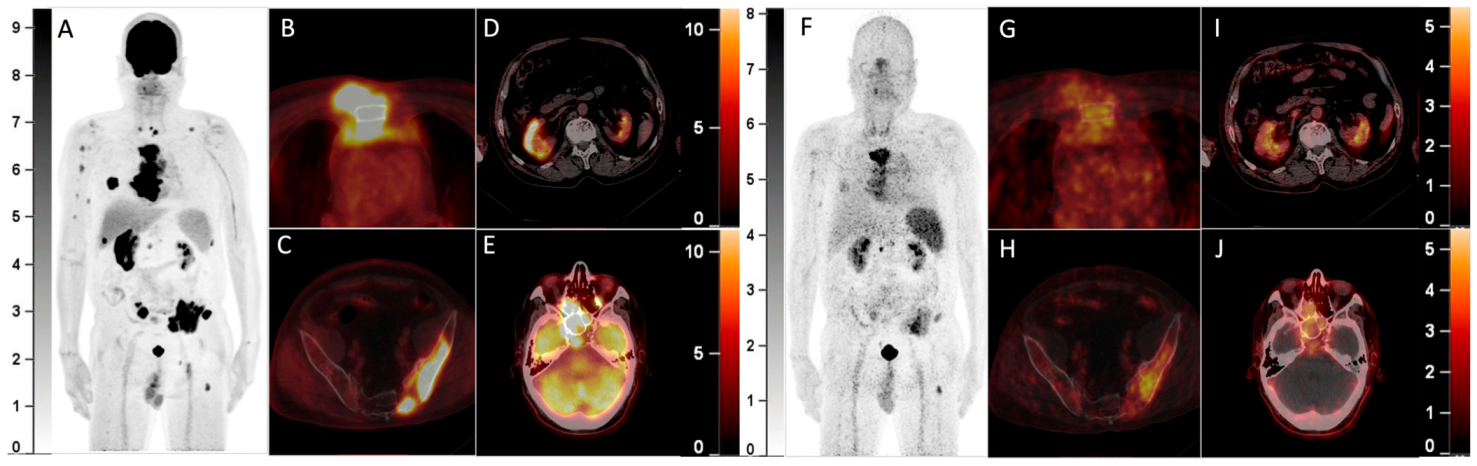
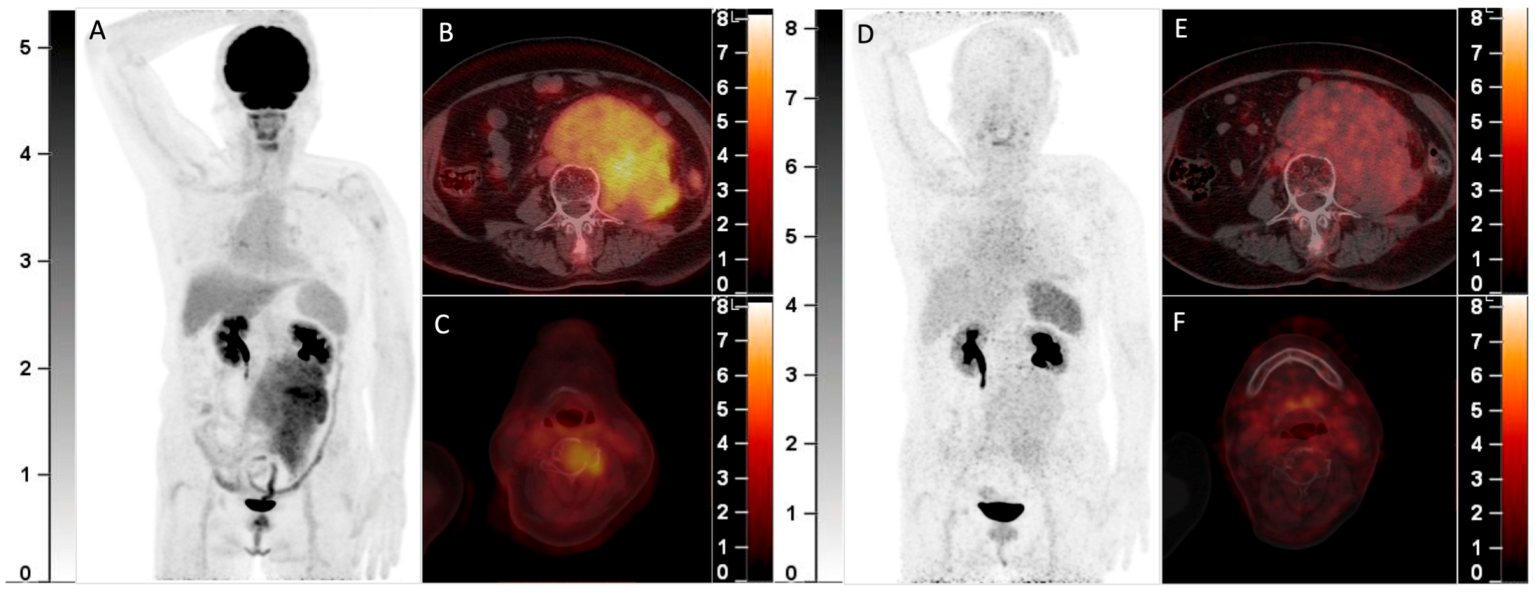
68Ga-Pentixafor PET/CT May Fail to Detect Recurrent Multiple Myeloma with Extramedullary Disease
Abstract
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapa, C.; Schreder, M.; Schirbel, A.; Samnick, S.; Kortüm, K.M.; Herrmann, K.; Kropf, S.; Einsele, H.; Buck, A.K.; Wester, H.J.; et al. [(68)Ga]Pentixafor-PET/CT for imaging of chemokine receptor CXCR4 expression in multiple myeloma-Comparison to [(18)F]FDG and laboratory values. Theranostics 2017, 7, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Cao, X.; Luo, Y.; Li, J.; Feng, J.; Li, F. Chemokine receptor-4 targeted PET/CT with (68)Ga-Pentixafor in assessment of newly diagnosed multiple myeloma: Comparison to (18)F-FDG PET/CT. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Philipp-Abbrederis, K.; Herrmann, K.; Knop, S.; Schottelius, M.; Eiber, M.; Lückerath, K.; Pietschmann, E.; Habringer, S.; Gerngroß, C.; Franke, K.; et al. In vivo molecular imaging of chemokine receptor CXCR4 expression in patients with advanced multiple myeloma. EMBO Mol. Med. 2015, 7, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Luo, Y.; Cao, X.; Ma, Y.; Li, F. Multiple myeloma presenting as a superscan on 68Ga-Pentixafor PET/CT. Clin. Nucl. Med. 2018, 43, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Alsayed, Y.; Ngo, H.; Runnels, J.; Leleu, X.; Singha, U.K.; Pitsillides, C.M.; Spencer, J.A.; Kimlinger, T.; Ghobrial, J.M.; Jia, X.; et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood 2007, 109, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Mishima, Y.; Sacco, A.; Moschetta, M.; Tai, Y.T.; Shi, J.; Zhang, Y.; Reagan, M.R.; Huynh, D.; Kawano, Y.; et al. CXCR4 regulates extra-medullary