Abstract
This critical review investigates the impact of SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4) mutations on survival outcomes in non-small cell lung cancer (NSCLC) through an analysis of 21 peer-reviewed articles. Survival analyses across this review demonstrated consistently worse outcomes for SMARCA4-mutated vs. SMARCA4 wild-type NSCLC patients, specifically emphasizing class 1 truncating mutations as an independent factor for poor overall survival. In addition, this review explores the clinicopathologic characteristics of SMARCA4 mutations and their impact on various treatment modalities, including immune checkpoint inhibitors (ICIs) both with and without Kirsten rat sarcoma viral oncogene homolog (KRAS) co-mutations. The potential ineffectiveness of ICI treatment in NSCLC is explored through the impact of SMARCA4/KRAS co-mutations on the tumor microenvironment. Moreover, this NSCLC review consistently reported statistically worse overall survival outcomes for SMARCA4/KRAS co-mutations than SMARCA4 wild-type/KRAS-mutated cohorts, extending across ICIs, chemo-immunotherapy (CIT), and KRAS G12C inhibitors. Designing prospective clinical SMARCA4-mutated or SMARCA4/KRAS co-mutated NSCLC trials to evaluate targeted therapies and immunotherapy may lead to a better understanding of how to improve cancer patients’ outcomes and survival rates.
Keywords:
SMARCA4; mutation; KRAS; co-mutation; KRAS/SMARCA4; NSCLC; non-small cell lung cancer; survival; treatment outcome; immunotherapy 1. Introduction
Over the past twenty years, the Food and Drug Administration (FDA) has approved a minimum of 20 new molecular entities that target oncogenic driver mutations in non-small cell lung cancer (NSCLC) [1]. These targeted therapies have led to some NSCLC studies reporting a median overall survival (OS) over three years in the metastatic setting [2]. While more than fifty percent of non-squamous NSCLC patients may have a mutation with an FDA-approved therapy, there remains a distinct unmet medical need for those with epigenetic dysregulation that may be more challenging to target [1,3]. This need is particularly vital given that the five-year survival rate for Stage IV NSCLC patients in 2020 was only 8.2% [4].
This critical review analyzes the current literature on the impact of mutations in the SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4) gene, focusing on the survival outcomes for patients with NSCLC. Typically, patients with SMARCA4-deficient NSCLC clinically present with adenocarcinoma, larger invasive tumor size, a smoking history, and fewer epidermal growth factor receptor (EGFR) mutations (p < 0.05) [5,6]. In addition, SMARCA4-mutated NSCLCs are associated with a higher likelihood of negative or low programmed cell death ligand-1 (PD-L1) expression (p =0.03) and higher tumor mutational burden (TMB) as compared to wild-type (WT) cases (p < 0.001) [7,8].
SMARCA4 mutations occur in approximately 5–7% of all human cancers and 7–11% of NSCLC cases [6,9,10,11]. Two classes of SMARCA4 alterations are linked to epigenetic dysregulation [12]. Class 1 mutations encompass truncating mutations, fusions, and homozygous deletions, typically associated with protein loss and loss of function. Class 2 mutations involve missense mutations, which are suggested to employ dominant-negative or gain-of-function effects. Some reports indicate a loss of function leading to reduced accessibility and diminished chromatin remodeling activity, mainly observed in lung cancer [11]. In a comprehensive study of 4813 tumors from NSCLC patients, Schoenfeld et al. discovered that 212 patients (4% of the total population) exhibited class 1 SMARCA4 alterations (52% of the SMARCA4 variants) [8]. In contrast, 195 patients (4% of the total population) had tumors with class 2 SMARCA4 alterations (48% of the SMARCA4 variants) [8]. Of particular interest, SMARCA4 mutations are associated with altered MYC gene expression, which is essential in regulating cell growth and proliferation [13].
SMARCA4 is a tumor suppressor gene located on chromosome 19p13.2 that encodes the Brahma-Related Gene 1 (BRG1) protein, which is one of two mutually exclusive critical DNA-dependent ATPases (Brahma is the other) that regulates gene expression by altering the chromatin structure [14]. In addition, the BRG1 protein contains a bromodomain that plays a critical role in gene transcription and is essential in recognizing acetylated lysine residues on N-terminal tails of histones [14].
BRG1 is part of the mammalian-type switch/sucrose non-fermenting (mSWI/SNF) chromatin-regulatory complex (CRC) [15]. The mSWI/SNF remodeling complex is essential in regulating chromatin structures and contains approximately 10–12 subunits [16]. Utilizing energy obtained through ATP hydrolysis, the catalytic subunits of human mSWI/SNF change nucleosome positioning. This alteration modulates the accessibility of transcriptional machinery to DNA, ultimately impacting the activation or repression of specific genes. The mSWI/SNF CRC protein subunits of interest are AT-Rich Interaction Domain 1A (ARID1A), SWI/SNF Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily B, Member 1 (SMARCB1), and BRG1. They are among the most studied CRCs and have higher rates of mutations in human cancers than other CRC subunits [17].
The most frequent co-mutations found in the Massachusetts General Hospital analysis of 29 SMARCA4 patients that lacked BRG1 protein expression affected TP53 (N = 17, 59%), STK11 (N = 15, 52%), KEAP1 (N = 12, 41%), and KRAS (N = 10, 34%) [6]. These co-mutation frequencies were validated in a more extensive Foundation Medicine data set with SMARCA4 co-mutations in TP53 (74%), CDKN2A (38%), STK11 (34%), KRAS (26%), and KEAP1 (15%) [6].
Further research is needed to unravel the intricate interplay between SMARCA4, chromatin remodeling, and tumorigenesis in non-small cell lung cancer. A comprehensive understanding of the nuanced interplay between SMARCA4 and KRAS co-occurring mutations in the context of NSCLC is paramount, as KRAS mutations commonly co-occur with various mutations and have been found to have a poor prognosis across several NSCLC studies [18]. This review aims to summarize the clinical studies that have elucidated the potential implications of SMARCA4 mutations and SMARCA4/KRAS co-mutations on survival outcomes when treating metastatic NSCLC patients across various treatments (i.e., chemotherapy and immune checkpoint inhibitors).
