Abstract
Toxic chemicals and epigenetic biomarkers associated with cancer have been used successfully in clinical diagnostic screening of feces and urine from individuals, but they have been underutilized in a global setting. We analyzed peer-reviewed literature to achieve the following: (i) compile epigenetic biomarkers of disease, (ii) explore whether research locations are geographically aligned with disease hotspots, and (iii) determine the potential for tracking disease-associated epigenetic biomarkers. Studies (n = 1145) of epigenetic biomarkers (n = 146) in urine and feces from individuals have established notable diagnostic potential for detecting and tracking primarily gastric and urinary cancers. Panels with the highest sensitivity and specificity reported more than once were SEPT9 (78% and 93%, respectively) and the binary biomarker combinations GDF15, TMEFF2, and VIM (93% and 95%), NDRG4 and BMP3 (98% and 90%), and TWIST1 and NID2 (76% and 79%). Screening for epigenetic biomarkers has focused on biospecimens from the U.S., Europe, and East Asia, whereas data are limited in regions with similar/higher disease incidence rates (i.e., data for New Zealand, Japan, and Australia for colorectal cancer). The epigenetic markers discussed here may aid in the future monitoring of multiple cancers from individual- to population-level scales by leveraging the emerging science of wastewater-based epidemiology (WBE).
1. Introduction
Suitable targets for tracking non-communicable diseases such as cancer include human genes that are enhanced or repressed by epigenetic changes (e.g., DNA methylation; histone modifications, such as methylation or acetylation; and regulation by non-coding RNAs, such as miRNAs and long non-coding RNAs [1]) induced by exposure to harmful chemicals and disease progression. For instance, DNA hypomethylation has been observed in fetal serum of smoking mothers [2]. DNA methylation, an epigenetic modification where a methyl group is added to cytosine bases, was first discovered to silence genes nearly 50 years ago [3]. Global hypomethylation of DNA was a molecular change observed early in epigenetic disease research and is thought to be a general biomarker of environmental exposures linked to chronic diseases, such as cancer [2].
As epigenetic changes are known to be cell- or tissue-specific, current epigenetic research has largely focused on pure cell lines or cells extracted from tissues. However, disease-specific genetic and epigenetic changes are also carried by expelled DNA known as cell-free DNA (cfDNA) [4]. Meanwhile, less invasive diagnostic screening methods using liquid biopsies (e.g., of urine, feces, or blood) are gaining momentum in research and have shown promise in translation into clinical practice [5]. The cfDNA present in liquid biopsy samples represents a novel and promising target for researchers to identify the origin, mechanisms of release, and biomarkers associated with a given disease [6]. The extent of methylation of cfDNA extracted from liquid biopsies, specifically from urine, has been quantified successfully to inform on the presence and progression of a variety of cancers [5]. The potential utility for non-invasive biopsy samples such as urine and feces to diagnose cancer cannot be understated from a patient care perspective.
While epigenetics is primarily studied at the individual level, population epigenetics is an emerging scientific frontier. Population epigeneticists seek to follow epigenetic changes visible at the population level over time and space. Zhao et al. has recommended a framework to illustrate population characteristics of DNA methylation by comparing methylated and unmethylated DNA in a population of known size [7]. Population epigenetics may be used to seek epidemiological information by collecting samples from individuals and then finding average trends within and between populations.
Here, we review the data available on the analysis of non-invasive liquid biopsy samples (i.e., urine and feces) from around the world for the detection of epigenetic biomarkers associated with disease. The goal was to establish a knowledge platform informing future research on tracking the incidence of cancer and other illnesses at the population level. First, we analyzed and ranked literature-sourced biomarker panels for their diagnostic sensitivity and specificity. Second, we used a geographic information system (GIS) to map the geospatial origin and cohort size of urine and feces samples that have been previously analyzed for epigenetic biomarkers associated with disease outcomes. Third, we then compared these geographic locales with information reported on the case incidence rate of specific cancers.
2. Materials and Methods
A systematic literature review and geographic trend analyses were performed to determine epidemiological patterns.
2.1. Literature Sources
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) methodology was used to review search criteria. This systematic review was conducted in Scopus for all publications prior to December 2021 (Supplemental Figure S1).
2.2. PRISMA Criteria
Keywords used to search for epigenetic biomarkers shed in urine or fecal liquid biopsy included “epigen* AND (fecal OR feces OR urine) AND cell AND biomarker”. Papers discussing biomarkers, e.g., urine-DNA methylation biomarkers, or cells, e.g., cell-based or classifier, were included in this analysis regardless of study sample size. Studies highlighting fecal transplants, blood plasma/serum liquid biopsy, chemical metabolite concentrations, or animal studies, e.g., rats and mice, were excluded.
