Abstract
Ulcerative colitis (UC) is an intractable disease associated with high morbidity and healthcare costs. Metabolites and gut microbes are areas of interest for mainstream and complementary and alternative medicine. We, therefore, aimed to contribute to the discovery of an integrative medicine for UC by comparing and analyzing gut microbes and metabolites in patients with UC and in healthy individuals. This was an observational case-control study. Blood and stool samples were collected from the participants, and metabolite and gut microbial studies were performed. Among metabolites, formate, glycolate, trimethylamine, valine, and pyruvate levels were significantly different between the two groups. Among gut microbes, the abundance of Bacteroidetes at the phylum level; Bacteroidia at the class level; Bacteroidales and Actinomycetales at the order level; Prevotellaceae, Acidaminococcaceae, and Leptotrichiaceae at the family level; and Prevotella, Roseburia, Paraprevotella, Phascolarctobacterium, Ruminococcus, Coprococcus, Clostridium_XIVB, Atopobium, and Leptotrichia at the genus level was also significantly different. Most of the metabolites and gut microbes significantly different between the two groups were involved in energy metabolism and inflammatory processes, respectively. The results of this study could be helpful for the identification of targets for integrative medicine approaches for UC.
1. Introduction
Ulcerative colitis (UC) is a chronic inflammatory bowel disease (IBD) characterized by inflammation localized in the mucosa or submucosal layer of the colon. The exact cause of this condition is unknown; however, it is thought to be caused by a complex interaction of elements, including the immune system, host genotype, and environment, especially the enteric commensal microbiota [1].
UC is an intractable disease that leads to high morbidity and healthcare costs [1]. Interest in the treatment of this intractable disease in both personalized and integrative medicine has been growing [2,3]. Genes are often mentioned in personalized medicine [4]. However, although genes can be useful for predicting disease potential, treatment targeted at changing the inherited gene itself is difficult for several reasons [5]. Therefore, gut microbes and metabolites may be better targets for personalized medicine. One study reported that even if the genes are the same, disease expression may differ depending on the gut microbes [6]. In addition, for integrative medicine, there must be a common field of communication among the various medical approaches, such as Western medicine and complementary and alternative medicine. Metabolites and gut microbes could be good candidates because various medical approaches have focused on them [7,8,9].
Clinical evidence suggests that metabolites and gut microbes play a role in the pathogenesis of IBD, including UC. For example, Lavelle et al. reported that metabolites derived from gut microbes are key actors in IBD [10], and Zitomerskty et al. reported that patients with IBD who undergo surgical diversion of the fecal stream recover their uninflamed healthy intestines, but the inflammation recurs when re-exposed to the microbial laden fecal stream [11]. Another study reported that antibiotics targeting anaerobic gut microbes have shown efficacy in treating IBD [12].
Whether metabolites and gut microbes cause or result from IBD remains controversial. However, considering the existing studies, it is clear that the regulation of gut microbes and metabolites could be a new target for integrative medicine treatment [13].
The purpose of this study was to attempt to discover new targets for personalized and integrative medicine approaches to UC by comparing and analyzing gut microbes and metabolites in patients with UC and healthy individuals. Although the number of participants was small, we report our findings because we obtained significant results.
2. Materials and Methods
2.1. Study Design
This was an observational study with a case-control design.
2.2. Subjects
2.2.1. Sample Size Calculation
As this was a pilot study and we were unable to find previous data that indicated the sample size required to produce significant findings, we relied on the recommendation made by Kieser and Wassmer that a sample size of 20–40 people be included in the pilot study [14]. From 10 December 2018 to 9 June 2020, posters in communities and hospitals were used to recruit the healthy control (HC) and UC groups. The HC group was age- and gender-matched with the UC group.
2.2.2. Inclusion and Exclusion Criteria for the UC Group
The inclusion criteria for the UC group were as follows: patients diagnosed with UC who were taking UC-related drugs (e.g., anti-inflammatory drugs) agreed to participate in this study, voluntarily signed informed consent, and consumed traditional Korean dishes, such as rice and seasoned vegetables.
The exclusion criteria were as follows: those diagnosed with diseases that could have affected the results of this study, such as diabetes mellitus and autoimmune diseases other than UC, those who had taken antibiotics or steroids within 6 months, those who were taking probiotics, those who regularly consumed alcohol and smoked, those from whom blood or stool samples could not be obtained, and those who were deemed inappropriate for participation in this study by the medical staff.
2.2.3. Inclusion and Exclusion Criteria for the HC Group
The inclusion criteria were as follows: those who consented to participate in this study freely signed informed consent, had no underlying disease, and were not taking any drugs. Those deemed inappropriate for participation in this study by the medical staff were excluded.
2.3. Variables
The variables were the metabolites extracted from the collected blood samples and gut microbes extracted from the collected stool samples.
2.3.1. Metabolite Analysis
Blood Collection Method
After 5 mL of blood was collected using the injection needle included in the blood collection kit, the blood was separated into 3.0- and 2.0-mL samples and placed in separate serum tubes and nonautologous-pooled human plasma containers, respectively. The serum and plasma were separated.