2. Materials and Methods
PubMed and the Web of Science were searched using the following keywords: “SMARCA4” and “NSCLC”. Boolean operators were used to connect specific search keywords. Peer-reviewed articles published in English between 2018 and 2023 were included, and duplicate records were removed. Articles were included if they contained any of the following concerning NSCLC patients with SMARCA4 mutations vs. SMARCA4 WT: OS hazard ratio (HR), progression-free survival (PFS) HR, median OS, or median PFS. Articles were excluded if they were solely pre-clinical or diagnostic. Articles were also excluded if the majority of SMARCA4 lung cancer patients were diagnosed with non-NSCLC (i.e., small-cell), an undifferentiated tumor, or sarcomatoid SMARCA4 NSCLC. Articles were loaded into Raayan [19] and reviewed by all authors. Discrepancies were discussed until a consensus was reached on included vs. excluded articles.
The search strategy was as follows: (smarca4 [Title/Abstract]) AND (nsclc [Title/Abstract]) Filters: Full text, Humans, from 2018–2023. The study selection process is outlined in the PRISMA flow diagram (Figure 1) [20].
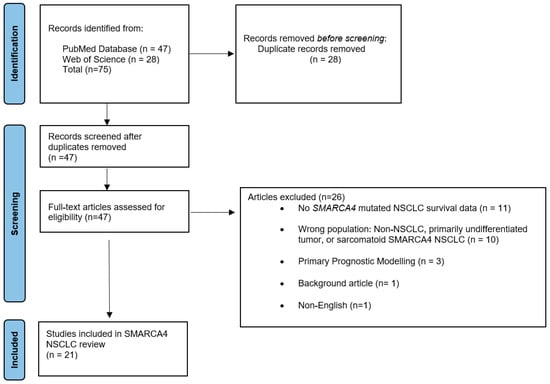
Figure 1.
PRISMA flow diagram illustrating the study selection process.
3. Results
3.1. Results and Description of Studies
In this critical review, 75 articles were identified by applying the search strategy. Twenty-eight articles were excluded and removed as duplicate records, and 47 articles were fully reviewed. Twenty-six articles were excluded due to the lack of SMARCA4-mutation NSCLC survival outcomes, primary prognostic modeling, lack of incorrect patient population (i.e., sarcomatoid histology), review articles, and being written in non-English.
Twenty-one studies were included and reviewed in this critical review of SMARCA4 NSCLC. The included articles focus on survival outcomes across various patient categories, clinicopathologic characteristics, and immune checkpoint inhibitors (ICIs) cancer treatment. Moreover, a separate subheading focusing on SMARCA4 and KRAS co-mutation survival outcomes in patients with NSCLC is included, as this has recently become a medically relevant topic in the lung cancer community. Table 1 summarizes the NSCLC mutation analyzed, treatment, outcome, and relevance of each study in this review.

Table 1.
Summary of articles included in this critical review. (A) SMARCA4 Mutations. (B) SMARCA4/KRAS Co-mutations.
3.2. SMARCA4 Mutations
Schoenfeld et al. examined the clinicopathologic characteristics in a multivariate NSCLC analysis and determined that both SMARCA4-mutated class 1 (N = 149) and class 2 (N = 143) had worse OS vs. WT (p < 0.001) using Kaplan–Meir methods [8]. In addition, class 1 SMARCA4-mutated patients had the worst OS compared to class 2 mutated or WT patients (N = 996) via a log-rank test analysis [8]. Furthermore, an ICI survival treatment analysis (N = 87) showed an OS improvement for SMARCA4-mutated patients receiving ICI treatment vs. without ICI treatment regardless of class 1 or class 2 mutation status (p = 0.01). However, there was no significant difference in PFS (p = 0.74) or OS (p = 0.35) for SMARCA4-mutated patients receiving ICIs when comparing class 1 vs. class 2 alterations in an unadjusted analysis [8].
Both studies from Alessi et al. further support SMARCA4 mutations as an independent factor that leads to worse outcomes for NSCLC patients [21,22]. In one of the most extensive NSCLC studies of patients with SMARCA4 mutations who received chemo-immunotherapy (CIT), Alessi et al. reported that patients with non-squamous histology who had SMARCA4 mutations (N = 114) vs. WT (N = 593) had significantly worse survival: PFS (2.7 versus 6.1 months, HR: 1.62; p < 0.001), and OS (8.1 versus 15.0 months, HR: 1.70; p < 0.001) from the start of CIT [22]. A multivariable analysis using inverse probability weighting was conducted to address potential selection bias due to the absence of Programmed Death Ligand-1 (PDL-1) reporting. The analysis revealed that confirmed SMARCA4 mutations independently correlated with reduced survival: PFS (SMARCA4-mutated HR: 1.61; p < 0.001), OS (SMARCA4-mutated HR: 1.66; p <0.001) to CIT [22].
A separate Alessi et al. study reported better OS in NSCLC patients with SMARCA4 WT (N = 1327) vs. SMARCA4-mutated patients (N = 163): SMARCA4 WT 25.0 months vs. SMARCA4-mutated 15.6 months (HR: 0.064; p < 0.001) [21]. However, in the immunotherapy-treated cohort of SMARCA4-mutated (N = 57) and WT patients (N = 475), which included single-agent checkpoint therapy or ICI in combination with a Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4) inhibitor, there were no significant differences in SMARCA4 WT vs. SMARCA4-mutated patients: PFS (3.1 vs. 2.1 months, HR: 0.93; p = 0.62) or OS (12.4 months vs. 11 months, HR: 0.83; p = 0.25) [21]. When getting more granular and looking at different types of SMARCA4 mutations class 1 (N = 26) vs. class 2 (N = 31), there was no statistical OS difference in outcomes after receiving PDL-1 therapy when comparing SMARCA4-mutated class 1 [nonsense, frameshift, and splice site] (6.7 months; p = 0.11) vs. WT patients (N = 275, 12.4 months) vs. SMARCA4-mutated class 2 missense mutations (11.9 months, HR: 1.03; p = 0.87) [21].
When examining class 1 or class 2 mutations regardless of the type of treatment, Fernando et al. compared patients with SMARCA4 WT (N = 2194) vs. SMARCA4 NSCLC mutations in four categories: (a) homozygous truncated (N = 102), (b) homozygous nontruncated (N = 101), (c) heterozygous truncated (N = 16), and (d) heterozygous nontruncated (N = 49] [11]. There was a significant OS difference when comparing SMARCA4-mutated homozygous truncated vs. WT (7.9 vs. 16.3 months, HR: 1.85; p < 0.0001) but no significant differences when comparing other SMARCA4-mutated categories vs. SMARCA4 WT [11].