2.3. Geographic Analysis
ArcGIS Pro 3.0.0 was used to analyze geospatial data. Qualitative and quantitative data reported by geographically linked studies were mapped to determine spatial trends. Cancer incidence rates were accessed from the Global Cancer Observatory database to compare study locations, sample size, and incidence rates at the country level.
2.4. Statistical Analysis
Biomarker accuracy was assessed using the combined frequency of sensitivity and specificity (log10 transformed). When converting these statistics using log10 transformation, threshold scores representing sensitivity and specificity can be added to a maximum of 2.0 for 100% accuracy. If threshold score calculation met or exceeded 1.5, then the gene or gene panel was considered viable as a diagnostic tool for the targeted disease. Sensitivity and specificity percentages for the same biomarker panel reported by more than one study were analyzed by calculating the average threshold scores, standard deviation, and subsequent 95% confidence interval.
3. Results
3.1. PRISMA Literature Review
Studies identified by the PRISMA search for epigenetic biomarkers in urine and feces have increased over time from 2003 to 2021 (Supplemental Figure S1). Of the 1145 studies screened, 146 articles were identified as relevant for qualitative and quantitative analysis (Supplemental Figure S2). Studies that analyzed epigenetic markers isolated from urine or feces for the diagnostic utility of disease were included in this review (Supplemental Table S1).
Most epigenetic biomarker studies of urine or fecal samples included 100 or fewer patients and fewer than 100 controls for sensitivity and specificity analysis of methylation panels (Figure 1). Additionally, Figure 1 indicates that the methylation status of DNA captured from feces is primarily used for colorectal (CRC) screening, while methylation levels in urine are mostly used to diagnose bladder cancer, prostate cancer, and kidney cancer. Average concentrations of genomic material analyzed in methylation assays were below 1 µg for RNA or DNA and below 5 µL for bisulfite-treated DNA. Extremely high concentrations were considered outliers and therefore excluded, as our analysis focused on the minimum concentrations used for successful methylation assays. Studies further shared concentrations averaging 49.29 genomic copies/reaction volume at a minimum of 10 copies/reaction volume and calculated concentrations averaging 0.02 µg/mL at a minimum of 0.0001 µg/mL (Figure 1).
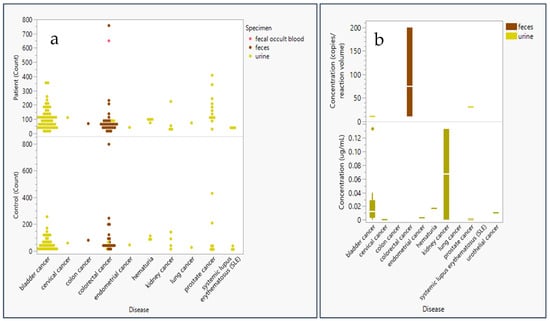
Figure 1.
Sample size and concentration of genomic material used in epigenetic panel studies employing liquid biopsy materials. (a) Patient and control count (sample size) in epigenetic panel screening studies * are plotted as a function of specimen type: urine (yellow), feces (brown), and fecal occult blood (pink). (b) Genetic (DNA or RNA) concentration analyzed for epigenetic markers in screening studies ** plotted in a box and whisker plot (distributed around the mean) by disease and specimen type: urine (yellow) and feces (brown) by either concentration in copies/reaction volume (n = 7) or ug/mL (n = 24). * Did not include controls in a study of feces for colorectal cancer due to its large study size of 9167 controls [8] ** Excluded study with extremely high copies/reaction volume (1014 genome equivalent copies) for a study extracting genomic content from fecal matter in colorectal patients [9] and a study extracting high µg/mL volume (1 µg/mL) from urine in kidney cancer [10].
3.2. Sensitivity and Specificity of Epigenetic Biomarkers Utilized in Clinical Screening of Urine and Feces
Studies reported statistics indicating the accuracy of the biomarkers chosen for their epigenetic panels. The percentage of tests in which a patient with the disease tested positive was recorded as sensitivity, whereas the percentage of tests in which a control subject tested negative was recorded as specificity. Studies with fewer false negatives are considered more accurate. If a study reports sensitivity + specificity above a threshold value of 1.5, further research or clinical translation are deemed warranted [11].
