36. [Google Scholar]
- Choi, J.; Soysouvanh, P.; Lee, S.; Hong, K.J. Review of the family Coccidae (Hemiptera: Coccomorpha) in Laos. Zootaxa 2018, 4460, 1–62. [Google Scholar] [CrossRef]
- Hodgson, C.J.; Williams, D.J. Revision of the soft scale genus Paralecanium (Hemiptera: Coccomorpha: Coccidae) with the introduction of three new genera and twenty new species. Zootaxa 2018, 4443, 1–162. [Google Scholar] [CrossRef]
- Mourad, A.K.; Mesbah, H.A.; Fata, A.A.; Moursi, K.S.; Abdel-Razak, S.I. Survey of scale insects of ornamental plants in Alexandria Governorate, Egypt. Meded. Rijksuniv. Gent. Fak. Landbouwkd. Toegep. Biol. Wet. 2001, 66, 571–580. [Google Scholar]
- Hodgson, C. A review of neococcid scale insects (Hemiptera: Sternorrhyncha: Coccomorpha) based on the morphology of the adult males. Zootaxa 2020, 4765, zootaxa.4765.1.1. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Arnoldi, F.G.; Ogoh, K.; Ohmiya, Y.; Viviani, V.R. Mitochondrial genome sequence of the Brazilian luminescent click beetle Pyrophorus divergens (Coleoptera: Elateridae): Mitochondrial genes utility to investigate the evolutionary history of Coleoptera and its bioluminescence. Gene 2007, 405, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Roskov, Y.; Ower, G.; Orrell, T.; Nicolson, D.; Bailly, N.; Kirk, P.M.; Bourgoin, T.; DeWalt, R.E.; Decock, W.; van Nieukerken, E.; et al. Species 2000 & ITIS Catalogue of Life, 2019 Annual Checklist, Species 2000; Naturalis: Leiden, The Netherlands, 2019; ISSN 2405-884X. Available online: www.catalogueoflife.org/annual-checklist/2019 (accessed on 25 April 2022).
- Zarkani, A.; Apriyanto, D.; Turanli, F.; Ercan, C.; Kaydan, M.B. A checklist of Indonesian scale insects (Hemiptera: Coccomorpha). Zootaxa 2021, 5016, 151–195. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Lu, C.; Huang, X. The first mitochondrial genome of scale insects (Hemiptera: Coccoidea). Mitochondrial DNA B Resour. 2019, 4, 2094–2095. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Huang, X.; Deng, J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020, 158, 854–864. [Google Scholar] [CrossRef]
- Gullan, P.J.; Kosztarab, M. Adaptations in scale insects. Annu. Rev. Entomol. 1997, 42, 23–50. [Google Scholar] [CrossRef]
- Fu, Z.Y.; An, J.Q.; Liu, W.; Zhang, H.P.; Yang, P. Genomic Analyses of the Fungus Paraconiothyrium sp. Isolated from the Chinese White Wax Scale Insect Reveals Its Symbiotic Character. Genes 2022, 13, 338. [Google Scholar] [CrossRef]
- Yang, P.; Yu, S.; Hao, J.; Liu, W.; Zhao, Z.; Zhu, Z.; Sun, T.; Wang, X.; Song, Q. Genome sequence of the Chinese white wax scale insect Ericerus pela: The first draft genome for the Coccidae family of scale insects. GigaScience 2019, 8, giz113. [Google Scholar] [CrossRef]
- Yang, P.; Zhu, J.Y.; Gong, Z.J.; Xu, D.L.; Chen, X.M.; Liu, W.W.; Lin, X.D.; Li, Y.F. Transcriptome analysis of the Chinese white wax scale Ericerus pela with focus on genes involved in wax biosynthesis. PLoS ONE 2012, 7, e35719. [Google Scholar] [CrossRef]
- Sun, T.; Wang, X.Q.; Zhao, Z.L.; Yu, S.H.; Yang, P.; Chen, X.M. A Lethal Fungus Infects the Chinese White Wax Scale Insect and Causes Dramatic Changes in the Host Microbiota. Sci. Rep. 2018, 8, 5324. [Google Scholar] [CrossRef]
- Yang, P.; Chen, X.M. Protein profiles of Chinese white wax scale, Ericerus pela, at the male pupal stage by high-throughput proteomics. Arch. Insect Biochem. Physiol. 2014, 87, 214–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.H.; Yang, P.; Sun, T.; Qi, Q.; Wang, X.-Q.; Xu, D.-L.; Chen, X.-M. Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. Springerplus 2016, 5, 791. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.A.; Okusu, A.; Normark, B.B. Molecular phylogenetics of Aspidiotini armored scale insects (Hemiptera: Diaspididae) reveals rampant paraphyly, curious species radiations, and multiple origins of association with Melissotarsus ants (Hymenoptera: Formicidae). Mol. Phylogenet. Evol. 2018, 129, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Chen, X.M.; Liu, W.W.; Feng, Y.; Sun, T. Transcriptome analysis of sexually dimorphic Chinese white wax scale insects reveals key differences in developmental programs and transcription factor expression. Sci. Rep. 2015, 5, 8141. [Google Scholar] [CrossRef]