Fernando et al. conducted a separate analysis when looking at the same four SMARCA4 mutation categories but for patients who received immunotherapy (nivolumab, pembrolizumab, atezolizumab, or durvalumab) at any time during their cancer treatment [11]. This ICI treatment analysis compared patients with SMARCA4 WT (N = 1069) vs. SMARCA4 NSCLC mutations in four categories: (a) homozygous truncated (N = 38), (b) homozygous nontruncated (N = 41), (c) heterozygous truncated (N = 5), and (d) heterozygous nontruncated (N = 23). Similar to the non-treatment-specific OS analysis, there was a significant OS difference when comparing SMARCA4-mutated homozygous truncated vs. SMARCA4-WT (9.9 vs. 19.5 months, HR: 1.62; p = 0.01), but no significant differences when comparing WT with the other SMARCA4-mutated categories in the ICI-treated patients [11].
In a different study with the largest number of SMARCA4 mutations in this review (N = 3305), the authors focused on clinicopathologic characteristics of patients with SMARCA4 mutations and BRG1-deficient NSCLC [6]. Even though this study had a large number of SMARCA4 patients, they only performed a SMARCA4 mutation PFS analysis (N = 16) with 11 patients on chemotherapy and five on CIT. These patients had extremely short PFS of only 38 and 35 days, respectively [6].
Several studies included OS multivariate analysis across stages to address the outcomes of patients with SMARCA4 mutations in the context of Stage III vs. Stage IV disease. One study conducted by Liang et al. revealed that individuals with SMARCA4 mutations experienced significantly poorer survival in both locally advanced (Stage III) (N = 21) and metastatic (Stage IV) (N = 69) scenarios when compared to those with WT status [5]. The median survival for SMARCA4-mutated vs. WT patients was notably lower in Stage III (23.73 vs. 29.43 months, p < 0.01) and Stage IV (11.93 vs. 28.23 months, p < 0.01) [5]. To support this finding in Stage III patients, Talvitie et al.’s multivariate analysis on SMARCA4-mutated patients (N = 31) also revealed worse OS vs. WT patients (HR:1.93; p = 0.04) [23]. Regardless of stage, both authors conducted a multivariate analysis showing that SMARCA4 WT (N = 221) patients had a better OS than mutated patients [(N = 105), 28.23 months vs. 12.17 months; p < 0.001] and SMARCA4-mutated patients had a worse OS than WT (HR: 3.522; p < 0.002) [5,23].
In association with SMARCA4 mutations and clinico-genomic biomarkers across two different studies, tumor mutational burden (TMB) and brain metastasis in Anaplastic Lymphoma Kinase (ALK)-positive NSCLC were evaluated [24,25]. In exploring the relevance of TMB, Xu et al. conducted an NSCLC adenocarcinoma multivariate analysis [24]. The four arms were as follows: (a) SMARCA4 WT, TMB Low (N = 761), (b) SMARCA4 WT, TMB High (N = 722), (c) SMARCA4-mutated, TMB Low (N = 26), and (d) SMARCA4-mutated, TMB High (N = 69) [24]. The results showed a statistically better overall survival for SMARCA4 WT, TMB High (p = 0.00019) [24]. In the context of brain metastasis, one study examined clinico-genomic outcomes in patients with (ALK)-positive NSCLC treated with alectinib [25]. They determined that SMARCA4-mutated patients (N = 3) with brain metastasis were the only group with co-mutations (SMARCA4/ALK) to do statistically worse across a multivariate analysis (HR: 8.76; p = 0.009).
When addressing SMARCA4 mutation OS differences in a multivariate analysis of biological sex, two OS results were mixed between the sexes [5,26]. Pan et al. conducted a retrospective multivariate analysis, which included a cohort across 21 medical centers. There were 44 NSCLC patients with SMARCA4 mutations, and men (N = 25) had a significantly worse OS than women (N = 19) (2.75 months vs. inestimable, HR:14.2; p = 0.02) [26]. The authors shared that one reason for the inestimable survival may be the low sample size of SMARCA4 NSCLC patients. In the second study, the authors conducted a multivariate analysis, which resulted in statistically worse OS for women (N = 88) vs. men (N = 25) (HR: 4.1; p = 0.04) [5].
There were two NSCLC analyses of the OAK study involving atezolizumab. In the first OAK study analysis, Wang et al. discovered that SMARCA4 was more likely to be seen in the negative PDL-1 expression group, but there was no statistical difference in OS when comparing atezolizumab vs. docetaxel in SMARCA4-mutated (N = 100) whether the patients were PDL-1 high or negative (HR = 0.67; p = 0.256) [27]. In addition, there was no difference in OS in SMARCA4-mutated NSCLC patients (N = 39) vs. SMARCA4 WT (N = 491) whether the patients were PDL-1 high or negative when treated with atezolizumab (HR = 1.185, p = 0.513) [27]. In a second OAK uni-variate analysis, SMARCA4-mutated patients (N = 18) receiving atezolizumab did not fare worse than SMARCA4 WT patients (N = 181) (HR: 1.70; p = 0.064) [28].
In the analysis by Velut et al., NSCLC patients with SMARCA4 mutations (N = 7) did worse than WT (N = 70), independent of whether they received ICI therapy or not (HR: 3.2; p = 0.006) [29]. Interestingly, OS rates at 1 year were not significantly different between SMARCA4-mutated and WT patients but dropped significantly at 2 (58% vs. 74%) and 5 years (37% vs. 53%). In addition, patients were significantly younger upon diagnosis with SMARCA4 mutation vs. without (61.9 years vs. 66.6 years; p = 0.01) [29]. A second single-institution analysis demonstrated a numerical, but not statistical, PFS benefit (6.3 vs. 3.9 months, HR: 0.64; p = 0.15) in SMARCA4 WT (N = 130) vs. SMARCA4-mutated (N = 16) patients receiving ICIs (87% received combination ICI-chemotherapy) [30].
