Deep Learning-Based DNA Methylation Detection in Cervical Cancer Using the One-Hot Character Representation Technique
Abstract
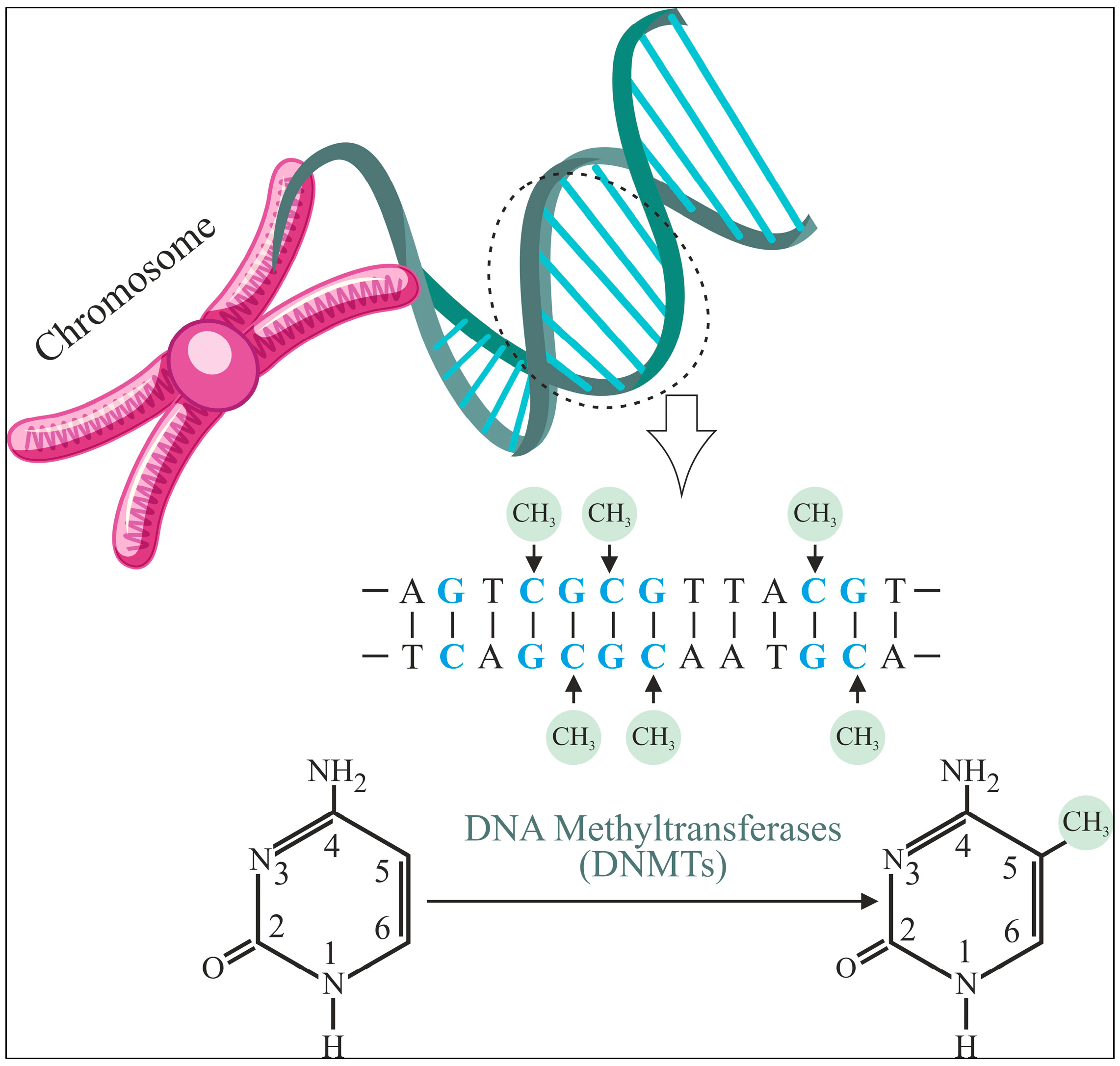
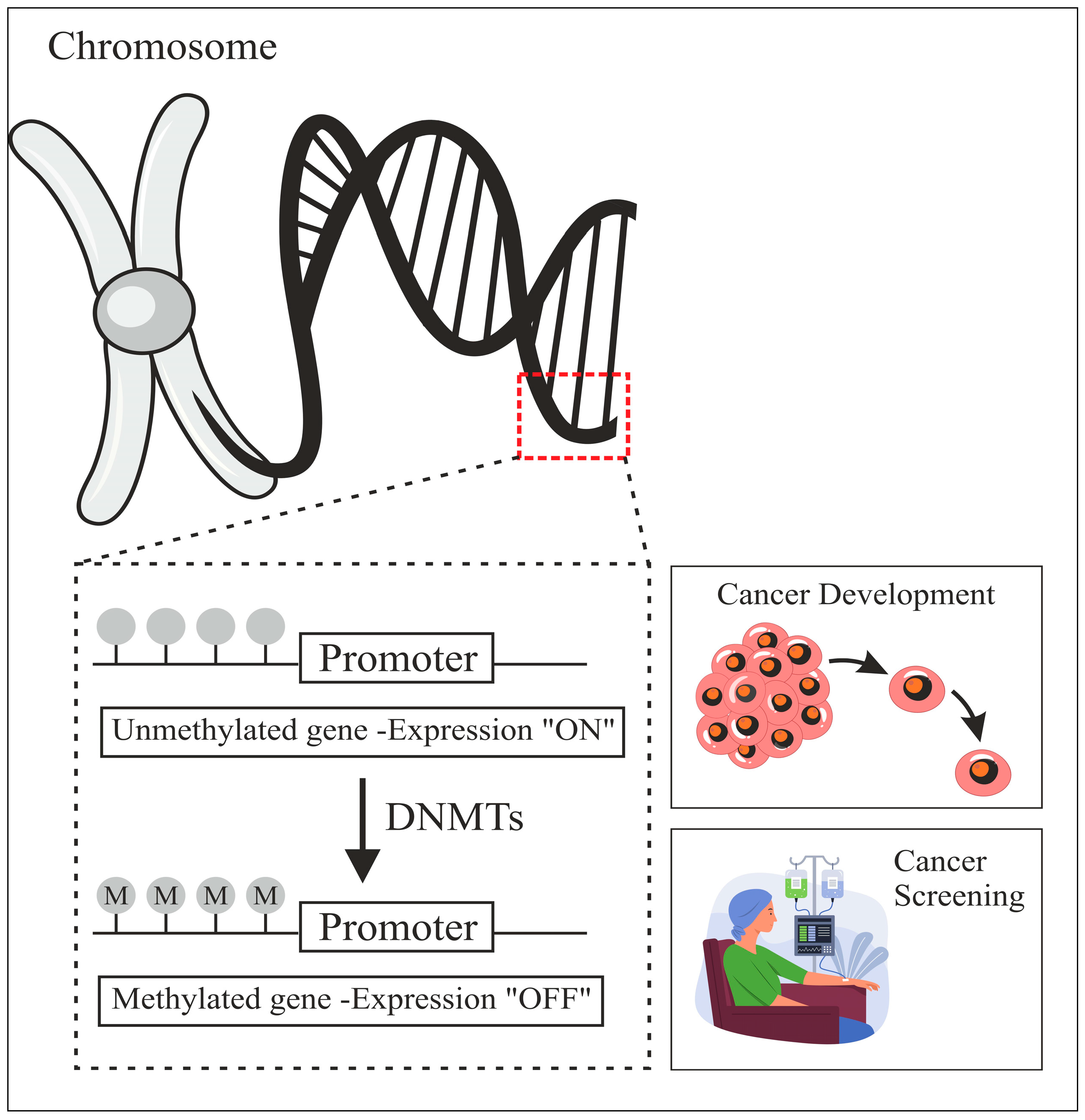
1. Introduction
Novel Contribution of the Study
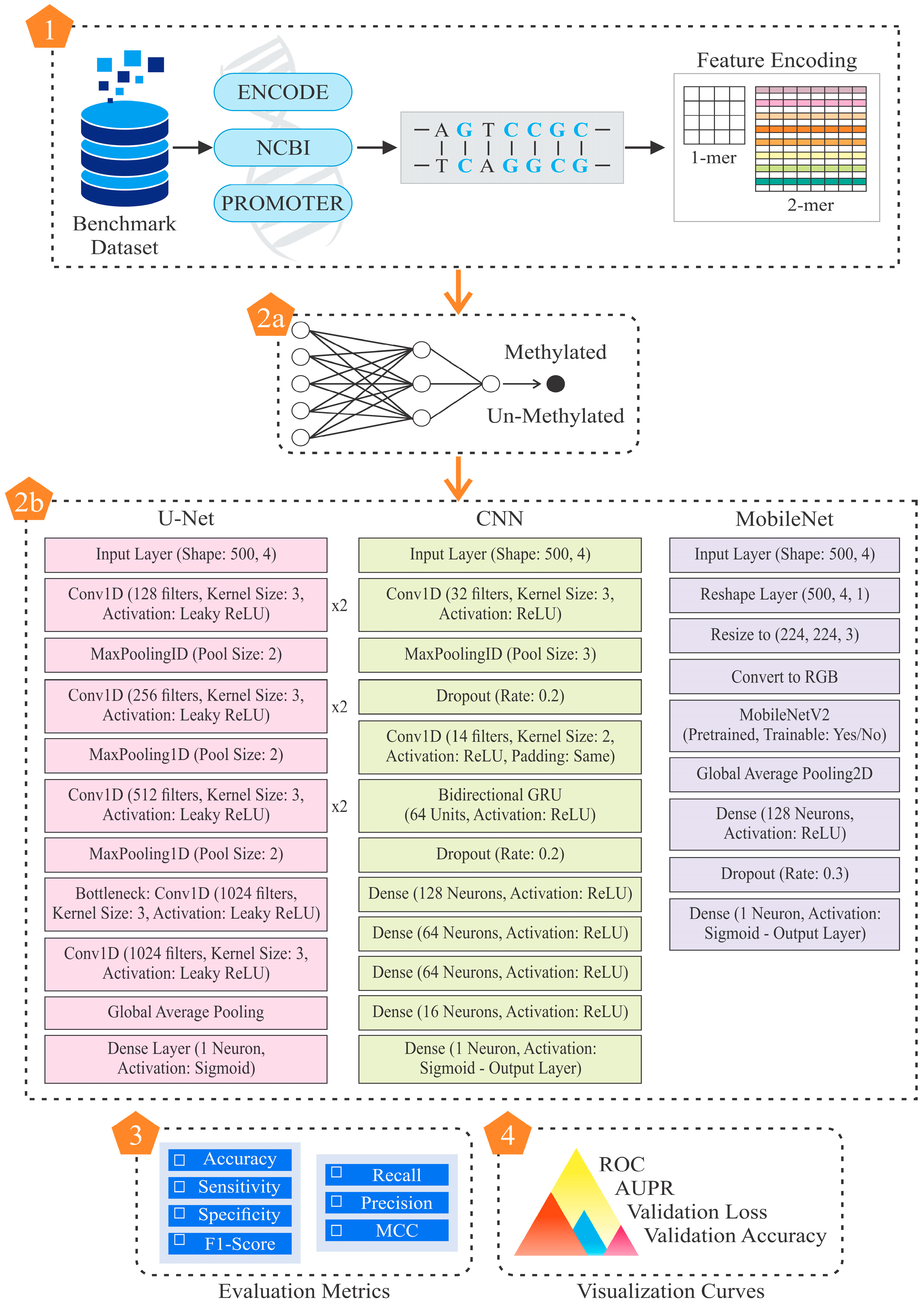
2. Materials and Methods
2.1. Benchmark Dataset
2.2. Feature Encoding Methods
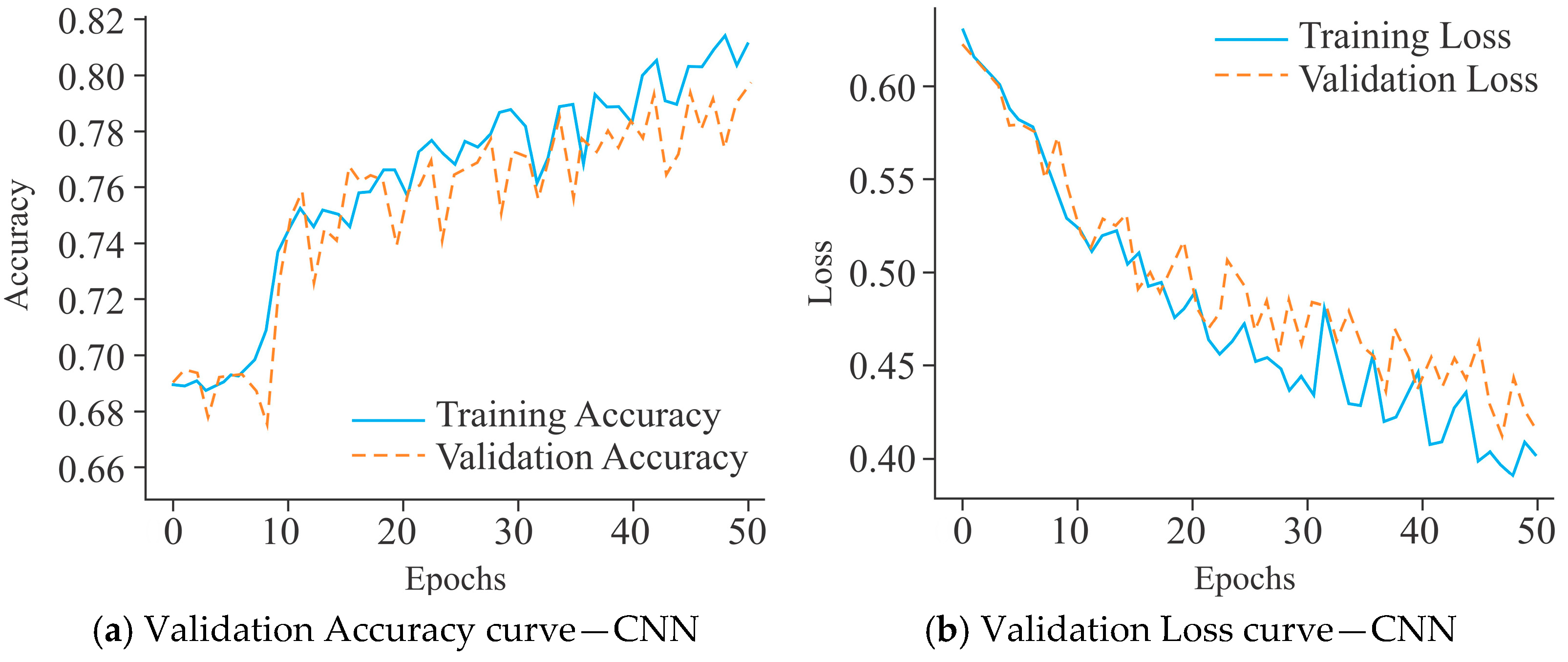
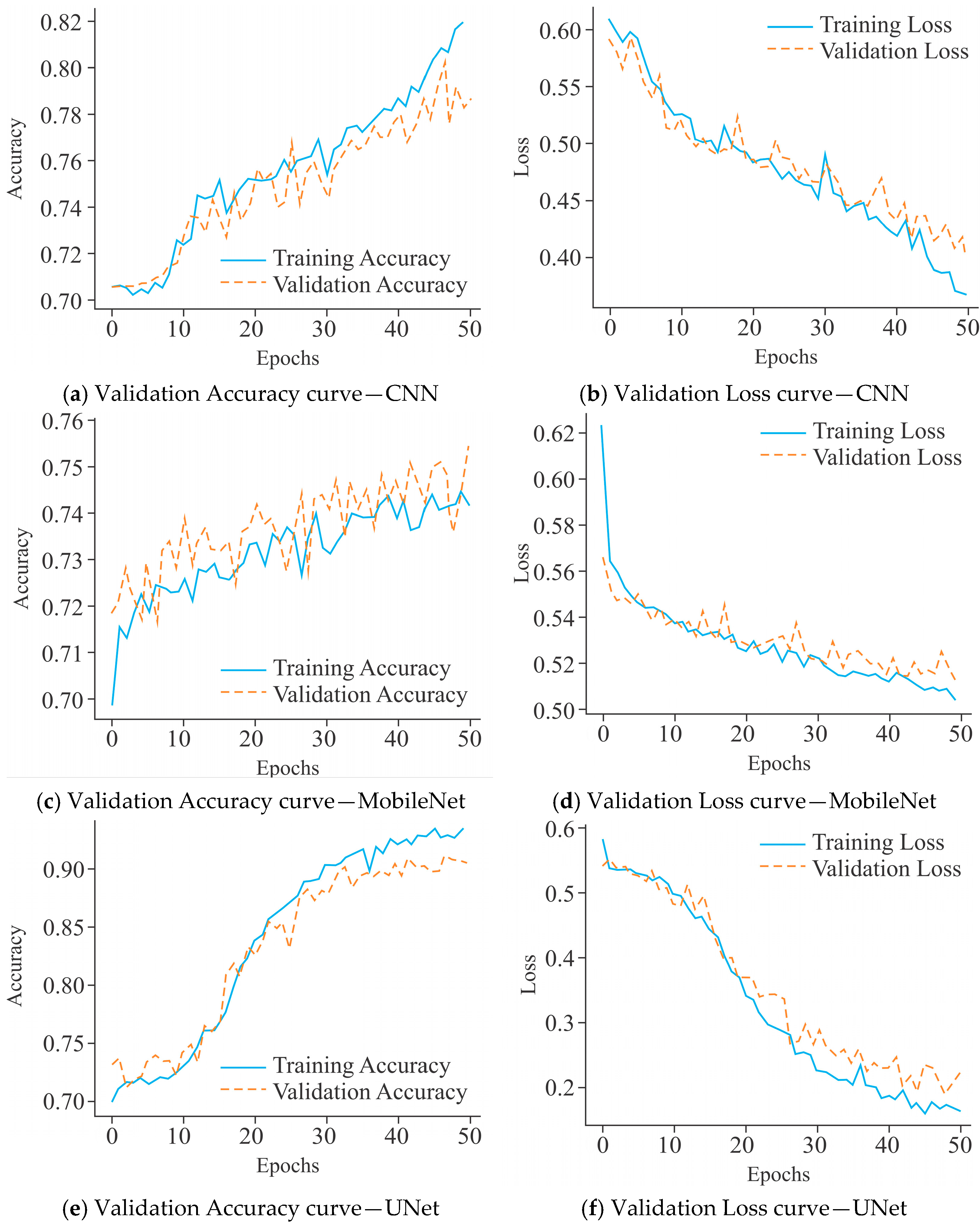
2.3. Model Training and Evaluation
2.4. Validation of Promoter-Region Genes
2.5. State-of-the-Art Methods
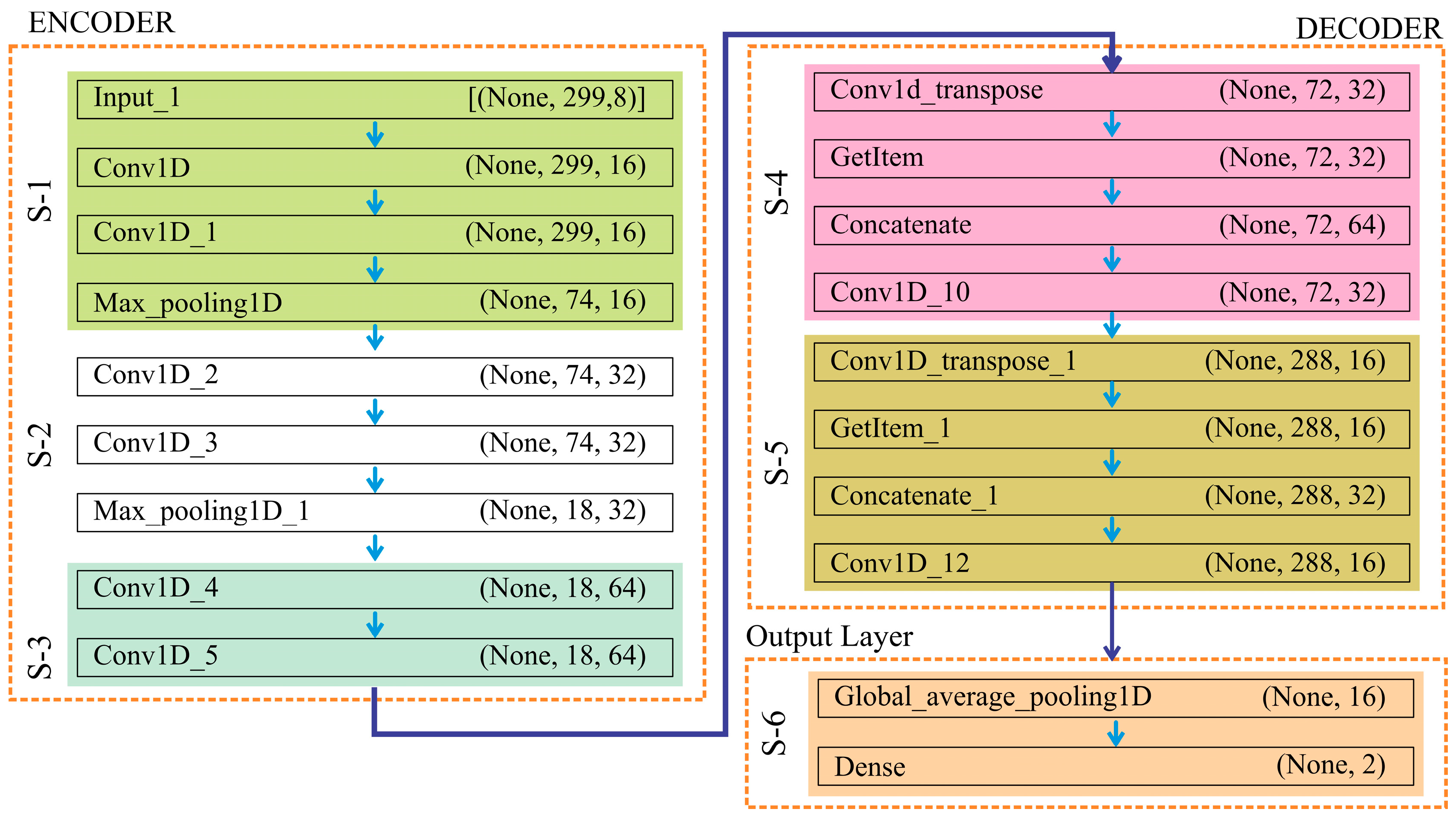
2.5.1. Proposed Convolutional Neural Network
2.5.2. Proposed Mobile Net
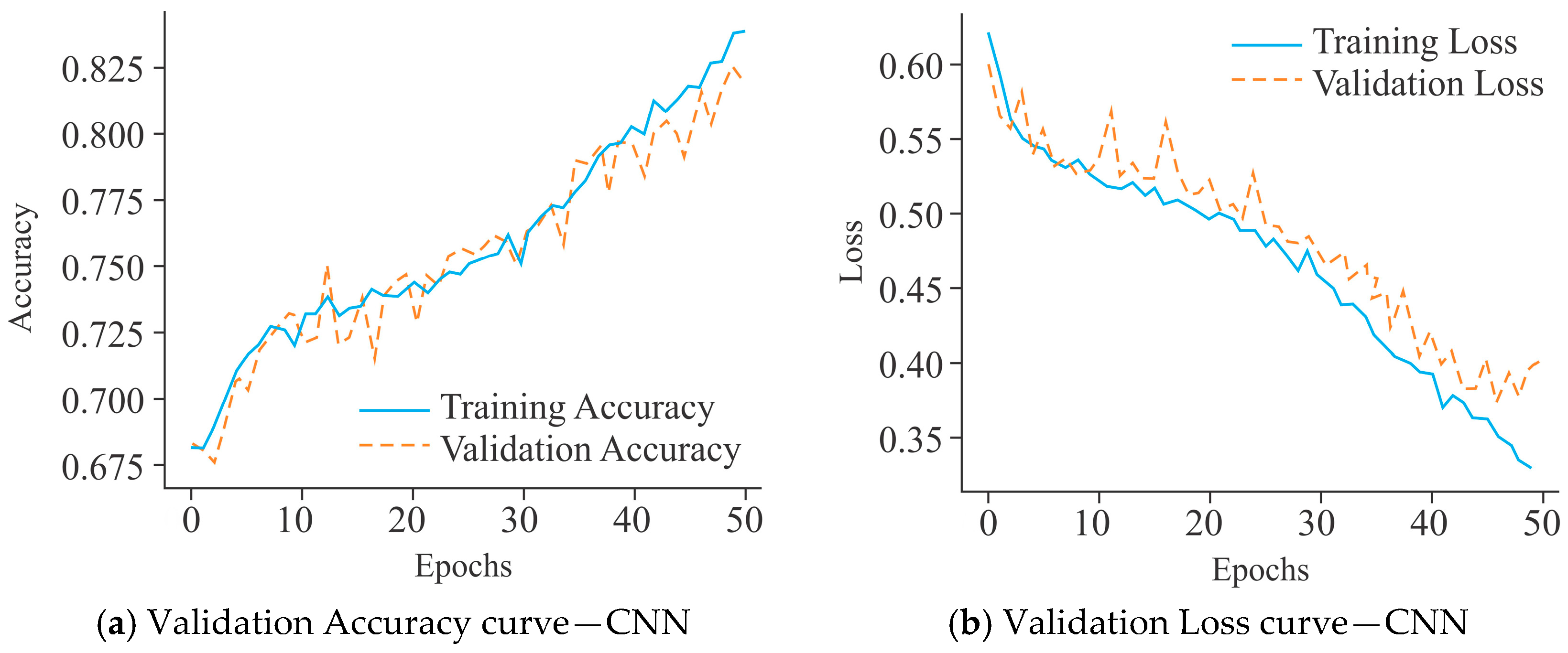
3. Results and Discussion
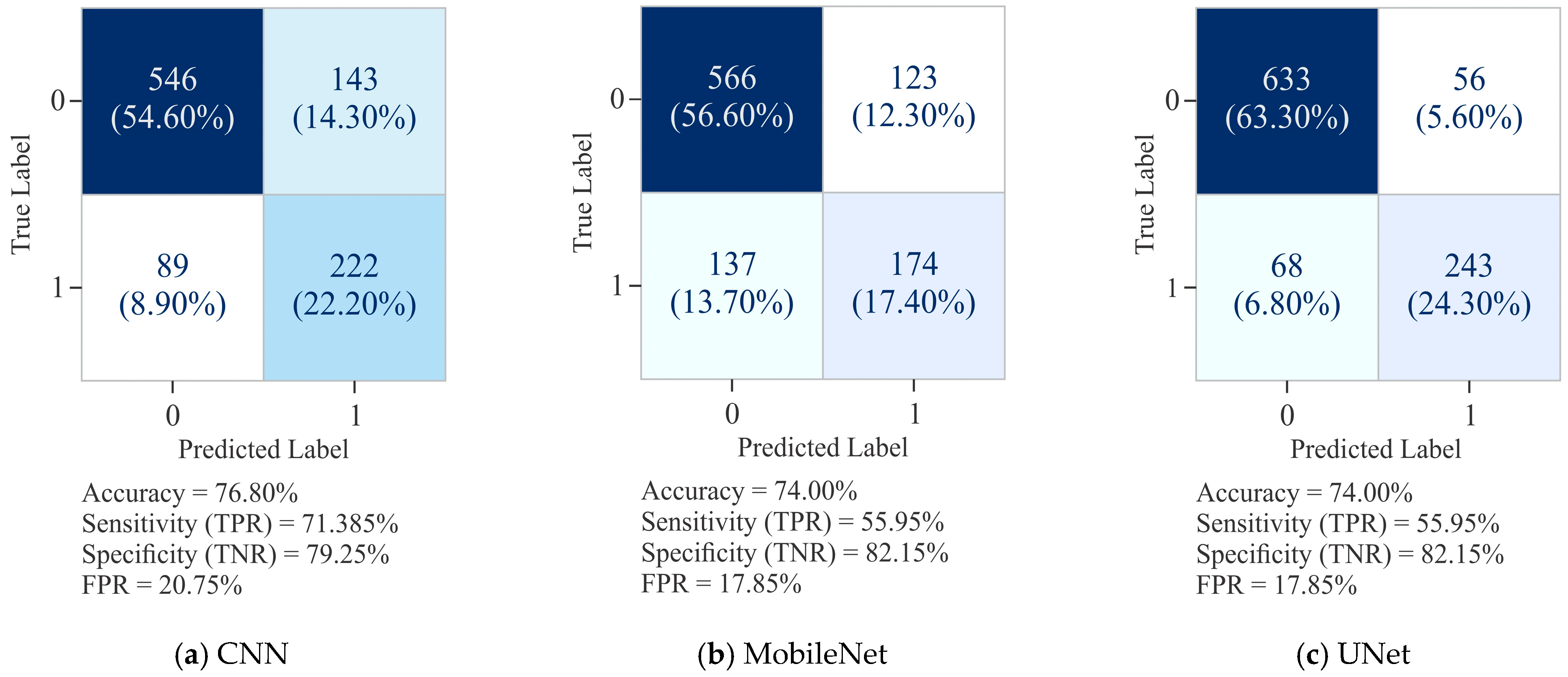
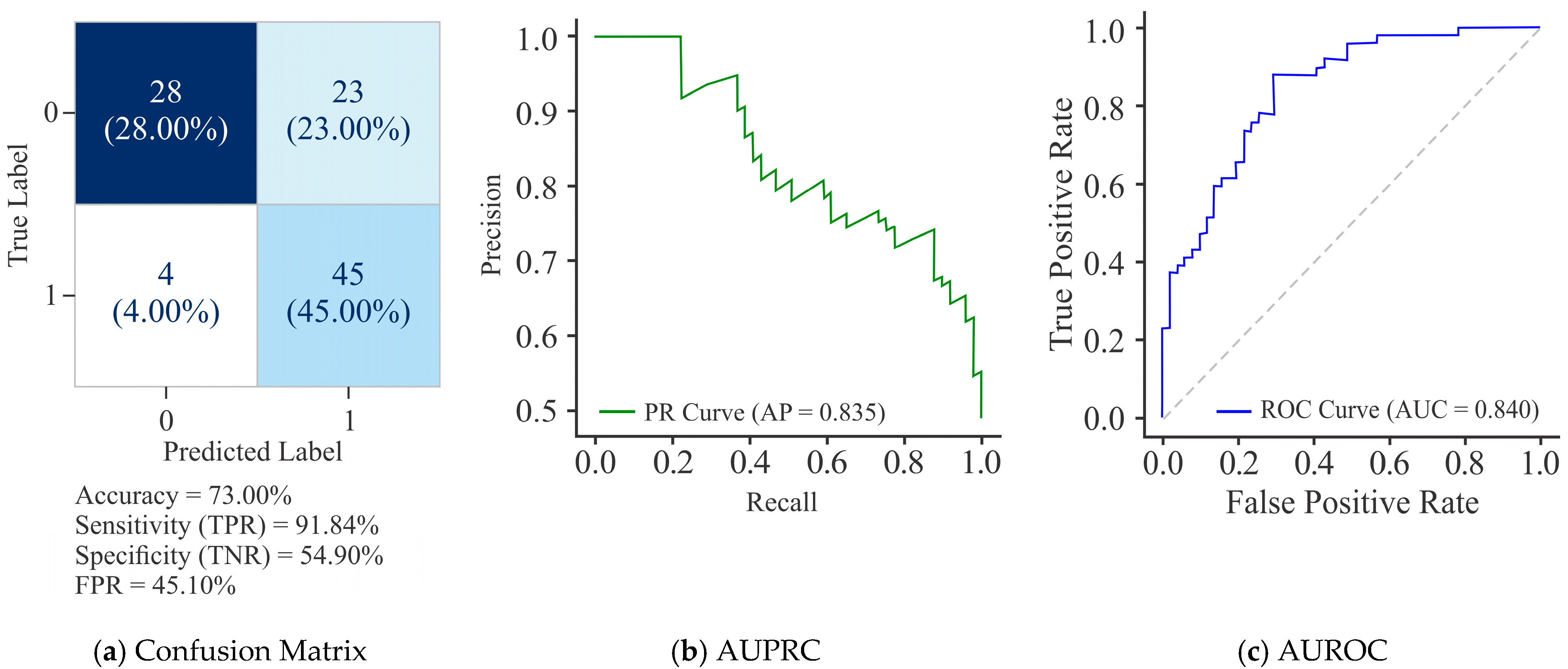