Metabolite Analysis Method
A total of 250 μL of serum was combined with 500 μL of saline solution (10% D2O for lock signal, NaCl 0.9%, 500 mM sodium phosphate buffer in D2O containing 0.05 trimethylsilylpropanoic acid [TSP] 0.05% for chemical shift calibration, and concentration reference, pH 7.0). After centrifuging the samples at 12,000× g for 10 min, 600 μL aliquots of the supernatant were transferred to 5-mm nuclear magnetic resonance (NMR) tubes for analysis. An ASCEND 800-MHz AVANCE III HD Bruker spectrometer was used, outfitted with a 5-mm CPTIC 1H-13C/15N/DZ-GRD Z1194227/0011 cryogenic probe. The NMR sequence (Carr-Purcell-Meiboom-Gill [CPMG] condition: total T2 relaxation time of 60, 4 K data points, 128 scans, four dummy scans, 8-s delay time) used was a CPMG spin-echo pulse. The Chenomx program performed baseline correction on the 1D data obtained from the NMR analysis. Binning was then performed in units of 0.05 ppm, followed by spectral alignment using the COW algorithm in MATLAB. SIMCA −P++ was used for the multivariate analysis of the data organized using MATLAB.
TSP was used as an internal standard for quality control. The TSP peak was used as a reference to correct for chemical shifts and quantify the metabolites.
Metabolite Pattern Analysis
The signal intensity of the spectrum was normalized concerning the TSP signal and then converted into an ASCII file. An orthogonal partial least-squares discriminant analysis (OPLS-DA) was performed on the UV scale to assess differences in metabolic patterns between the HC and UC groups.
2.3.2. Gut Microbe Analysis
Meal Adjustment Guide
The day before stool collection, participants were instructed not to drink alcohol or eat extremely fatty foods.
Stool Collection and Specimen Delivery
A stool (4 mg) was placed in the stool collection kit. The outside of the kit was labeled to help distinguish specimens. The specimens were then frozen at −20 °C and transferred to the laboratory for analysis.
Gut Microbe Analysis
A library was designed to enable Illumina sequencing by constructing a hybrid primer that selectively amplified the V3–V4 region of the 16S rRNA gene (the standard for identifying bacteria), and an adaptor sequence was recognized by the Illumina sequencer. According to Illumina’s MisSeq platform guide, the complete sequencing library mixture was sequenced using 300-bp paired-end sequencing. The bacteria were identified using quantitative insights into the microbial ecology pipeline after trimming the sequencing data. Greengenes was used as the bacterial identification library. A total of 20 samples that passed quality control were used in the analysis. Alpha diversity, which examines the diversity distribution of gut microbes, was compared, and a non-metric multidimensional scaling (NMDS) was performed using the Bray-Curtis distance for pattern analysis.
2.3.3. Statistical Analysis
Data collected from participants were coded and analyzed using the SPSS for Windows (version 20.0) statistical software program. The Shapiro-Wilk test was used for continuous variables to check the normality of the data. An independent t-test or Mann-Whitney U-test was used to compare the levels of blood metabolites and gut microbes in the stool between the UC and HC groups. To control for confounding factors, independent t-tests or Mann-Whitney U-tests were performed for both the sex and age groups. p values < 0.05 were considered statistically significant.
3. Results
3.1. Subject Characteristics
Ten patients with UC and 10 healthy individuals were recruited between 10 December 2018 and 26 February 2020. There were no significant differences in demographic characteristics, such as sex and age, between the two groups (see Table 1 for more information).

Table 1.
Demographic characteristics and medical history of enrolled subjects.
3.2. Metabolite Analysis
Metabolites in the UC and HC groups were clearly differentiated using principal component analysis (R2X = 0.563, Q2 = 0.378, Figure 1) and OPLS-DA (R2Y = 0.551, Q2 = 0.266, Figure 2). According to cross-validation with a 100-permutation test, the established model was considered reliable (Figure 3). Green R2 values and blue Q2 values to the left were lower than the original points to the right, and the regression line of the Q2 points intersected the vertical axis below zero (R2 = 0.377, Q2 = −0.157). The corresponding regression coefficients for the included metabolites sorted by their variable importance in the OPLS-DA model are shown in Figure 4. Among the metabolites analyzed, the levels of formate, glycolate, trimethylamine, valine, and pyruvate were significantly different between the two groups (p < 0.05). Formate, glycolate, trimethylamine, and valine levels were significantly lower, while pyruvate levels were significantly higher in the UC group than in the HC group (Figure 5).
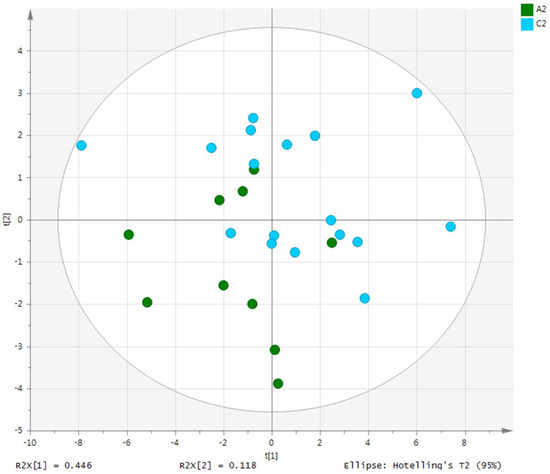
Figure 1.