Three additional studies demonstrated significantly worse OS outcomes for NSCLC patients with SMARCA4 mutations than wild-type patients across a multivariate analysis. In the first study, Yang et al.’s analysis resulted in worse OS for mutated (N = 5) vs. WT (N = 32) patients (4.5 months vs. 13.3 months, HR: 2.86; p = 0.031) [31]. The second multivariate analysis obtained a worse PFS (3 vs. 8 months; p = 0.007) for SMARCA4-mutated (N = 4) vs. WT patients (N = 99), and the results concluded that SMARCA4 mutations were independently associated with a worse OS (HR: 3.098; p = 0.038) [32]. The last SMARCA4 mutation analysis highlighted that the differences in all-cause mortality remained significant between the adenocarcinoma NSCLC patients that were mutated (N = 21) vs. wild-type (N = 204) (HR: 2.06; p = 0.003) [33].
3.3. SMARCA4/KRAS Co-Mutations
Seven NSCLC publications contained SMARCA4/KRAS co-mutation survival outcomes [8,21,22,33,34,35,36]. Two of these studies had specific SMARCA4/KRAS point mutation outcomes (i.e., KRAS G12C and G12D), and one analyzed survival with co-mutations in patients across clinical characteristics (i.e., brain and liver mets) [34,35,36]. Moreover, two different co-mutated analyses were performed by Alessi et al., highlighting robust results across many types and combinations of ICIs and chemotherapy [21,22]. Across various treatments, Schoenfeld et al.’s multivariate SMARCA4/KRAS co-mutation analysis resulted in worse statistical OS regardless of class 1 (N = 58) or 2 alterations (N = 52) (class 1: HR: 1.59 and class 2: HR: 2.75 vs. WT; p < 0.001) [8].
Negrao et al. produced the only publication focusing on NSCLC patients with co-mutations and KRAS G12C inhibitors (sotorasib or adagrasib). All NSCLC patients in their study had KRAS G12C mutations [35]. SMARCA4/KRAS G12C co-mutated patients (N = 18) had significantly worse PFS than SMARCA4 WT/KRAS G12C-mutated patients (N = 213) (1.6 vs. 5.4 months, HR: 3.04; p < 0.001) and had a significantly worse OS (4.9 months vs. 11.8 months, HR: 3.07; p < 0.001) [35]. In relation to these survival outcomes, SMARCA4 was one of the three tumor suppressor genes significantly enriched in the early progressing subgroup (p = 0.001) [35].
The second study that contained granular co-mutation outcomes (KRAS G12D) demonstrated statistically worse PFS for NSCLC patients; PFS: SMARCA4/KRAS G12D-co-mutated (N = 8) vs. SMARCA4 WT/KRAS G12D-mutated (N = 49) (1.5 vs. 4.0 months; p = 0.0039) [34]. However, OS was only numerically worse but not statistically significant: SMARCA4/KRAS G12D-co-mutated vs. SMARCA4 WT/KRAS G12D-mutated (6.1 vs. 17.3 months; p = 0.4202) [34].
Boiarsky et al. demonstrated that more than two times the number of patients had co-mutated SMARCA4/KRAS NSCLC Stage IV disease upon diagnosis (14%, N = 214) vs. Stage I (5%, N = 83) (p = 0.038), and these co-mutated patients had worse outcomes across patterns of metastatic spread (i.e., the brain) [36]. For example, OS was significantly worse for SMARCA4/KRAS-co-mutated (N = 33) patients with brain metastasis vs. SMARCA4 WT/KRAS-mutated (N = 231) patients (7.4 months vs. 15.0 months, HR: 2.1; p = 0.0003) [36]. This HR was similar to SMARCA4/KRAS-co-mutated (N = 25) patients with liver metastasis vs. SMARCA4 WT/KRAS-mutated (N = 155) patients (5.2 months vs. 13.2 months, HR: 2.1; p = 0.00015) [36].
SMARCA4-mutated patients had worse CIT treatment outcomes than SMARCA4 WT patients in both KRAS WT and KRAS-mutated non-squamous NSCLC [22]. In reference to how co-mutated SMARCA4/KRAS non-squamous patients (N = 44) fared vs. SMARCA4 WT/KRAS-mutated patients (N = 232), they fared even worse: PFS: SMARCA4/KRAS-mutated vs. SMARCA4 WT/KRAS-mutated (2.2 vs. 6.2 months, HR: 2.39; p < 0.001), OS: SMARCA4/KRAS-mutated vs. SMARCA4 WT/KRAS-mutated (6.6 vs. 14.6 months, HR: 2.52; p < 0.001) [22]. In the second Alessi et al. analysis with single-agent ICI or ICI-combination with CTLA-4, KRAS-mutant NSCLC was the most common co-mutation in the SMARCA4 mutant subset (N = 17) [21]. The SMARCA4/KRAS-co-mutated patients had significantly shorter PFS (1.4 versus 4.1 months, HR: 0.25; p < 0.001) and OS (3 versus 15.1 months, HR: 0.29; p < 0.001) compared with NSCLC patients with SMARCA4 WT/KRAS mutations (N = 159). Thus, the presence of a SMARCA4/KRAS co-mutation may confer a worse outcome to ICIs [21].
The last analysis from Liu et al. demonstrated significantly worse OS for all four SMARCA4/KRAS-mutated adenocarcinoma NSCLC cohorts across ICI-treated and non-ICI-treated NSCLC patients [37]. In Cohort A, SMARCA4/KRAS-co-mutated vs. SMARCA4 WT/KRAS-mutated patients had worse OS (15.73 vs. 19.73 months) in a non-immunotherapy cohort in The Cancer Genome Atlas (TCGA) (HR: 2.32, p = 0.047). In Cohort B, SMARCA4/KRAS-co-mutated vs. SMARCA4 WT/KRAS-mutated patients had worse OS (5.2 vs. 6.5 months) in a non-immunotherapy MSK-CT cohort (HR: 1.95, p = 0.015). In Cohort C, SMARCA4/KRAS co-mutated vs. SMARCA4 WT/KRAS-mutated patients (1.73 vs. 4.22 months) had worse PFS in an immunotherapy MSK-IO cohort (HR: 2.15; p = 0.048). In Cohort D, SMARCA4/KRAS-co-mutated vs. SMARCA4 WT/KRAS-mutated patients had worse OS in the immunotherapy Wake Forest cohort (HR: 11.98, p = 0.0018) [37].