- Zhang, H.-P.; Liu, W.; An, J.-Q.; Yang, P.; Guo, L.-H.; Li, Y.-Q.; Lv, J.; Yu, S.-H. Transcriptome analyses and weighted gene coexpression network analysis reveal key pathways and genes involved in the rapid cold resistance of the Chinese white wax scale insect. Arch. Insect Biochem. Physiol. 2021, 107, e21781. [Google Scholar] [CrossRef]
- Lung, T.N. The Culturing of Ericerus pela (Chavannes) in China and the Transmission of Chinese Wax into Europe. Agric. Hist. China 2004, 4, 18–23. [Google Scholar]
- Yang, K. Reanalysis of the Origin of Cera Chinensis Utilization in China. Stud. Hist. Nat. Sci. 2020, 39, 2. [Google Scholar]
- Zhao, J.J.; Deng, L.; Tan, T.W.; Wang, F. Wax esters synthesis from rapeseed oil by immobilized lipase. Chem. Ind. Eng. Prog. 2007, 26, 1311–1315. [Google Scholar]
- Wang, Y.; Chen, J.; Jiang, L.Y.; Qiao, G.X. Hemipteran mitochondrial genomes: Features, structures and implications for phylogeny. Int. J. Mol. Sci. 2015, 16, 12382–12404. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, L.; Baumann, P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol. Biol. 2004, 4, 25. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Al Arab, M.; Höner Zu Siederdissen, C.; Tout, K.; Sahyoun, A.H.; Stadler, P.F.; Bernt, M. Accurate annotation of protein-coding genes in mitochondrial genomes. Mol. Phylogenet. Evol. 2017, 106, 209–216. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Mohajeri, A.; Nobandegani, F.F. Detection and evaluation of hydrogen bond strength in nucleic acid base pairs. J. Phys. Chem. A 2008, 112, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.L.; Claessens, A.; Otto, T.D.; Kekre, M.; Fairhurst, R.M.; Rayner, J.C.; Kwiatkowski, D. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017, 45, 1889–1901. [Google Scholar]
- Domes, K.; Maraun, M.; Scheu, S.; Cameron, S.L. The complete mitochondrial genome of the sexual oribatid mite Steganacarus magnus: Genome rearrangements and loss of tRNAs. BMC Genom. 2008, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Okimoto, R.; Wolstenholme, D.R. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm: Towards a minimal tRNA adaptor. EMBO J. 1990, 9, 3405–3411. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef]
- Watanabe, Y.; Suematsu, T.; Ohtsuki, T. Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 2014, 5, 109. [Google Scholar] [CrossRef]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Chandra, K.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef]
- Liu, H.L.; Chen, Q.D.; Chen, S.; Pu, D.Q.; Chen, Z.T.; Liu, Y.Y.; Liu, X. The highly rearranged mitochondrial genomes of three economically important scale insects and the mitochondrial phylogeny of Coccoidea (Hemiptera: Sternorrhyncha). PeerJ 2020, 8, e9932. [Google Scholar] [CrossRef]


| Species | Accession Number | Family | Suborder | Order |
|---|---|---|---|---|
| Didesmococcus koreanus | NC_057479 | Coccidae | Sternorrhyncha | Hemiptera |
| Aclerda takahashii | MW839575 | Aclerdidae | Sternorrhyncha | Hemiptera |
| Ceroplastes japonicus | MK847519 | Coccidae | Sternorrhyncha | Hemiptera |
| Ericerus pela | ON055488 | Coccidae | Sternorrhyncha | Hemiptera |
| Cervaphis quercus | NC_024926 | Aphididae | Sternorrhyncha | Hemiptera |
| Aphis glycines | MK111111 | Aphididae | Sternorrhyncha | Hemiptera |
| Brevicoryne brassicae | MT900510 | Aphididae | Sternorrhyncha | Hemiptera |
| Aleurocanthus camelliae | KU761949 | Aleyrodidae | Sternorrhyncha | Hemiptera |
| Bemisia afer | KR819174 | Aleyrodidae | Sternorrhyncha | Hemiptera |