3.1. Comparative Assessment Showing the Influence of All Three Models on the HeLa Cell Line with Different Sample Sizes Using Varied Encoding Techniques and Window Sizes on the Basis of Evaluation Metrics
3.2. Assessing the Influence of All Three Selected Models on Different Cell Lines Using Various Sample Sizes
3.2.1. Assessing the Performance of the Proposed Model in Comparison to CNN and MobileNet on 5000 CG Sites of the HepG2 Cell Line
3.2.2. Performance Assessment of the Proposed Model in Comparison to CNN and MobileNet on 10,000 CG Sites of the HepG2 Cell Line
3.2.3. Assessing the Performance of the Proposed Model with CNN and MobileNet on 20,000 CG Sites of the HepG2 Cell Line
3.3. Validation Using CGs Present in the Promoter Region
4. Conclusions, Limitations, and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. The Pseudocode of the UNet Model Is as Follows
| Variables: | |
| : | |
| : | |
| : | |
| : | |
| Input: as | |
| Output: Classification labels | |
| Encoder Block | |
| Step 1: | |
| = (, | |
| = () | |
| = () | |
| Step 2: | |
| = ( | |
| = () | |
| = () | |
| Step 3: | |
| = ( | |
| = ( | |
| Decoder Block | |
| Step 4: | |
| = (, | |
| 2 = () | |
| = () | |
| Step 5: | |
| = () | |
| = () | |
| = () | |
| Output Layer | |
| Step 6: | |
| = () | |
| Step 7: | |
| Apply a final dense layer with activation function | |
| () | |
References
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef] [PubMed]
- Handa, V.; Jeltsch, A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005, 348, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.A.; Hitz, B.C.; Sloan, C.A.; Chan, E.T.; Davidson, J.M.; Gabdank, I.; Hilton, J.A.; Jain, K.; Baymuradov, U.K.; Narayanan, A.K.; et al. The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 2018, 46, 794–801. [Google Scholar] [CrossRef]
- Choi, J.; Chae, H. methCancer-gen: A DNA methylome dataset generator for user-specified cancer type based on conditional variational autoencoder. BMC Bioinform. 2020, 21, 181. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ma, D.; Zhao, S. DNA methylation changes in cervical cancers. In Cancer Epigenetics: Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; pp. 155–176. [Google Scholar] [CrossRef]
- Clarke, M.A.; Wentzensen, N.; Mirabello, L.; Ghosh, A.; Wacholder, S.; Harari, A.; Lorincz, A.; Schiffman, M.; Burk, R.D. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2125–2137. [Google Scholar] [CrossRef]
- Gene (Internet) (2004) Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 1 September 2023).