4. Discussion
This SMARCA4 NSCLC critical review focuses on survival outcomes across clinicopathological characteristics, KRAS co-mutations, and treatment with ICIs within twenty-one peer-reviewed articles. Eleven of thirteen (85%) SMARCA4 mutation survival analyses in this review demonstrated significantly worse overall survival for SMARCA4-mutated NSCLC patients regardless of cancer treatment type. There were somewhat mixed results across SMARCA4-mutated treatment analysis for patients receiving ICIs and ICIs in combination with chemotherapy or CTLA-4 treatment. Three of the seven ICI analyses had worse OS for SMARCA4-mutated patients, three had no OS differences, and one outlier study highlighted that the patients with SMARCA4 mutations did better on ICIs regardless of the class of SMARCA4 mutation. Regarding SMARCA4/KRAS co-mutations, all seven treatment analyses (four with ICIs, two with non-ICIs, and one with KRAS G12C inhibitors) indicated that co-mutated SMARCA4/KRAS NSCLC patients consistently had significantly worse OS than SMARCA4 WT/KRAS-mutated cohorts [8,21,22,34,35,36,37].
4.1. SMARCA4 Molecular Sub-Types and Genomic Features
In the largest SMARCA4-mutated survival analysis in this review (N = 407), the authors determined that class 1 truncating mutations were the strongest independent factor for significantly worse OS for NSCLC patients [8]. Class 1 mutations are typically associated with BRG1 protein loss compared to class 2 mutations, not losing any BRG1 protein expression (81% vs. 0%, p < 0.001) [8]. This lack of protein expression was also seen in an analysis completed at Massachusetts General Hospital (MGH), where 84% of patients with class 1 truncating mutations lacked BRG1 protein expression [6]. However, class 2 mutations (missense, nontruncating) also had worse OS, indicating that protein function could be impacted even though protein expression was evident [8].
As the only outlier in this review, SMARCA4-mutated NSCLC patients treated with ICI therapy had a longer OS (p = 0.01) [8]. In contrast, this ICI treatment OS benefit is not seen as appreciable when looking at SMARCA4 WT vs. class 1 or 2 SMARCA4 mutations. Alessi et al. point out that Schoenfeld’s results may differ from their extensive SMARCA4 ICI therapy analysis due to imbalances in unreported baseline clinicopathological characteristics in the Schoenfeld publication, which could have altered the OS [21].
As further support, both Alessi et al. studies highlight SMARCA4 mutations as an independent factor in a significantly worse outcome for NSCLC patients [21,22]. However, different conclusions arose in their two separate publication analyses of ICI treatment: (1) based on whether it was used in the first line with chemotherapy (CIT), or (2) in pre-treated patients as a single agent (ICI) or ICI/CTLA4 combination therapy. In the first-line setting, SMARCA4 mutations were associated with significantly worse OS to first-line CIT treatment in non-squamous NSCLC (SMARCA4-mutated 8.1 months vs. SMARCA4 WT 15.0 months (p < 0.001) [22]. In the pre-treatment setting, the authors did not find an association between SMARCA4 mutational status and ICI treatment on PFS or OS, or whether the patient had a class 1 or 2 SMARCA4 mutation [21].
When looking more granularly at truncated vs. nontruncated SMARCA4 mutations or homozygous vs. heterozygous, there was a significantly worse OS when comparing SMARCA4 homozygous truncated vs. WT (7.9 vs. 16.3 months, HR: 1.85; p < 0.0001) but no significant differences when comparing other SMARCA4-mutated categories vs. WT [11]. This significant OS difference was also demonstrated when looking specifically at ICI treatment when comparing homozygous truncated vs. WT (9.9 vs. 19.5 months, HR: 1.62; p = 0.01), but no significant differences were obtained when comparing WT with the other SMARCA4-mutated categories in the ICI-treated patients [11].
TMB is a genomic feature being analyzed in NSCLC across various ICI studies [38]. It is being reviewed as a possible biomarker for the use of ICIs in SMARCA4-mutated NSCLC [21]. Xu et al. showed a worse OS for NSCLC patients that are SMARCA4-mutated (TMB Low or TMB High) as compared to SMARCA4 WT/TMB High (p = 0.00019) [24].
4.2. Clinicopathological Features
Multiple studies, including Liang et al. and Talvitie et al., conducted OS multivariate analyses across different stages, revealing consistently poorer survival for patients with SMARCA4 mutations [5,23]. Liang et al.’s study demonstrated significantly worse OS in both locally advanced (Stage III) and metastatic (Stage IV) SMARCA4-mutated scenarios compared to WT status (Stage III (23.73 vs. 29.43 months, p < 0.01) and Stage IV (12.17 vs. 28.23 months, p < 0.01) [5]. Talvitie et al.’s analysis in Stage III patients further supported worse OS for SMARCA4-mutated patients [23]. Regardless of which stage, multivariate analyses consistently showed worse OS for SMARCA4-mutated patients, highlighting the impact of these mutations on survival outcomes.
This SMARCA4 review illustrated statistically significant OS differences based on brain metastasis and mixed results based on biological sex [5,25,26]. In the context of brain metastases, Miao et al. investigated clinicogenomic outcomes in ALK-positive NSCLC patients treated with alectinib, finding that SMARCA4-mutated patients with brain metastases were the only group with co-mutations to exhibit statistically worse outcomes in a multivariate analysis (HR: 8.76; p = 0.009) [25]. In two multivariate analyses of the impact of biological sex on SMARCA4 mutation OS differences, one analysis demonstrated significantly worse survival for men, and the other demonstrated worse survival for women [5,26]. Pan et al. reported that male NSCLC patients with SMARCA4 mutations had a markedly worse OS than females (HR: 12.64; p = 0.002), while Liang et al.’s study highlighted a worse OS for women across a cohort of Stage IV SMARCA4-mutated patients (HR:14.2; p = 0.02) [5,26]. Although women had a statistically significant worse OS in the multivariate analysis, the authors share that one of the reasons for the possible large female survival discrepancy was due to lower numbers of females than males in the study (88 vs. 133) [5].