PCA score plot derived from the 1H-NMR spectra of serum from the ulcerative colitis (UC) patient group (n = 10) and healthy control (HC) group (n = 10). PCA, principal component analysis; NMR, nuclear magnetic resonance; A2, ulcerative colitis group; C2, healthy control group.
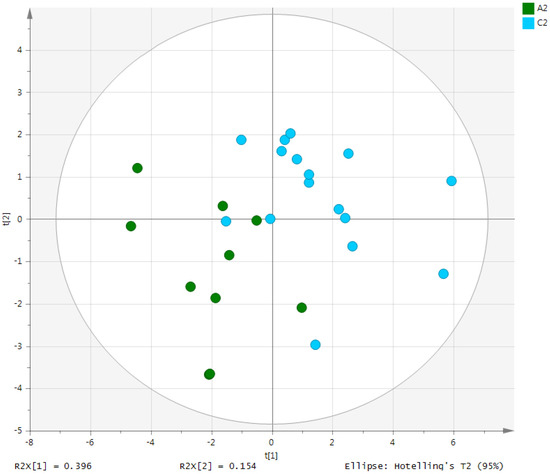
Figure 2.
OPLS-DA score plot derived from the 1H-NMR spectra of serum from the ulcerative colitis (UC) patient group (n = 10) and healthy control (HC) group (n = 10). OPLS-DA, orthogonal partial least-squares discriminant analysis; NMR, nuclear magnetic resonance; A2, ulcerative colitis patient group; C2, healthy control group.
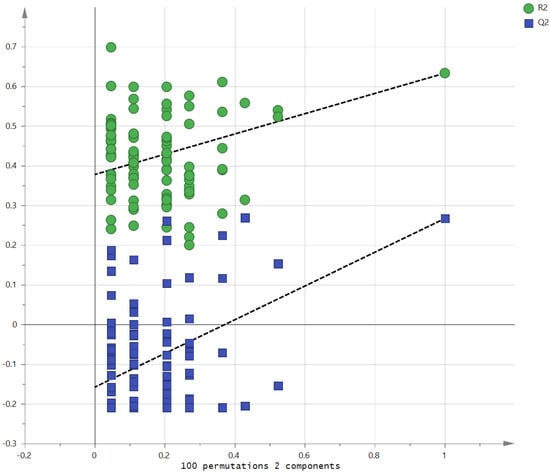
Figure 3.
Validation of the OPLS model using the 100-permutation test.
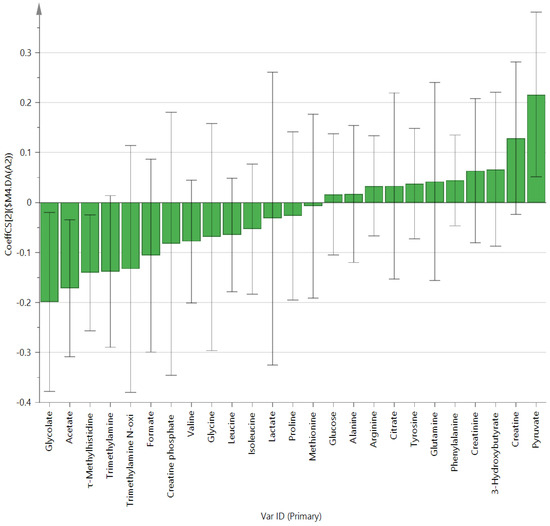
Figure 4.
OPLS-DA coefficient plot of all metabolites in patients with ulcerative colitis.