- Wu, C.; Yang, H.; Li, J.; Geng, F.; Bai, J.; Liu, C.; Kao, W. Prediction of DNA methylation site status based on fusion deep learning algorithm. In Proceedings of the 2022 5th International Conference on Advanced Electronic Materials, Computers and Software Engineering (AEMCSE), Wuhan, China, 22–24 April 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 180–183. [Google Scholar] [CrossRef]
- Khwaja, M.; Kalofonou, M.; Toumazou, C. A deep belief network system for prediction of DNA methylation. In Proceedings of the 2017 IEEE Biomedical Circuits and Systems Conference (BioCAS), Turin, Italy, 19–21 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Zheng, H.; Wu, H.; Li, J.; Jiang, S.W. CpGIMethPred: Computational model for predicting methylation status of CpG islands in human genome. BMC Med. Genom. 2013, 6, 1–12. [Google Scholar] [CrossRef]
- Pavlovic, M.; Ray, P.; Pavlovic, K.; Kotamarti, A.; Chen, M.; Zhang, M.Q. DIRECTION: A machine learning framework for predicting and characterizing DNA methylation and hydroxymethylation in mammalian genomes. Bioinformatics 2017, 33, 2986–2994. [Google Scholar] [CrossRef]
- The McDonnell Genome Institute; Zou, L.S.; Erdos, M.R.; Taylor, D.L.; Chines, P.S.; Varshney, A.; Parker, S.C.J.; Collins, F.S.; Didion, J.P. BoostMe accurately predicts DNA methylation values in whole genome bisulfite sequencing of multiple human tissues. BMC Genom. 2018, 19, 390. [Google Scholar] [CrossRef]
- Seger, C. An Investigation of Categorical Variable Encoding Techniques in Machine Learning: Binary Versus One-Hot and Feature Hashing. 2018. Available online: https://www.diva-portal.org/smash/record.jsf?pid=diva2%3A1259073&dswid=-4068 (accessed on 15 June 2025).
- Choong, A.C.H.; Lee, N.K. Evaluation of convolutionary neural networks modeling of DNA sequences using ordinal versus one-hot encoding method. In Proceedings of the 2017 International Conference on Computer and Drone Applications (IConDA), Kuching, Malaysia, 9–11 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 60–65. [Google Scholar]
- Testas, A. Support vector machine classification with Pandas, scikit-learn, and PySpark. In Distributed Machine Learning with PySpark: Migrating Effortlessly from Pandas and Scikit-Learn; Apress: Berkeley, CA, USA, 2023; pp. 259–280. [Google Scholar] [CrossRef]
- Marom, N.D.; Rokach, L.; Shmilovici, A. Using the confusion matrix for improving ensemble classifiers. In Proceedings of the IEEE 26-th Convention of Electrical and Electronics Engineers in Israel, Eilat, Israel, 17–20 November 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 000555–000559. [Google Scholar]
- Varoquaux, G.; Colliot, O. Evaluating machine learning models and their diagnostic value. In Machine Learning for Brain Disorders; Humana: New York, NY, USA, 2023; pp. 601–630. [Google Scholar] [CrossRef]
- Fan, J.; Upadhye, S.; Worster, A. Understanding receiver operating characteristic (ROC) curves. Can. J. Emerg. Med. 2006, 8, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.H.; Yi, L.Z.; Pan, J. Tuning model parameters in class-imbalanced learning with precision-recall curve. Biom. J. 2019, 61, 652–664. [Google Scholar] [CrossRef]
- Mohr, F.; van Rijn, J.N. Fast and informative model selection using learning curve cross-validation. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 9669–9680. [Google Scholar] [CrossRef] [PubMed]
- Yim, A.; Chung, C.; Yu, A. Matplotlib for Python Developers: Effective Techniques for Data Visualization with PYTHON; Packt Publishing Ltd.: Birmingham, UK, 2018; Available online: https://books.google.co.in/books?hl=en&lr=&id=G99YDwAAQBAJ&oi=fnd&pg=PP1&dq=%E2%80%A2%09Yim+A,+Chung+C,+Yu+A+(2018)+Matplotlib+for+Python+Developers:+Effective+techniques+for+data+visualization+with+Python.+Packt+Publishing+Ltd,+&ots=tz91xvaP0d&sig=of_oLdvOw9PYtV7LEOHI84YLW9U&redir_esc=y#v=onepage&q=%E2%80%A2%09Yim%20A%2C%20Chung%20C%2C%20Yu%20A%20(2018)%20Matplotlib%20for%20Python%20Developers%3A%20Effective%20techniques%20for%20data%20visualization%20with%20Python.%20Packt%20Publishing%20Ltd%2C&f=false (accessed on 11 July 2025).
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007, 35, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wu, F.; Liu, Y.; Zhao, Q.; Tang, H. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin. Epigenetics 2019, 11, 1–15. [Google Scholar] [CrossRef]
- Yao, T.; Yao, Y.; Chen, Z.; Peng, Y.; Zhong, G.; Huang, C.; Li, J.; Li, R. CircCASC15-miR-100-mTOR may influence the cervical cancer radioresistance. Cancer Cell Int. 2022, 22, 165. [Google Scholar] [CrossRef]
- Chen, R.; Gan, Q.; Zhao, S.; Zhang, D.; Wang, S.; Yao, L.; Yuan, M.; Cheng, J. DNA methylation of miR-138 regulates cell proliferation and EMT in cervical cancer by targeting EZH2. BMC Cancer 2022, 22, 488. [Google Scholar] [CrossRef]
- Sugimoto, J.; Schust, D.J.; Sugimoto, M.; Jinno, Y.; Kudo, Y. Controlling Trophoblast Cell Fusion in the Human Placenta—Transcriptional Regulation of Suppressyn, an Endogenous Inhibitor of Syncytin-1. Biomolecules 2023, 13, 1627. [Google Scholar] [CrossRef]
- Wu, J. Introduction to convolutional neural networks. Natl. Key Lab. Nov. Softw. Technol. 2017, 5, 495. Available online: https://cs.nju.edu.cn/wujx/paper/CNN.pdf (accessed on 17 July 2025).