In the context of SMARCA4 NSCLC patients treated with immune checkpoint inhibitors (ICIs), one study found that those with SMARCA4 mutations had worse outcomes than WT patients regardless of ICI treatment or not (HR = 3.2; p = 0.006), and two separate OAK trial NSCLC analyses showed no OS differences with atezolizumab (ICI) vs. docetaxel in pre-treated NSCLC SMARCA4-mutated patients [27,28,29]. Three additional studies demonstrated significantly worse OS outcomes for NSCLC patients with SMARCA4 mutations vs. WT patients across a multivariate analysis and various standards of care treatments [30,31,32].
4.3. SMARCA4/KRAS Co-Mutations
All seven SMARCA4/KRAS co-mutation NSCLC publications in this review showed either a significantly worse PFS or OS as compared to SMARCA4 WT/KRAS mutation analyses across standard-of-care cancer treatments or within a specific treatment analysis (i.e., KRAS G12C inhibitor, ICIs, or non-ICIs) [8,21,22,34,35,36,37]. Two of these studies presented granular survival outcomes specific to two KRAS isoforms (G12C and G12D) [34,35]. Finally, one analysis demonstrated significantly worse survival for SMARCA4/KRAS-co-mutated patients as they presented at a higher rate of Stage IV NSCLC with a significantly higher propensity of brain or liver metastasis [36]. These higher rates of metastatic spread led to significantly worse survival for co-mutated SMARCA4/KRAS vs. SMARCA4 WT/KRAS-mutated patients who either had brain or liver metastasis (i.e., brain metastasis 7.4 months vs. 15.0 months, HR: 2.1; p = 0.0003) [36].
SMARCA4/KRAS co-mutations are among the most common co-mutations within this review. Among the largest co-mutation analyses within the review, SMARCA4/KRAS was co-mutated at a rate of 36% in NSCLC [8]. Within this multi-variate analysis and across various treatments, the SMARCA4/KRAS co-mutation analysis by Schoenfeld et al. resulted in worse OS regardless of class 1 or 2 alteration [class 1: HR: 1.59, and class 2: HR: 2.75] vs. WT (p < 0.001) [8]. Cooper et al.’s PFS co-mutation SMARCA4/KRAS G12D analysis demonstrated significantly worse outcomes for co-mutated patients vs. SMARCA4 WT/KRAS G12D-mutated patients (1.5 vs. 4.0 months; p = 0.0039) [34]. However, as the outlier in this critical review focusing on KRAS G12D, the SMARCA4/KRAS G12C co-mutation OS analysis did not result in a significant difference as compared to SMARCA4 WT/KRAS-mutated NSCLC patients (6.1 vs. 17.3 months; p = 0.42) [34].
4.4. SMARCA4/KRAS Co-Mutation Treatment Analysis
Four studies reported six SMARCA4/KRAS co-mutation OS survival treatment analyses [21,22,35,37]. All six treatment analyses across various treatment types (i.e., ICI, non-ICI, or KRAS G12C inhibitor) resulted in a significantly worse OS (p < 0.05) for SMARCA4/KRAS-co-mutated NSCLC patients. Negrao et al. had the only publication focusing on KRAS G12C NSCLC patients with co-mutations and KRAS G12C inhibitors (sotorasib or adagrasib) [35]. NSCLC patients with SMARCA4/KRAS G12C co-mutations vs. SMARCA4 WT/KRAS G12C mutations had significantly worse OS (4.9 months vs. 11.8 months, HR: 3.07; p < 0.001). In relation to these survival outcomes, SMARCA4 was one of the three tumor suppressor genes significantly enriched in the early progressing subgroup (p = 0.001) [35].
Across two publications by Alessi et al., whether the co-mutated NSCLC patients were treated in the first line with CIT or in ≥ 2nd line with single-agent ICI in combination with CTLA-4, the patients had statistically worse OS [21,22]. Co-mutated SMARCA4/KRAS non-squamous CIT-treated patients had worse OS compared to SMARCA4 WT/KRAS-mutated OS (6.6 vs. 14.6; p < 0.001) [22]. In the second Alessi et al. analysis with single-agent ICIs or an ICI combination with CTLA-4, co-mutated NSCLC patients had significantly worse OS compared with NSCLC patients with SMARCA4 WT/KRAS mutations (3 months versus 15.1 months, HR: 0.29; p < 0.001) [21]. Thus, the authors concluded that SMARCA4/KRAS co-mutation may confer worse NSCLC survival outcomes to ICIs [21].
Liu et al.’s ICIs and non-ICIs treatment adenocarcinoma NSCLC analysis was associated with significantly worse OS or PFS across all four SMARCA4/KRAS co-mutated vs. SMARCA4 WT/KRAS-mutated cohorts [37]. Cohort A and B SMARCA4KRAS-mutated patients were associated with worse OS when treated with non-ICIs (p = 0.047 and p = 0.015). In Cohort C and Cohort D, SMARCA4KRAS-mutated patients treated with immunotherapy had worse PFS and OS (Cohort C (PFS): p = 0.048 and Cohort D (OS): p = 0.0018) [37].
There are potential reasons why current treatments are not highly effective in treating SMARCA4/KRAS NSCLC co-mutations, as the tumor microenvironment (TME) plays an important role in metastatic NSCLC [39]. Regarding ICI therapy, Liu et al. used a signature gene panel to evaluate the level of immune cell type across each NSCLC individual [37]. They found that SMARCA4/KRAS-co-mutated patients had a significantly lower estimated proportion of Cluster of Differentiation 8 (CD8+) T-cells (p = 0.015) and activated CD4+ memory T cells (p = 0.0035) than SMARCA4 WT/KRAS-mutated subjects [37]. This suggests that CD8+ T-cell function may be impacted by regulatory T cells (Tregs), which can lead to a more immunosuppressive tumor microenvironment landscape [39]. Beyond ICI monotherapy and looking at CIT, SMARCA4 mutations can activate Nuclear Factor Erythroid 2-Related Factor 2 (NRF2) signaling, leading to the up-regulation of cytoprotective genes and enabling platinum therapy resistance [40].
5. Future Direction
The majority of SMARCA4-mutated NSCLC patient analyses within this review demonstrated a worse overall survival as compared to SMARCA4 wild-type patients. Mutations in this gene should be explored as a prospective biomarker across ICI, CIT, and KRAS G12C inhibitor clinical trials. As each SMARCA4/KRAS co-mutated treatment analysis in this review showed a statistically worse OS or PFS than SMARCA4 WT/KRAS-mutated patients, a prospective comprehensive immune cell analysis could be considered for SMARCA4-mutated and co-mutated SMARCA4/KRAS patients to understand better how these mutations and co-mutations impact the tumor microenvironment. Utilization of this prospective biomarker approach and immune cell data analysis could inform the design of future NSCLC clinical trials.