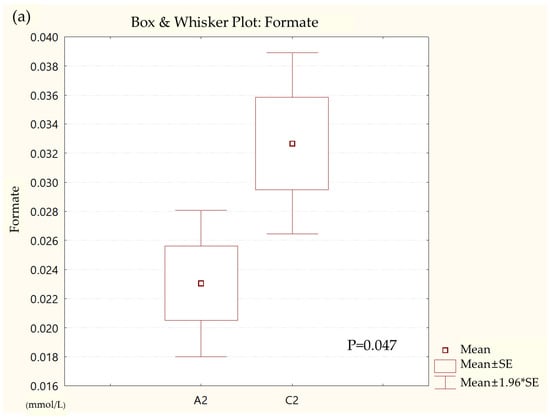
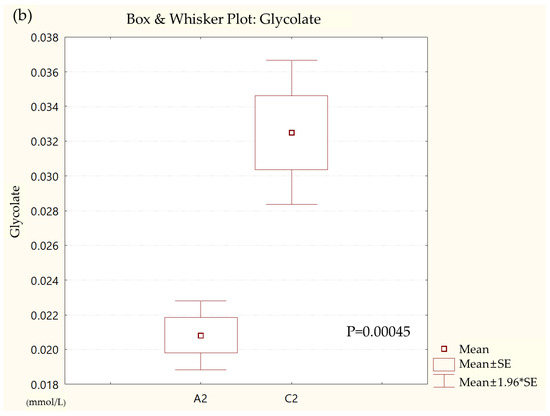
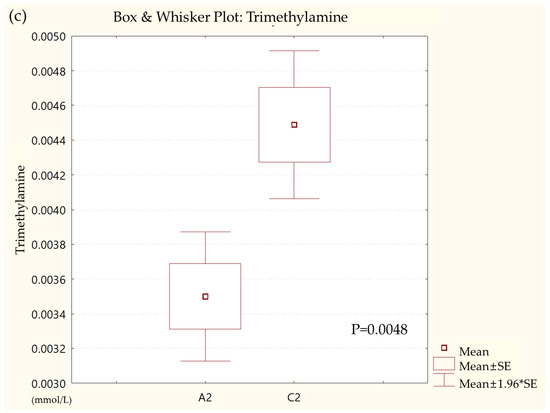
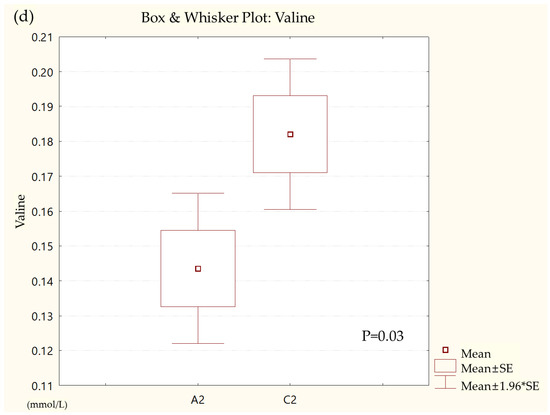
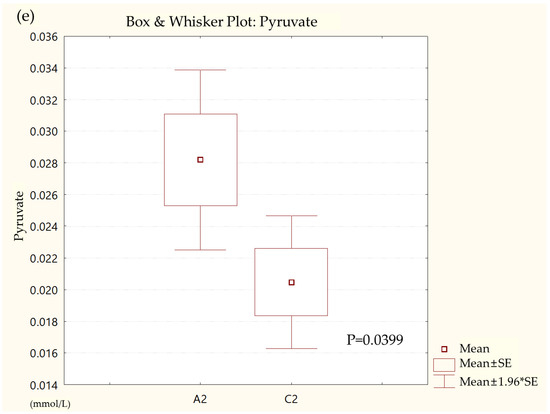
Figure 5.
Box and whisker plot of (a) formate, (b) glycolate, (c) trimethylamine, (d) valine, and (e) pyruvate in ulcerative colitis (UC) patient group and healthy control (HC) group. A2, ulcerative colitis patient group; C2, healthy control group.
3.3. Gut Microbe Analysis
The alpha diversity comparison between the two groups revealed that the UC group had significantly lower Chao1 levels, indicating lower diversity of gut microbes in this group than in the HC group (p = 0.013) (Figure 6).
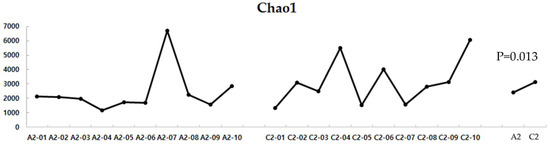
Figure 6.
Alpha diversity of the UC and HC groups. UC, ulcerative colitis; HC, healthy control; A2, ulcerative colitis patient group; C2, healthy control group.
The NMDS based on the Bray-Curtis distance revealed that the two groups had different gut microbial patterns, but no discernable patterns were evident (Figure 7).
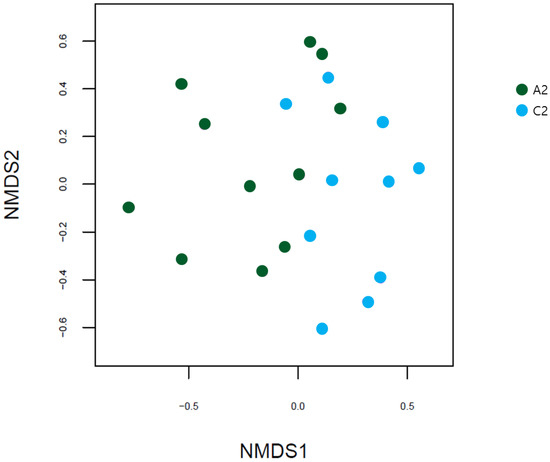
Figure 7.
NMDS plots based on Bray-Curtis distances between the UC and HC groups. NMDS, non-metric multidimensional scaling; UC, ulcerative colitis; HC, healthy control; A2, ulcerative colitis patient group; C2, healthy control group.
Significant differences in the distribution of the gut microbiota composition between the two groups were observed in Bacteroidetes at the phylum level; Bacteroidia at the class level; Bacteroidales and Actinomycetales at the order level; Prevotellaceae, Acidaminococcaceae, and Leptotrichiaceae at the family level; and Prevotella, Roseburia, Paraprevotella, Phascolarctobacterium, Ruminococcus, Coprococcus, Clostridium_XIVB, Atopobium, and Leptotrichia at the genus level (Table 2). Gut microbiota compositions at the phylum and genus levels for the UC and HC groups are shown in Figure 8 and Figure 9, respectively.