myeloma through epithelial-mesenchymal-transition-like transcriptional activation. Cell Rep. 2015, 12, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Stessman, H.A.F.; Mansoor, A.; Zhan, F.; Janz, S.; Linden, M.A.; Baughn, L.B.; Van Ness, B. Reduced CXCR4 expression is associated with extramedullary disease in a mouse model of myeloma and predicts poor survival in multiple myeloma patients treated with bortezomib. Leukemia 2013, 27, 2075–2077. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.M. Myeloma as a model for the process of metastasis: Implications for therapy. Blood 2012, 120, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Bladé, J.; Fernández de Larrea, C.; Rosiñol, L. Extramedullary disease in multiple myeloma in the era of novel agents. Br. J. Haematol. 2015, 169, 763–765. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Q.; Luo, Y.; Cao, X.; Li, J.; Li, F. 68Ga-Pentixafor PET/CT May Fail to Detect Recurrent Multiple Myeloma with Extramedullary Disease. Diagnostics 2023, 13, 871. https://doi.org/10.3390/diagnostics13050871
Pan Q, Luo Y, Cao X, Li J, Li F. 68Ga-Pentixafor PET/CT May Fail to Detect Recurrent Multiple Myeloma with Extramedullary Disease. Diagnostics. 2023; 13(5):871. https://doi.org/10.3390/diagnostics13050871
Chicago/Turabian StylePan, Qingqing, Yaping Luo, Xinxin Cao, Jian Li, and Fang Li. 2023. "68Ga-Pentixafor PET/CT May Fail to Detect Recurrent Multiple Myeloma with Extramedullary Disease" Diagnostics 13, no. 5: 871. https://doi.org/10.3390/diagnostics13050871
APA StylePan, Q., Luo, Y., Cao, X., Li, J., & Li, F. (2023). 68Ga-Pentixafor PET/CT May Fail to Detect Recurrent Multiple Myeloma with Extramedullary Disease. Diagnostics, 13(5), 871. https://doi.org/10.3390/diagnostics13050871