6. Limitations
The vast majority of mutation and treatment analyses (i.e., single agent and combination approaches) in this review are retrospective and vary in SMARCA4 mutation sample size. The scope of the publications was limited to those in the English language published from 2018 through 2023. KRAS was the primary co-mutated gene included, rather than analyzing other possible co-mutations with SMARCA4 (e.g., KEAP1/STK11). Given these limitations, this review’s strength is that it is one of the largest SMARCA4 overall survival NSCLC reviews completed to date.
7. Conclusions
In one of the largest reviews to date of SMARCA4-mutated overall survival data in NSCLC, two consistent trends emerged from the twenty-one publications in scope: First, SMARCA4 NSCLC mutated patients had statistically worse OS than SMARCA4 WT patients. Secondly, NSCLC patients with SMARCA4/KRAS co-mutations tended to have significantly worse overall survival than SMARCA4 WT/KRAS-mutated patients, regardless of treatment type, within each treatment analysis. Moreover, designing prospective clinical SMARCA4-mutated or SMARCA4/KRAS-co-mutated NSCLC trials to evaluate targeted therapies and immunotherapy may lead to a better understanding of how to improve cancer patients’ outcomes and survival rates. These findings may have implications for future research and clinical practice.
Author Contributions
Conceptualization, P.M.; methodology, P.M.; validation, P.M.; formal analysis, P.M.; investigation, P.M.; resources, P.M. and D.S.I.; data curation, P.M. and L.B.; writing—original draft preparation, P.M.; writing—review and editing, P.M., L.B. and D.S.I.; supervision, L.B. and D.S.I.; project administration, L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval was not sought for this literature review.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
A special thank you to A.M. for all her support. Also, thank you to Janice Withycombe for her help in the initial editing of the manuscript.
Conflicts of Interest
P.M. is an employee of and a stockholder in Novartis®. L.B. and D.I. declare no conflicts of interest.
References
- Compilation of CDER-NME-New Biologic Approvals from 1985–2022. Available online: https://www.fda.gov/media/135308/download (accessed on 1 February 2024).
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. FLAURA Investigators Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Surveillance Epidemiology and End Results (SEER). Cancer Stat Facts: Lung and Bronchus Cancer. Available online: https://seer.cancer.gov/statfacts/html/lungb.html (accessed on 1 March 2024).
- Liang, X.; Gao, X.; Wang, F.; Li, S.; Zhou, Y.; Guo, P.; Meng, Y.; Lu, T. Clinical characteristics and prognostic analysis of SMARCA4-deficient non-small cell lung cancer. Cancer Med. 2023, 12, 14171–14182. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Schrock, A.B.; Kem, M.; Jessop, N.; Lee, J.; Ali, S.M.; Ross, J.S.; Lennerz, J.K.; Shaw, A.T.; Mino-Kenudson, M.; et al. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J. Thorac. Oncol. 2020, 15, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, G.; Spurr, L.F.; Li, Y.; Ricciuti, B.; Recondo, G.; Umeton, R.; Nishino, M.; Sholl, L.M.; Meyerson, M.L.; Cherniack, A.D.; et al. Clinicopathological and genomic correlates of programmed cell death ligand 1 (PD-L1) expression in non-squamous non-small-cell lung cancer. Ann. Oncol. 2020, 31, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Bandlamudi, C.; Lavery, J.A.; Montecalvo, J.; Namakydoust, A.; Rizvi, H.; Egger, J.; Concepcion, C.P.; Paul, S.; Arcila, M.E.; et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin. Cancer Res. 2020, 26, 5701–5708. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Graziano, S.L.; Lin, D.; Elvin, J.A.; Vergilio, J.A.; Killian, J.K.; Ngo, N.; Ramkissoon, S.; Severson, E.; Hemmerich, A.; Duncan, D.; et al. SMARCA4 deficient non-small cell lung cancer (NSCLC): A comprehensive genomic profiling (CGP) study. Ann. Oncol. 2019, 30, v652–v653. [Google Scholar] [CrossRef]
- Fernando, T.M.; Piskol, R.; Bainer, R.; Sokol, E.S.; Trabucco, S.E.; Zhang, Q.; Trinh, H.; Maund, S.; Kschonsak, M.; Chaudhuri, S.; et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat. Commun. 2020, 11, 5551. [Google Scholar] [CrossRef]
- Mardinian, K.; Adashek, J.J.; Botta, G.P.; Kato, S.; Kurzrock, R. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol. Cancer Ther. 2021, 20, 2341–2351. [Google Scholar] [CrossRef]
- Hodges, H.C.; Stanton, B.Z.; Cermakova, K.; Chang, C.Y.; Miller, E.L.; Kirkland, J.G.; Ku, W.L.; Veverka, V.; Zhao, K.; Crabtree, G.R. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 2018, 25, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Armon, S.; Hofman, P.; Ilié, M. Perspectives and Issues in the Assessment of SMARCA4 Deficiency in the Management of Lung Cancer Patients. Cells 2021, 10, 1920. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Orlando, K.A.; Nguyen, V.; Raab, J.R.; Walhart, T.; Weissman, B.E. Remodeling the cancer epigenome: Mutations in the SWI/SNF complex offer new therapeutic opportunities. Expert Rev. Anticancer Ther. 2019, 19, 375–391. [Google Scholar] [CrossRef] [PubMed]