Table 2.
Gut microbiota compositions according to taxonomic level in UC and HC groups.
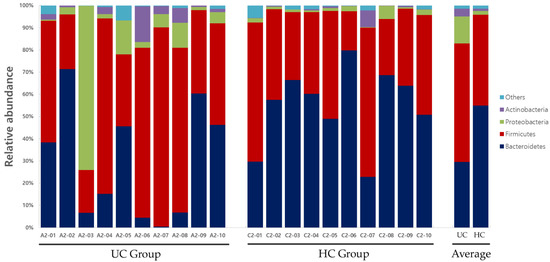
Figure 8.
Gut microbiota composition at the phylum level in UC and HC groups. UC, ulcerative colitis; HC, healthy control.
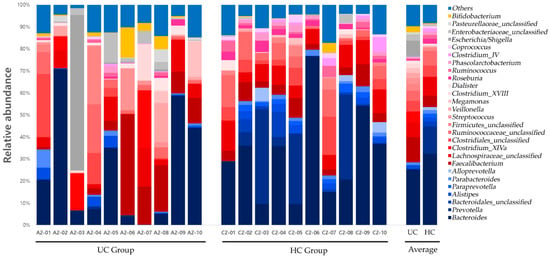
Figure 9.
Gut microbiota composition at the genus level in UC and HC groups. UC, ulcerative colitis; HC, healthy control.
4. Discussion
We compared metabolites and gut microbiota between 10 patients with UC and 10 healthy individuals. The extracted metabolite mixture was analyzed via NMR spectroscopy. Afterward, a Fourier transform on the NMR data was done. The phase was adjusted to obtain a spectrum and perform baseline correction. The signal intensity of the spectrum was normalized concerning the TSP signal and then converted into an ASCII file. The converted values were analyzed using multivariate analysis. Among metabolites, univariate analysis showed formate, glycolate, trimethylamine, valine, and pyruvate levels were significantly different between the two groups. In the multivariate analysis, there were also significant differences in acetate and τ-methylhistidine between groups. Among gut microbes, the abundance of Bacteroidetes at the phylum level; Bacteroidia at the class level; Bacteroidales and Actinomycetales at the order level; Prevotellaceae, Acidaminococcaceae, and Leptotrichiaceae at the family level; and Prevotella, Roseburia, Paraprevotella, Phascolarctobacterium, Ruminococcus, Coprococcus, Clostridium_XIVB, Atopobium, and Leptotrichia at the genus level was also significantly different. The roles that these metabolites and gut microbes play are listed in Table 3.

Table 3.
Description of metabolites and gut microbes that significantly differed between UC and HC groups.
Van Kessel and El Aidy reported that gut microbial products are metabolites [56], and Wang et al. reported that inflammation regulates energy metabolism under physiological and pathological conditions [57]. This is consistent with the results of this study, which found that most of the metabolites and gut microbes that were significantly different between the UC and HC groups were related to energy metabolism and inflammatory processes, respectively.
Metabolites and gut microbes are areas of interest for both mainstream and complementary and alternative medicine. For example, studies have shown that herbal medicines cause metabolite change [7] and interact with gut microbes [8], and studies have shown that Western medicine also focuses on the relationship between disease and metabolites and gut microbes [9].
The fact that both mainstream medicine and complementary and alternative medicine are focusing on metabolites and gut microbes could have vast implications, particularly since one of the reasons that integrative medicine treatment is difficult to implement is the lack of common interests [58]. Considering these points and the results of this study, metabolites and gut microbes could be excellent targets for integrative medicine treatment.
This study had several limitations. First, because this was a pilot study, the number of participants analyzed was small. Thus, it is difficult to conclude that the results of this study reflect the characteristics of all patients with UC. However, the reliability of the results is not considered low because, despite the small number of patients, significant results were obtained that are consistent with previous research findings. Second, this study did not compare differences based on detailed information on the subjects’ diets. However, it was the same for the broad framework of traditional Korean dishes. Therefore, the possibility that diet affected the results of this study is considered insignificant. Third, this study did not evaluate the detailed correlations between the metabolites and gut microbes that showed a significant difference between the two groups. However, it was confirmed that they are commonly related to energy metabolism and inflammation. Fourth, we could not determine the names of the gut microbes that showed a significant difference between the two groups at the species level. However, we were able to confirm the lack of gut microbial diversity at the species level in the UC group through alpha diversity analysis. Fifth, it was unclear whether the patients with UC in this study were in the active or remission stage. However, it is presumed that the patients with UC included in this study were in the remission stage since those taking antibiotics and steroids, primarily used for active UC [59], were excluded. Sixth, although feces are closely related to the gut, only serum metabolites were analyzed in our study. However, considering a study by Seo [60] noted a significant difference in the metabolites in serum rather than those of the feces between chronic colitis and normal mouse models, it cannot be said that the analysis of serum metabolites in this study was incorrect.