The panel with the highest sensitivity and specificity were SOX1, TJP2, MYOD, HOXA9_1, and HOXA9_2 [12], reporting at 100% for both sensitivity and specificity (Figure 2). The panels that were tested in more than one study with the highest sensitivity and specificity included the GDF15, TMEFF2, and VIM panel at 92.5% (95% CI: 96.7–88.3) and 95.0% (95% CI: 109.1–80.9), respectively [13,14]. In addition, the proprietary undisclosed (“unspecified”) panel for the product Bladder EpiCheck reported at 73.1% (95% CI: 101.1–45.2) and 87.4% (95% CI: 89.4–85.5), respectively [15,16,17]. The NDRG4 and BMP3 panel reported 95.2% (95% CI: 103.2–87.1) and 88.3% (95% CI: 93.1–83.5) [8,18]. A test with SEPT9 alone reported 78.2% (95% CI: 92.7–63.6) and 93.3% (95% CI: 96.7–89.9) [19,20]. Finally, the TWIST1 and NID2 panel reported 76.1% (95% CI: 101.1–51.1) and 79.4% (95% CI: 106.2–52.5) [21,22,23,24,25,26].
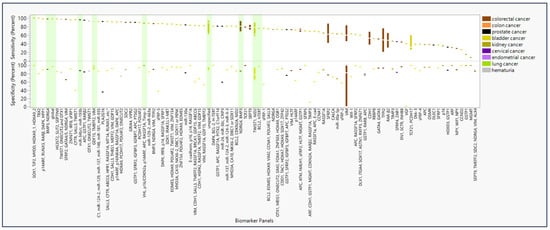
Figure 2.
Sensitivity and specificity of panels of epigenetic biomarker detectable in urine and feces and diseases of diagnostic interest. Percent sensitivity and specificity reported in studies (n = 95) are color-coded by disease type (n = 9) that screened each epigenetic biomarker panel * in liquid biopsy of urine and feces. Bars highlighted in green represent biomarker panels and individual biomarkers with the highest sensitivity and specificity given cancer type or replicability. * Panel “SALL3, CFTR, ABCC6, HPR1, RASSF1A, MT1A, RUNX3 … etc.” also includes biomarkers ITGA4, BCL2, ALX4, MYOD1, DRM, CDH13, BMP3B, CCNA1, RPRM, MINT1, and BRCA1 in the full panel.
For biomarker panels informing on CRC, the highest sensitivity (98%) and specificity (90%) were found for BMP3 and NDRG4 (Table 1) [18]. For prostate cancer, the highest sensitivity (94.3%) and specificity (84.4%) were found, respectively, for miR-34b/c and miR-193b, monitored in separate individual assays [27]. For bladder cancer, a perfect score (100%) was found for both sensitivity and specificity in the SOX1, TJP2, MYOD, HOXA9_1, and HOXA9_2 panel [12]. For kidney cancer, a maximum sensitivity and specificity of 88% and 100%, respectively, were found for VHL, p16/CDKN2a, p14ARF, APC, RASSF1A, and Timp-3 [10]. However, only 10 panels have been studied more than once, many yielding sensitivity and specificity ranges below the optimal levels summarized here. Biomarkers with high sensitivity and specificity in multiple panels include RUNX3, SOX1, IRF8, and DAPK; these were used specifically for diagnosis of bladder cancer (Supplemental Figure S3). Biomarkers selected for multiple studies produced sensitivity and specificity results of greater variability and lower quality.

Table 1.
Ranked biomarker panels by disease, sensitivity, and specificity.
Nearly all studies of panels with epigenetic biomarker testing urine or feces from patients and controls reported over 80% values for both sensitivity and specificity (Figure 3). Eleven studies reported sensitivity and specificity above 90% for both. In fact, 73 of the 94 studies reported sensitivity and specificity values exceeding the threshold value of 1.5. The most common sensitivity + specificity score reported was approximately 1.8. When biomarkers are assessed individually, nearly all studies report over 60% for both panel sensitivity and panel specificity (Supplemental Figure S4). Biomarkers disaggregated from their panels were represented in 106 studies, reporting 80% or higher for both sensitivity and specificity. Nevertheless, any specific panel is only represented in four or fewer of the 94 studies that met the inclusion criteria of this literature review.
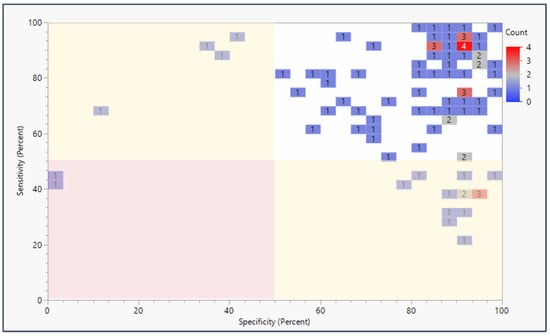
Figure 3.