- St Pierre, R.; Kadoch, C. Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 2017, 42, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Manolakos, P.; Ward, L.D. A Critical Review of the Prognostic and Predictive Implications of KRAS and STK11 Mutations and Co-Mutations in Metastatic Non-Small Lung Cancer. J. Pers. Med. 2023, 13, 1010. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Alessi, J.V.; Ricciuti, B.; Spurr, L.F.; Gupta, H.; Li, Y.Y.; Glass, C.; Nishino, M.; Cherniack, A.D.; Lindsay, J.; Sharma, B.; et al. SMARCA4 and Other SWItch/Sucrose Non Fermentable Family Genomic Alterations in NSCLC: Clinicopathologic Characteristics and Outcomes to Immune Checkpoint Inhibition. J. Thorac. Oncol. 2021, 16, 1176–1187. [Google Scholar] [CrossRef]
- Alessi, J.V.; Elkrief, A.; Ricciuti, B.; Wang, X.; Cortellini, A.; Vaz, V.R.; Lamberti, G.; Frias, R.L.; Venkatraman, D.; Fulgenzi, C.A.; et al. Clinicopathologic and Genomic Factors Impacting Efficacy of First-Line Chemoimmunotherapy in Advanced NSCLC. J. Thorac. Oncol. 2023, 18, 731–743. [Google Scholar] [CrossRef]
- Talvitie, E.M.; Liljeroos, L.; Vilhonen, H.; Orte, K.; Leivo, I.; Kallajoki, M.; Taimen, P. Comprehensive genomic profiling of Finnish lung adenocarcinoma cohort reveals high clinical actionability and SMARCA4 altered tumors with variable histology and poor prognosis. Neoplasia 2022, 32, 100832. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, H.C.; Yang, L.; Yang, G.; Liang, L.; Yang, Y.; Tang, H.; Bao, H.; Wu, X.; Shao, Y.; et al. Mutational landscape of SWI/SNF complex genes reveal correlation to predictive biomarkers for immunotherapy sensitivity in lung adenocarcinoma patients. ESMO Open 2023, 8, 101585. [Google Scholar] [CrossRef] [PubMed]
- Miao, E.; Eichholz, J.E.; Lebow, E.S.; Flynn, J.; Zhang, Z.; Walch, H.; Hubbeling, H.; Beal, K.; Moss, N.S.; Yu, K.K.; et al. Characterization of Central Nervous System Clinico-Genomic Outcomes in ALK-Positive Non-Small Cell Lung Cancer Patients with Brain Metastases Treated with Alectinib. Lung Cancer 2023, 178, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Jiang, C.; Zhang, Z.; Achacoso, N.; Solorzano-Pinto, A.V.; Tse, P.; Chung, E.; Suga, J.M.; Thomas, S.; Habel, L.A. Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies. Cancers 2023, 15, 2665. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shan, Q.; Guo, J.; Han, X.; Zhao, C.; Li, H.; Wang, Z. PDL1 high expression without TP53, KEAP1 and EPHA5 mutations could better predict survival for patients with NSCLC receiving atezolizumab. Lung Cancer 2021, 151, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, D.; Mazzotta, M.; Scalera, S.; Terrenato, I.; Sperati, F.; D’Ambrosio, L.; Pallocca, M.; Corleone, G.; Krasniqi, E.; Pizzuti, L.; et al. KEAP1-driven co-mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann. Oncol. 2020, 31, 1746–1754. [Google Scholar] [CrossRef] [PubMed]
- Velut, Y.; Decroix, E.; Blons, H.; Alifano, M.; Leroy, K.; Petitprez, F.; Boni, A.; Garinet, S.; Biton, J.; Cremer, I.; et al. SMARCA4-deficient lung carcinoma is an aggressive tumor highly infiltrated by FOXP3+ cells and neutrophils. Lung Cancer 2022, 169, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Li, W.; Bai, H.; Duan, J.; Wang, Z.; Du, X.; Yu, R.; Wang, Y.; Wang, M.; Zhu, Y.; et al. Correlations of switch/sucrose nonfermentable complex mutations with clinical outcomes in advanced non-small cell lung cancer. Thorac. Cancer 2022, 13, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Tang, M.; Cui, L.; Bai, J.; Yu, J.; Gao, J.; Nie, X.; Li, X.; Xia, X.; Yi, X.; et al. Prognostic and predictive impact of molecular tumor burden index in non-small cell lung cancer patients. Thorac. Cancer 2023, 14, 3097–3107. [Google Scholar] [CrossRef]
- Wang, C.X.; Yan, J.; Lin, S.; Ding, Y.; Qin, Y.R. Mutant-allele dispersion correlates with prognosis risk in patients with advanced non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 8545–8555. [Google Scholar] [CrossRef]
- La Fleur, L.; Falk-Sörqvist, E.; Smeds, P.; Berglund, A.; Sundström, M.; Mattsson, J.S.; Brandén, E.; Koyi, H.; Isaksson, J.; Brunnström, H.; et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019, 130, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Muzikansky, A.; Lennerz, J.; Narinesingh, F.; Mino-Kenudson, M.; Hung, Y.P.; Piotrowska, Z.; Dagogo-Jack, I.; Sequist, L.V.; Gainor, J.F.; et al. Clinicopathologic Characteristics and Outcomes for Patients with KRAS G12D-Mutant NSCLC. JTO Clin. Res. Rep. 2022, 3, 100390. [Google Scholar] [CrossRef] [PubMed]
- Negrao, M.V.; Araujo, H.A.; Lamberti, G.; Cooper, A.J.; Akhave, N.S.; Zhou, T.; Delasos, L.; Hicks, J.K.; Aldea, M.; Minuti, G.; et al. Comutations and KRASG12C Inhibitor Efficacy in Advanced NSCLC. Cancer Discov. 2023, 13, 1556–1571. [Google Scholar] [CrossRef] [PubMed]
- Boiarsky, D.; Lydon, C.A.; Chambers, E.S.; Sholl, L.M.; Nishino, M.; Skoulidis, F.; Heymach, J.; Luo, J.; Awad, M.; Janne, P.; et al. Molecular markers of metastatic disease in KRAS-mutant lung adenocarcinoma. Ann. Oncol. 2023, 34, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ahmed, T.; Petty, W.J.; Grant, S.; Ruiz, J.; Lycan, T.W.; Topaloglu, U.; Chou, P.; Miller, L.D.; Hawkins, G.A.; et al. SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: A multi-cohort analysis. Mol. Oncol. 2021, 15, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Wang, X.; Alessi, J.V.; Rizvi, H.; Mahadevan, N.R.; Li, Y.Y.; Polio, A.; Lindsay, J.; Umeton, R.; Sinha, R.; et al. Association of high tumor mutation burden in non-small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol. 2022, 8, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wu, K.; Liu, Q.; Han, N.; Zhang, L.; Chu, Q.; Chen, Y. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J. Hematol. Oncol. 2016, 9, 83. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).