To the best of our knowledge, most existing studies have either analyzed metabolites or gut microbes alone. However, in this study, both metabolites and gut microbes were collected from the same subjects and compared. Our data confirmed that the metabolites and gut microbes that significantly differed between the UC and HC groups were mostly related to energy metabolism and inflammation processes. If significant differences are confirmed through large-scale studies comparing metabolites and gut microbes before and after various treatments, such as with herbal medicine or Western medicine, diet, and fecal transplantation, the results could be used in developing new targets for integrative medicine approaches for UC.
Author Contributions
Conceptualization, C.-H.K. and S.L.; methodology, C.-H.K.; software, S.K.; validation, S.L.; formal analysis, H.C.; investigation, C.-H.K.; resources, K.-H.K.; data curation, G.-H.K.; writing—original draft preparation, C.-H.K.; writing—review and editing, S.L.; visualization, Y.-U.L.; supervision, S.L.; project administration, S.L.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP), grant number 2017R1A5A2015805.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Saint Carollo Hospital, Suncheon-si, Jeollanam-do, Republic of Korea (protocol code SCH2018-0116, 23 May 2019) and registered in the Clinical Research Information Service (CRIS) of the Korea National Institute of Health (NIH), Republic of Korea (KCT0003976, 23 May 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the subjects to publish this paper.
Data Availability Statement
The data in this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Reiff, C.; Kelly, D. Inflammatory Bowel Disease, Gut Bacteria and Probiotic Therapy. Int. J. Med. Microbiol. 2010, 300, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Schee Genannt Halfmann, S.; Mählmann, L.; Leyens, L.; Reumann, M.; Brand, A. Personalized Medicine: What’s in It for Rare Diseases? Adv. Exp. Med. Biol. 2017, 1031, 387–404. [Google Scholar] [CrossRef]
- Czerska, I.; Skweres-Kuchta, M. Integrative Medicine as a New Treatment Model and the Future of Health Care Systems in the World in the Context of Rare Diseases. Eur. Res. Stud. 2021, 24, 800–809. [Google Scholar] [CrossRef]
- Schork, N.J.; Nazor, K. Integrated Genomic Medicine: A Paradigm for Rare Diseases and Beyond. Adv. Genet. 2017, 97, 81–113. [Google Scholar] [CrossRef] [PubMed]
- Gyngell, C.; Bowman-Smart, H.; Savulescu, J. Moral Reasons to Edit the Human Genome: Picking up from the Nuffield Report. J. Med. Ethics. 2019, 45, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Bao, Q.; Di, S.; Zhao, Y.; Zhao, S.; Zhang, H.; Lian, F.; Tong, X. The Interaction between the Gut Microbiota and Herbal Medicines. Biomed. Pharmacother. 2019, 118, 109252. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut Microbiota-Derived Metabolites as Key Actors in Inflammatory Bowel Disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Zitomersky, N.L.; Atkinson, B.J.; Franklin, S.W.; Mitchell, P.D.; Snapper, S.B.; Comstock, L.E.; Bousvaros, A. Characterization of Adherent Bacteroidales from Intestinal Biopsies of Children and Young Adults with Inflammatory Bowel Disease. PLoS ONE 2013, 8, e63686. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and Noninvasive Metabonomic Characterization of Inflammatory Bowel Disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- Kieser, M.; Wassmer, G. On the Use of the Upper Confidence Limit for the Variance from a Pilot Sample for Sample Size Determination. Biom. J. 1996, 38, 941–949. [Google Scholar] [CrossRef]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate Metabolism in Health and Disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Faber, F.; Bäumler, A.J. The Impact of Intestinal Inflammation on the Nutritional Environment of the Gut Microbiota. Immunol. Lett. 2014, 162, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Cramer, S.D.; Kennedy, M.; Assimos, D.G.; Holmes, R.P. Glycolate and Glyoxylate Metabolism in HepG2 Cells. Am. J. Physiol. Cell Physiol. 2004, 287, C1359–C1365. [Google Scholar] [CrossRef]
- Marengo, S.R.; Romani, A.M. Oxalate in Renal Stone Disease: The Terminal Metabolite That Just Won’t Go Away. Nat. Clin. Pract. Nephrol. 2008, 4, 368–377. [Google Scholar] [CrossRef]