| Pealius machili | NC_060433 | Aleyrodidae | Sternorrhyncha | Hemiptera |
| Acizzia uncatoides | NC_038146 | Psyllidae | Sternorrhyncha | Hemiptera |
| Cacopsylla burckhardti | OK574466 | Psyllidae | Sternorrhyncha | Hemiptera |
| Cacopsylla coccinea | NC_027087 | Psyllidae | Sternorrhyncha | Hemiptera |
| Magicicada tredecassini | MH937658 | Cicadidae | Auchenorrhyncha | Hemiptera |
| Lethocerus deyrollei | KU237288 | Belostomatidae | Heteroptera | Hemiptera |
| Species | Accession Number | A + T% |
|---|---|---|
| Ceroplastes japonicus | MK847519 | 85.15% |
| Aclerda takahashii | MW839575 | 84.51% |
| Didesmococcus koreanus | NC_057479 | 82.54% |
| Saissetia coffeae | MN863803 | 84.72% |
| Unaspis yanonensis | MT611525 | 86.57% |
| Ceroplastes rubens | MT677923 | 87.52% |
| Drosicha corpulenta | MK251061 | 87.21% |
| Nipponaclerda biwakoensis | MN193722 | 81.3% |
| Planococcus citri | MT611526 | 82.77% |
| Ericerus pela | ON055488 | 88.21% |
| Gene | Start | Stop | Length | Start/Stop Codons | A + T% |
|---|---|---|---|---|---|
| nad4L | 191 | 460 | 270 | ATT/TAA | 88.52 |
| nad4 | 462 | 1757 | 1296 | ATA/TAA | 89.74 |
| trnH | 1765 | 1820 | 56 | ||
| nad5 | 1786 | 3417 | 1632 | ATA/TAA | 89.58 |
| trnE | 3514 | 3543 | 30 | ||
| trnN | 3605 | 3662 | 58 | ||
| nad3 | 3779 | 4120 | 342 | ATT/TAG | 90.06 |
| trnG | 4118 | 4176 | 59 | ||
| cox3 | 4201 | 4956 | 756 | ATG/TAA | 88.63 |
| atp6 | 4964 | 5578 | 615 | ATG/TAA | 88.94 |
| trnD | 5716 | 5771 | 56 | ||
| trnK | 5767 | 5835 | 69 | ||
| cox2 | 5795 | 6484 | 690 | ATG/TAA | 85.22 |
| trnR | 6459 | 6516 | 58 | ||
| trnL2 | 6503 | 6571 | 69 | ||
| cox1 | 6582 | 8117 | 1536 | ATA/TAA | 81.57 |
| trnM | 8151 | 8215 | 65 | ||
| Control region | 8215 | 11163 | 2949 | ||
| trnV | 10491 | 10563 | 73 | ||
| trnY | 10885 | 10938 | 54 | ||
| atp8 | 11428 | 11619 | 192 | AGA/AAA | 85.94 |
| trnL1 | 11786 | 11845 | 60 | ||
| rrnS | 11911 | 12509 | 599 | 85.81 | |
| trnS1 | 12380 | 12425 | 46 | ||
| trnA | 12642 | 12694 | 53 | ||
| trnC | 12862 | 12936 | 75 | ||
| rrnL | 13076 | 13808 | 733 | 87.03 | |
| trnW | 13110 | 13160 | 51 | ||
| nad1 | 13830 | 14738 | 909 | ATA/TAA | 87.68 |
| trnS2 | 14773 | 14822 | 50 | ||
| cob | 14822 | 15901 | 1080 | ATG/TAA | 85.56 |
| trnT | 15922 | 15974 | 53 | ||
| nad2 | 16113 | 17018 | 906 | ATT/TAA | 90.73 |
| trnI | 17052 | 17116 | 65 | ||
| trnQ | 17237 | 17295 | 59 | ||
| trnP | 17367 | 17425 | 59 | ||
| trnF | 17396 | 17450 | 55 | ||
| nad6 | 17451 | 146 | 462 | ATA/TAA | 93.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.-Q.; Yu, S.-H.; Wei, S.-J.; Zhang, H.-P.; Shi, Y.-C.; Zhao, Q.-Y.; Fu, Z.-Y.; Yang, P. The Complete Mitochondrial Genome of the Chinese White Wax Scale Insect, Ericerus pela Chavannes (Hemiptera: Coccidae), with Novel Gene Arrangement and Truncated tRNA Genes. Insects 2023, 14, 290. https://doi.org/10.3390/insects14030290
An J-Q, Yu S-H, Wei S-J, Zhang H-P, Shi Y-C, Zhao Q-Y, Fu Z-Y, Yang P. The Complete Mitochondrial Genome of the Chinese White Wax Scale Insect, Ericerus pela Chavannes (Hemiptera: Coccidae), with Novel Gene Arrangement and Truncated tRNA Genes. Insects. 2023; 14(3):290. https://doi.org/10.3390/insects14030290
Chicago/Turabian StyleAn, Jia-Qi, Shu-Hui Yu, Shu-Jun Wei, Hong-Ping Zhang, Yuan-Chong Shi, Qiu-Yu Zhao, Zuo-Yi Fu, and Pu Yang. 2023. "The Complete Mitochondrial Genome of the Chinese White Wax Scale Insect, Ericerus pela Chavannes (Hemiptera: Coccidae), with Novel Gene Arrangement and Truncated tRNA Genes" Insects 14, no. 3: 290. https://doi.org/10.3390/insects14030290
APA StyleAn, J.-Q., Yu, S.-H., Wei, S.-J., Zhang, H.-P., Shi, Y.-C., Zhao, Q.-Y., Fu, Z.-Y., & Yang, P. (2023). The Complete Mitochondrial Genome of the Chinese White Wax Scale Insect, Ericerus pela Chavannes (Hemiptera: Coccidae), with Novel Gene Arrangement and Truncated tRNA Genes. Insects, 14(3), 290. https://doi.org/10.3390/insects14030290