- Zhao, R.; Yan, R.; Wang, J.; Mao, K. Learning to monitor machine health with convolutional bi-directional LSTM networks. Sensors 2017, 17, 273. [Google Scholar] [CrossRef]
- Muhammad, L.J.; Algehyne, E.A.; Usman, S.S. Predictive supervised machine learning models for diabetes mellitus. SN Comput. Sci. 2020, 1, 240. [Google Scholar] [CrossRef] [PubMed]
- Alsayed, A.O.; Rahim, M.S.M.; AlBidewi, I.; Hussain, M.; Jabeen, S.H.; Alromema, N.; Hussain, S.; Jibril, M.L. Selection of the right undergraduate major by students using supervised learning techniques. Appl. Sci. 2021, 11, 10639. [Google Scholar] [CrossRef]
- Algehyne, E.A.; Jibril, M.L.; Algehainy, N.A.; Alamri, O.A.; Alzahrani, A.K. Fuzzy neural network expert system with an improved Gini index random forest-based feature importance measure algorithm for early diagnosis of breast cancer in Saudi Arabia. Big Data Cogn. Comput. 2022, 6, 13. [Google Scholar] [CrossRef]
- Chang, H.-C.; Yu, L.-W.; Liu, B.-Y.; Chang, P.-C. Classification of the implant-ridge relationship utilizing the MobileNet architecture. J. Dent. Sci. 2024, 19, 411–418. [Google Scholar] [CrossRef]
- Zeng, H.; Gifford, D.K. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017, 45, e99. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhu, M.; Ren, Z.; Zhao, Q.; Wang, P.; He, C.K.; Zhang, M.; Peng, X.; Wu, B.; Feng, R.; et al. Deep learning algorithm reveals two prognostic subtypes in patients with gliomas. BMC Bioinform. 2022, 23, 417. [Google Scholar] [CrossRef]
- Fu, L.; Peng, Q.; Chai, L. Predicting DNA methylation states with hybrid information based deep-learning model. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1721–1728. [Google Scholar] [CrossRef]
- Mallik, S.; Seth, S.; Bhadra, T.; Zhao, Z. A linear regression and deep learning approach for detecting reliable genetic alterations in cancer using DNA methylation and gene expression data. Genes 2020, 11, 931. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, H.; Yang, Z.; Wang, D. Optimizing the Prognostic Model of Cervical Cancer Based on Artificial Intelligence Algorithm and Data Mining Technology. Wirel. Commun. Mob. Comput. 2022, 1, 5908686. [Google Scholar] [CrossRef]
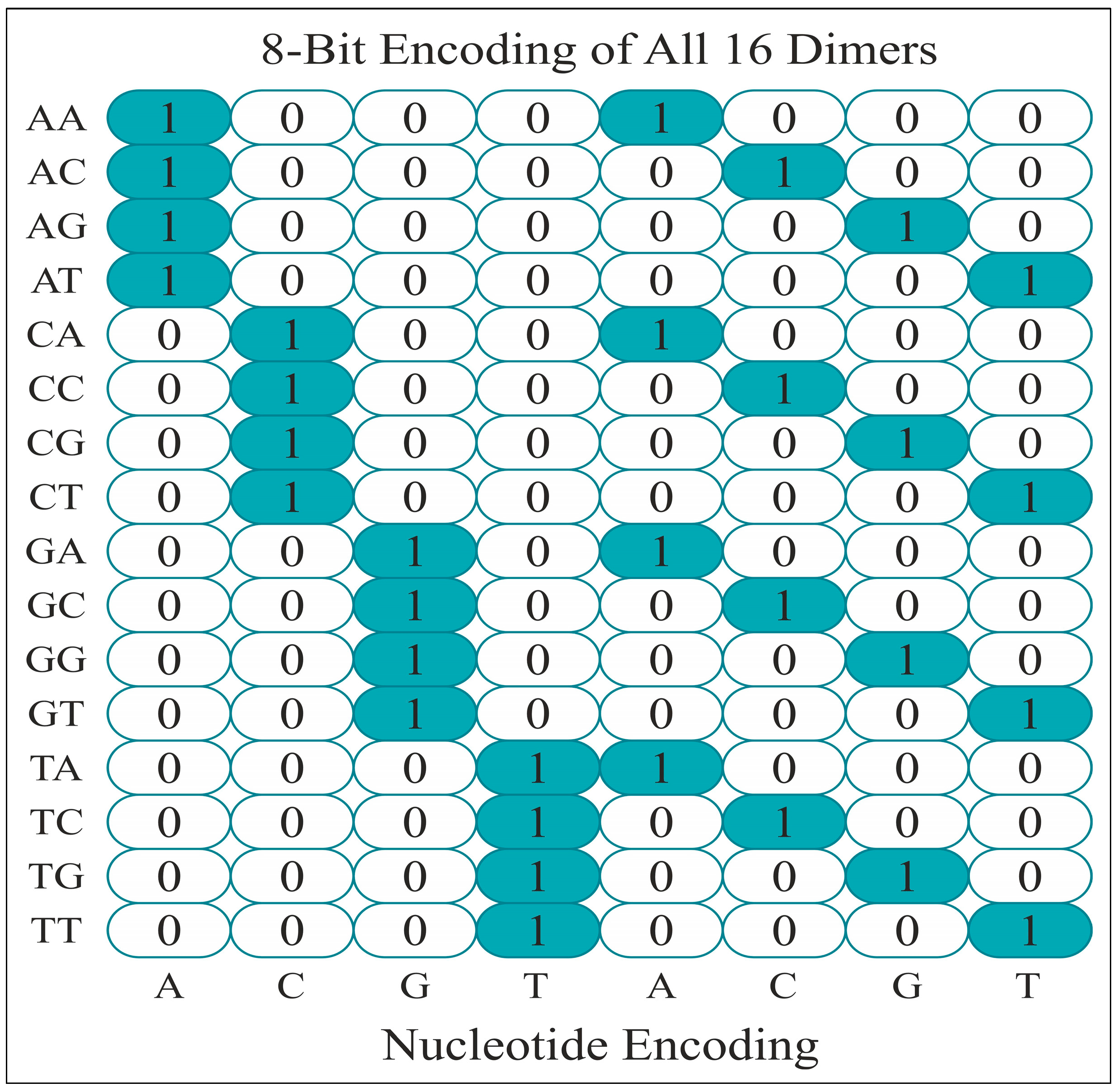





| Base Sequence | A | C | G | T | A | C | G | T | |
|---|---|---|---|---|---|---|---|---|---|
| 1. | CA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2. | AC | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3. | CA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 4. | AC | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 5. | CA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 6. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 7. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 8. | AT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 9. | TA | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| 10. | AG | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 11. | GT | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| 12. | TA | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| 13. | AT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 14. | TA | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| 15. | AC | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 16. | CA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 17. | AT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 18. | TC | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| 19. | CA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 20. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 21. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 22. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 23. | AA | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 24. | AT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 25. | TG | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| 26. | GA | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| 27. | AT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 28. | TT | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| 29. | TT | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| S. No. | Author | Dataset | Pre-Processing | Model | ACC% | SN% | SP% | MCC% | AUROC |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Wu et al., 2022 [9] | GEO, GSE152204 | CNN + RNN + One-hot encoding | ResNet | 84.90 | - | - | - | - |
| 2. | Ma et al., 2022 [39] | miRNA + mRNA datasets for cervical cancer | Functional Normalization | Cox Proportional Hazard Regression Analysis model | - | - | - | - | 83.30 |
| 3. | Mallik et al., 2020 [38] | Uterine Cervical cancer dataset from NCBI | Voom Normalization and Limma | FFNN | 90.69 | 73.97 | 97.63 | 78.23 | 85.80 |
| 4. | Fu et al., 2019 [37] | Cell lines: GM12878 and K562 | Convolutional layers + One-hot encoding | - | - | - | - | 97.70 | |
| 5. | Tian et al., 2022 [36] | Non-cancerous: H1-ESC; Cancerous: white matter of brain, lung tissue and colon tissue | One-hot encoding | CNN | >93.20 | >95.00 | 85.00 | - | 96.00 |
| 6. | Zeng et al., 2017 [35] | 50 human cancer cell lines | 90.00 | - | - | - | 85.40 | ||
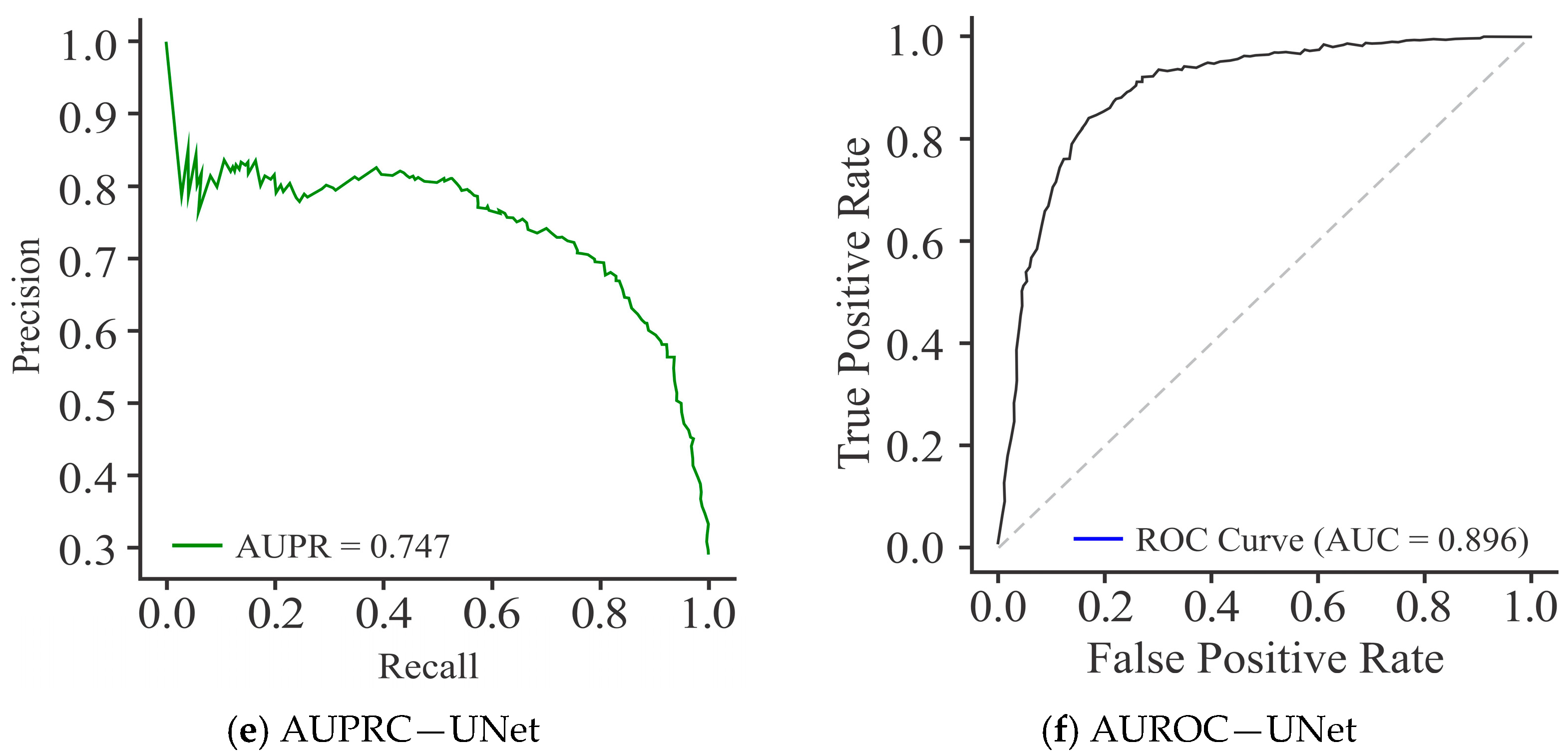
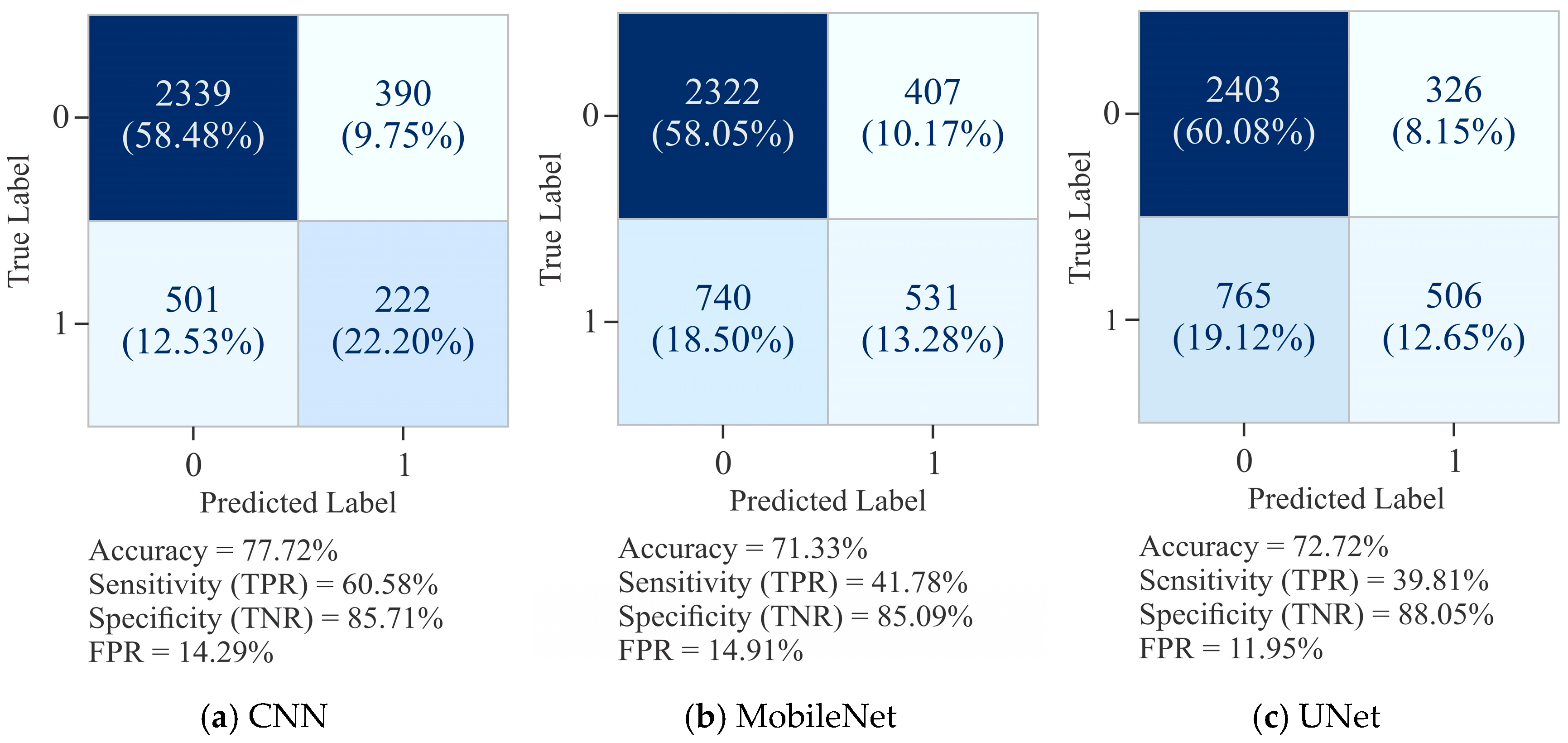
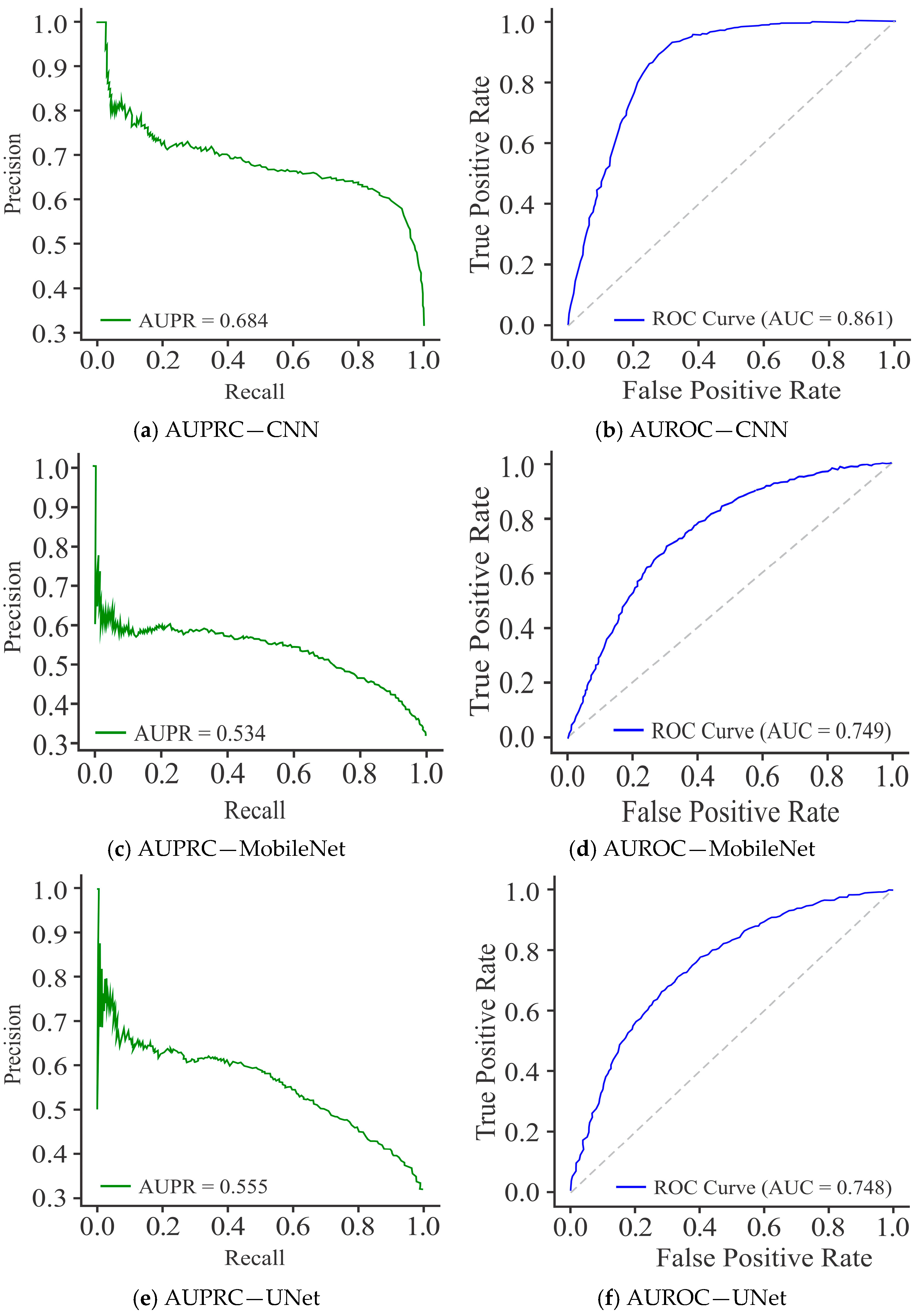
| 7. | Proposed method | HeLa, HepG2 | Dimer encoding | UNet | 91.60 | 96.71 | 87.32 | 83.72 | 96.53 |
| S. No. | Window Size | Encoding | Sample Size | Model | ACC% | SN% | SP% | MCC% | Precision Score | F-1 Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 100 | Monomer | 5000 | UNet | 89.40 | 86.40 | 91.91 | 78.62 | 89.95 | 88.14 |
| 2. | CNN | 76.00 | 66.45 | 84.01 | 51.52 | 77.69 | 71.63 | |||
| 3. | MobileNet | 69.80 | 61.84 | 76.47 | 38.80 | 68.78 | 65.13 | |||
| 4. | 10,000 | UNet | 83.15 | 84.13 | 82.27 | 66.31 | 80.96 | 82.51 | ||
| 5. | CNN | 71.05 | 70.37 | 71.66 | 41.99 | 68.98 | 69.67 | |||
| 6. | MobileNet | 70.30 | 65.35 | 74.45 | 39.96 | 68.19 | 66.74 | |||
| 7. | 20,000 | UNet | 81.77 | 80.92 | 82.60 | 63.54 | 81.88 | 81.40 | ||
| 8. | CNN | 74.68 | 79.81 | 69.69 | 49.71 | 71.89 | 75.64 | |||
| 9. | MobileNet | 68.00 | 46.49 | 86.03 | 35.77 | 73.61 | 56.99 | |||
| 10. | 100 | Dimer | 5000 | UNet | 87.30 | 84.65 | 89.52 | 74.37 | 87.13 | 85.87 |
| 11. | CNN | 76.20 | 76.97 | 75.55 | 52.35 | 72.52 | 74.68 | |||
| 12. | MobileNet | 70.00 | 73.68 | 66.91 | 40.46 | 65.12 | 69.14 | |||
| 13. | 10,000 | UNet | 83.90 | 86.14 | 81.90 | 67.93 | 81.00 | 83.49 | ||
| 14. | CNN | 75.65 | 88.78 | 63.89 | 53.91 | 68.77 | 77.51 | |||
| 15. | MobileNet | 68.80 | 74.60 | 63.60 | 38.30 | 64.74 | 69.32 | |||
| 16. | 20,000 | UNet | 77.88 | 90.56 | 65.55 | 57.83 | 71.86 | 80.13 | ||
| 17. | CNN | 76.55 | 82.14 | 71.12 | 53.54 | 73.42 | 77.54 | |||
| 18. | MobileNet | 70.20 | 67.76 | 72.24 | 39.98 | 67.17 | 67.47 | |||
| 19. | 200 | Monomer | 5000 | UNet | 87.80 | 79.82 | 94.49 | 75.74 | 92.39 | 85.65 |
| 20. | CNN | 72.60 | 46.93 | 94.12 | 47.47 | 86.99 | 60.97 | |||