- Caudarella, R.; Rizzoli, E.; Pironi, L.; Malavolta, N.; Martelli, G.; Poggioli, G.; Gozzetti, G.; Miglioli, M. Renal Stone Formation in Patients with Inflammatory Bowel Disease. Scanning Microsc. 1993, 7, 371–379, discussion 379. [Google Scholar]
- Chhibber-Goel, J.; Gaur, A.; Singhal, V.; Parakh, N.; Bhargava, B.; Sharma, A. The Complex Metabolism of Trimethylamine in Humans: Endogenous and Exogenous Sources. Expert Rev. Mol. Med. 2016, 18, e8. [Google Scholar] [CrossRef] [PubMed]
- Papa, A.; Danese, S.; Urgesi, R.; Grillo, A.; Guglielmo, S.; Roberto, I.; Bonizzi, M.; Guidi, L.; De Vitis, I.; Santoliquido, A.; et al. Early Atherosclerosis in Patients with Inflammatory Bowel Disease. Eur. Rev. Med. Pharmacol. Sci. 2006, 10, 7–11. [Google Scholar] [PubMed]
- Murín, R.; Mohammadi, G.; Leibfritz, D.; Hamprecht, B. Glial Metabolism of Valine. Neurochem. Res. 2009, 34, 1195–1203. [Google Scholar] [CrossRef]
- Lupton, J.R.; Brooks, J.; Butte, N.; Caballero, B.; Flatt, J.; Fried, S. Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids; National Academy Press: Washington, DC, USA, 2002; Volume 5, pp. 589–768. [Google Scholar]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of Pyruvate Metabolism and Human Disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut Microbiota Metabolism of Dietary Fiber Influences Allergic Airway Disease and Hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Jiang, Z.; Wang, C.; Li, N.; Bo, L.; Zha, Y.; Bian, J.; Zhang, Y.; Deng, X. Acetate attenuates inflammasome activation through GPR43-mediated Ca2+-dependent NLRP3 ubiquitination. Exp Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sara, D.; Kathleen, M.; Jeroen, R.; Kristin, V.; Séverine, V. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef]
- Milan, H. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef]
- Wang, D.; Ma, X.; Guo, S.; Wang, Y.; Li, T.; Zou, D.; Song, H.; Yang, W.; Ge, Y. Effect of Huangqin Tang on Urine Metabolic Profile in Rats with Ulcerative Colitis Based on UPLC-Q-Exactive Orbitrap MS. Evid. Based Complement. Altern. Med. 2020, 2020, 1874065. [Google Scholar] [CrossRef]
- Kim, C.H.; Jung, J.; Lee, Y.U.; Kim, K.H.; Kang, S.; Kang, G.H.; Chu, H.; Kim, S.Y.; Lee, S. Comparison of Metabolites and Gut Microbes between Patients with Parkinson’s Disease and Healthy Individuals-A Pilot Clinical Observational Study (STROBE Compliant). Healthcare 2022, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Comstock, L.E. Niche-Specific Features of the Intestinal Bacteroidales. J. Bacteriol. 2008, 190, 736–742. [Google Scholar] [CrossRef]
- Waksman, S.A.; Schatz, A.; Reynolds, D.M. Production of Antibiotic Substances by Actinomycetes. Ann. N. Y. Acad. Sci. 2010, 1213, 112–124. [Google Scholar] [CrossRef]
- Ganji, L.; Alebouyeh, M.; Shirazi, M.H.; Eshraghi, S.S.; Mirshafiey, A.; Ebrahimi Daryani, N.; Zali, M.R. Dysbiosis of Fecal Microbiota and High Frequency of Citrobacter, Klebsiella spp., and Actinomycetes in Patients with Irritable Bowel Syndrome and Gastroenteritis. Gastroenterol. Hepatol. Bed Bench. 2016, 9, 325–330. [Google Scholar]
- Yap, G.; Hong, P.; Lee, B. Microflora of the Intestine. Encycl. Food Microbiol. 2014, 2, 634–665. [Google Scholar]
- Chatterjee, K.; Banerjee, S. Microbiome and Motor Neuron Diseases. Prog. Mol. Biol. Transl. Sci. 2020, 176, 111–122. [Google Scholar] [CrossRef]
- Liu, B.; Piao, X.; Niu, W.; Zhang, Q.; Ma, C.; Wu, T.; Gu, Q.; Cui, T.; Li, S. Kuijieyuan Decoction Improved Intestinal Barrier Injury of Ulcerative Colitis by Affecting TLR4-Dependent PI3K/AKT/NF-κB Oxidative and Inflammatory Signaling and Gut Microbiota. Front. Pharmacol. 2020, 11, 1036. [Google Scholar] [CrossRef]
- Acharya, C.; Bajaj, J.S. Altered Microbiome in Patients with Cirrhosis and Complications. Clin. Gastroenterol. Hepatol. 2019, 17, 307–321. [Google Scholar] [CrossRef]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016, 7, 1081. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Eisenberg, T.; Fawzy, A.; Nicklas, W.; Semmler, T.; Ewers, C. Phylogenetic and Comparative Genomics of the Family Leptotrichiaceae and Introduction of a Novel Fingerprinting MLVA for Streptobacillus moniliformis. BMC Genom. 2016, 17, 864. [Google Scholar] [CrossRef]
- Larsen, J.M. The Immune Response to Prevotella Bacteria in Chronic Inflammatory Disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.; Griffiths, A.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef]
- Zhu, C.; Song, K.; Shen, Z.; Quan, Y.; Tan, B.; Luo, W.; Wu, S.; Tang, K.; Yang, Z.; Wang, X. Roseburia intestinalis Inhibits Interleukin-17 Excretion and Promotes Regulatory T Cells Differentiation in Colitis. Mol. Med. Rep. 2018, 17, 7567–7574. [Google Scholar] [CrossRef]
- Mills, E.; O’Neill, L.A. Succinate: A Metabolic Signal in Inflammation. Trends Cell Biol. 2014, 24, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Meng, L.; Ai, D.; Hou, N.; Li, H.; Shuai, X.; Peng, X. Acetic Acid Alleviates the Inflammatory Response and Liver Injury in Septic Mice by Increasing the Expression of TRIM40. Exp. Ther. Med. 2019, 17, 2789–2798. [Google Scholar] [CrossRef]
- Knights, D.; Lassen, K.G.; Xavier, R.J. Advances in Inflammatory Bowel Disease Pathogenesis: Linking Host Genetics and the Microbiome. Gut 2013, 62, 1505–1510. [Google Scholar] [CrossRef]
- Tedelind, S.; Westberg, F.; Kjerrulf, M.; Vidal, A. Anti-Inflammatory Properties of the Short-Chain Fatty Acids Acetate and Propionate: A Study with Relevance to Inflammatory Bowel Disease. World J. Gastroenterol. 2007, 13, 2826–2832. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kamada, N. Host-Microbial Cross-Talk in Inflammatory Bowel Disease. Immune Netw. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- La Reau, A.J.; Suen, G. The Ruminococci: Key Symbionts of the Gut Ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Shaw, K.A.; Bertha, M.; Hofmekler, T.; Chopra, P.; Vatanen, T.; Srivatsa, A.; Prince, J.; Kumar, A.; Sauer, C.; Zwick, M.E.; et al. Dysbiosis, Inflammation, and Response to Treatment: A Longitudinal Study of Pediatric Subjects with Newly Diagnosed Inflammatory Bowel Disease. Genome Med. 2016, 8, 75. [Google Scholar] [CrossRef]
- Bernstein, C.N.; Forbes, J.D. Gut Microbiome in Inflammatory Bowel Disease and Other Chronic Immune-Mediated Inflammatory Diseases. Inflamm. Intest. Dis. 2017, 2, 116–123. [Google Scholar] [CrossRef]
- Carretta, M.D.; Quiroga, J.; López, R.A.; Hidalgo, M.A.; Burgos, R.A. Participation of Short-Chain Fatty Acids and Their Receptors in Gut Inflammation and Colon Cancer. Front. Physiol. 2021, 12, 662739. [Google Scholar] [CrossRef] [PubMed]
- Labus, J.S.; Osadchiy, V.; Hsiao, E.Y.; Tap, J.; Derrien, M.; Gupta, A.; Tillisch, K.; Le Nevé, B.; Grinsvall, C.; Ljungberg, M.; et al. Evidence for an Association of Gut Microbial Clostridia with Brain Functional Connectivity and Gastrointestinal Sensorimotor Function in Patients with Irritable Bowel Syndrome, Based on Tripartite Network Analysis. Microbiome 2019, 7, 45. [Google Scholar] [CrossRef]
- Jørandli, J.W.; Thorsvik, S.; Skovdahl, H.K.; Kornfeld, B.; Sæterstad, S.; Gustafsson, B.I.; Sandvik, A.K.; van Beelen Granlund, A. The Serotonin Reuptake Transporter Is Reduced in the Epithelium of Active Crohn’s Disease and Ulcerative Colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G761–G768. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zang, S.Q.; Wei, J.; Yu, H.C.; Yang, Z.; Wu, H.M.; Kang, Y.; Tao, H.; Yang, M.F.; Jin, L.; et al. High-Throughput Sequencing Provides Insights into Oral Microbiota Dysbiosis in Association with Inflammatory Bowel Disease. Genomics 2021, 113, 664–676. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, S.P.; El Aidy, S. Bacterial Metabolites Mirror Altered Gut Microbiota Composition in Patients with Parkinson’s Disease. J. Parkinsons Dis. 2019, 9, S359–S370. [Google Scholar] [CrossRef]
- Wang, H.; Ye, J. Regulation of Energy Balance by Inflammation: Common Theme in Physiology and Pathology. Rev. Endocr. Metab. Disord. 2015, 16, 47–54. [Google Scholar] [CrossRef]
- Sugito, R.; Son, D. Obstacles to the Use of Complementary and Alternative Medicine by Primary Care Physicians: Preliminary Study. Trad. Kampo Med. 2019, 6, 173–177. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yoshimura, N.; Saniabadi, A.R.; Saito, Y. Selective Granulocyte and Monocyte Adsorptive Apheresis as a First-Line Treatment for Steroid Naïve Patients with Active Ulcerative Colitis: A Prospective Uncontrolled Study. Dig. Dis. Sci. 2004, 49, 565–571. [Google Scholar] [CrossRef]
- Seo, S.H. Effect of Jakyakgamcho-Tang on Inflammatory Bowel Disease Using GC/MS-Based Metabolic Profiling Analysis. Ph.D. Dissertation, Dongshin University, Naju-Si, Korea, 2019. Available online: http://www.riss.kr/link?id=T15092756&outLink=K (accessed on 12 December 2018).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).