| 21. | MobileNet | 68.70 | 55.70 | 79.60 | 36.52 | 69.59 | 61.88 | |||
| 22. | 10,000 | UNet | 87.25 | 85.08 | 89.19 | 74.41 | 87.58 | 86.31 | ||
| 23. | CNN | 77.85 | 92.59 | 64.64 | 58.99 | 70.11 | 79.80 | |||
| 24. | MobileNet | 65.20 | 87.28 | 46.69 | 36.52 | 57.85 | 69.58 | |||
| 25. | 20,000 | UNet | 85.62 | 84.78 | 86.45 | 71.24 | 85.87 | 85.32 | ||
| 26. | CNN | 80.90 | 91.63 | 70.48 | 63.41 | 75.09 | 82.54 | |||
| 27. | MobileNet | 69.30 | 62.50 | 75.00 | 37.83 | 67.70 | 64.99 | |||
| 28. | 200 | Dimer | 5000 | UNet | 90.20 | 86.84 | 93.01 | 80.25 | 91.24 | 88.99 |
| 29. | CNN | 81.70 | 78.29 | 84.56 | 63.04 | 80.95 | 79.60 | |||
| 30. | MobileNet | 70.50 | 54.61 | 83.82 | 40.49 | 73.89 | 62.80 | |||
| 31. | 10,000 | UNet | 86.35 | 84.97 | 87.58 | 72.60 | 85.97 | 85.47 | ||
| 32. | CNN | 76.25 | 93.65 | 60.66 | 56.85 | 68.08 | 78.84 | |||
| 33. | MobileNet | 69.10 | 78.84 | 60.38 | 39.69 | 64.06 | 70.68 | |||
| 34. | 20,000 | UNet | 86.10 | 87.32 | 84.92 | 72.23 | 84.90 | 86.09 | ||
| 35. | CNN | 82.67 | 91.07 | 74.52 | 66.40 | 77.64 | 83.82 | |||
| 36. | MobileNet | 71.00 | 72.59 | 69.67 | 42.09 | 66.73 | 69.54 | |||
| 37. | 300 | Monomer | 5000 | UNet | 64.20 | 46.05 | 79.41 | 27.14 | 65.22 | 53.98 |
| 38. | CNN | 80.70 | 79.82 | 81.43 | 61.17 | 78.28 | 79.04 | |||
| 39. | MobileNet | 69.80 | 58.33 | 79.41 | 38.77 | 70.37 | 63.79 | |||
| 40. | 10,000 | UNet | 65.20 | 71.53 | 59.53 | 31.18 | 61.29 | 66.02 | ||
| 41. | CNN | 80.60 | 92.17 | 70.24 | 63.41 | 73.50 | 81.78 | |||
| 42. | MobileNet | 66.60 | 41.67 | 87.50 | 33.20 | 73.64 | 53.22 | |||
| 43. | 20,000 | UNet | 65.20 | 71.53 | 59.53 | 31.18 | 61.29 | 66.02 | ||
| 44. | CNN | 83.27 | 93.25 | 73.58 | 68.03 | 77.42 | 84.60 | |||
| 45. | MobileNet | 69.60 | 67.98 | 70.96 | 38.87 | 66.24 | 67.10 | |||
| 46. | 300 | Dimer | 5000 | UNet | 91.60 | 96.71 | 87.32 | 83.72 | 86.47 | 91.30 |
| 47. | CNN | 85.40 | 83.40 | 87.13 | 70.62 | 84.87 | 84.13 | |||
| 48. | MobileNet | 71.10 | 77.56 | 68.29 | 42.30 | 51.54 | 61.92 | |||
| 49. | 10,000 | UNet | 88.00 | 83.60 | 92.85 | 76.48 | 92.80 | 87.96 | ||
| 50. | CNN | 84.80 | 78.44 | 93.01 | 70.98 | 93.54 | 85.33 | |||
| 51. | MobileNet | 69.70 | 65.77 | 74.27 | 39.98 | 74.81 | 70.00 | |||
| 52. | 20,000 | UNet | 87.08 | 89.60 | 84.62 | 74.27 | 84.99 | 87.23 | ||
| 53. | CNN | 83.63 | 78.24 | 91.14 | 68.43 | 92.50 | 84.77 | |||
| 54. | MobileNet | 71.00 | 72.59 | 69.54 | 42.09 | 66.73 | 69.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apoorva; Handa, V.; Batra, S.; Arora, V. Deep Learning-Based DNA Methylation Detection in Cervical Cancer Using the One-Hot Character Representation Technique. Diagnostics 2025, 15, 2263. https://doi.org/10.3390/diagnostics15172263
Apoorva, Handa V, Batra S, Arora V. Deep Learning-Based DNA Methylation Detection in Cervical Cancer Using the One-Hot Character Representation Technique. Diagnostics. 2025; 15(17):2263. https://doi.org/10.3390/diagnostics15172263
Chicago/Turabian StyleApoorva, Vikas Handa, Shalini Batra, and Vinay Arora. 2025. "Deep Learning-Based DNA Methylation Detection in Cervical Cancer Using the One-Hot Character Representation Technique" Diagnostics 15, no. 17: 2263. https://doi.org/10.3390/diagnostics15172263
APA StyleApoorva, Handa, V., Batra, S., & Arora, V. (2025). Deep Learning-Based DNA Methylation Detection in Cervical Cancer Using the One-Hot Character Representation Technique. Diagnostics, 15(17), 2263. https://doi.org/10.3390/diagnostics15172263