Sensitivity and specificity of epigenetic biomarker panels applied to liquid biopsy (urine and feces) by publication count. Percent sensitivity and specificity are plotted against each other in a heatmap to show publication counts for each sensitivity/specificity range reported by each epigenetic biomarker panel screening study. The red highlighted section represents sensitivity + specificity below the preferred threshold value of 1.5, the yellow sections highlight studies meeting the 1.5 threshold, and the green section shows the studies above the 1.5 threshold.
In addition, sensitivity and specificity reporting was compared to the sample (cohort) size of each study in Supplemental Figure S5. No significant patterns were identified, showing that studies with high sensitivity and specificity ranged significantly in sample size, from four-patient studies reporting 100% sensitivity and 80% specificity for TBX2 [28] to a 207-patient study reporting 98% sensitivity and 90% specificity for the BMP3 and NDRG4 panel [18].
3.3. Geographic Locations of Patient Cohorts Clinically Analyzed for Epigenetic Biomarkers in Urine and Feces Compared to Cancer Hotspots Globally
Studies of epigenetic markers in human feces and urine thus far have been confined to regions of North America, Europe, and East Asia (Figure 4). Only four studies were reported from countries in the Middle East and North African regions, while populations in South Asia and Sub-Saharan Africa are not represented in this review due to a lack of search results from these regions. The largest studies were performed for colorectal cancer in Tianjin, China, (n = 650) [19] and in a shared study between Canada and the United States (n = 757 patients; n = 9167 controls) [8]. Additionally, studies for bladder cancer with the highest numbers of participants were in Vienna, Austria, (n = 357) [15] and a study combining Nijmegen and Hengelo, the Netherlands; Barcelona, Spain; Wurzberg and Sindelfingen, Germany; and Kfar, Israel (n = 353) [17]. Prostate cancer was also studied in large sample sizes in 18 clinics across the United States (n = 342 patients, n = 430 controls) [9] and in a combined study across Norwich, England; Toronto, Canada; and Dublin, Ireland (n = 408) [19]. However, other studies with relatively small sample sizes were reported for colorectal cancer in Hong Kong, China, (n = 20 patients, n = 30 controls) [76] and Cagliari, Italy, (n = 10) [70]. Likewise, studies for bladder cancer with small sample sizes were reported in Aachen, Germany (n = 20 patients; n = 5 controls) [55]; Maryland, United States (n = 20 patients; n = 20 controls) [64]; California, United States (n = 20 patients; n = 20 controls) [44]; and Rotterdam, the Netherlands (n = 4) [28].
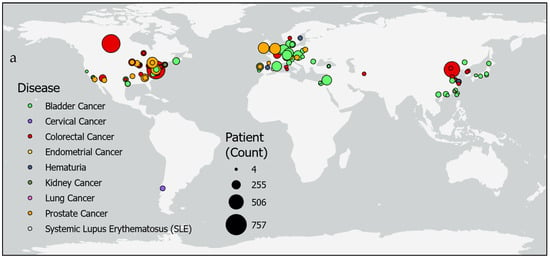
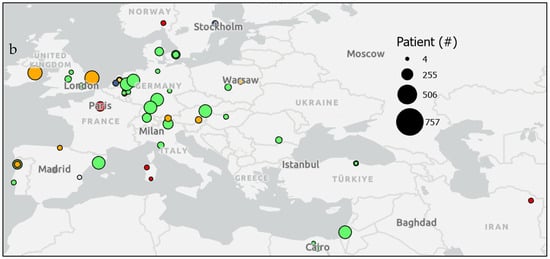
Figure 4.
Urine or fecal epigenetic biomarker panel study patient counts by location. Maps show patient counts (sized circles) in epigenetic biomarker panel screening studies of various diseases (colors) (a) globally and (b) in regions in Europe and the Middle East.
In Supplemental Figure S6, incidence rates per 100,000 people are compared geographically for various types of cancer reported to the Global Cancer Observatory. While cancer incidence rates tend to be higher in males than females, regions with the highest rates are not always shared between the sexes. The highest incidence rates for colorectal cancer in males are reported in Canada, Japan, and Italy (70.3, 62.0, and 58.7, respectively), whereas colorectal cancer in females is highest in Canada, New Zealand, and Japan (41.9, 36.1, and 35.3). While incidence rates for bladder cancer top the charts in Italy, Spain, and France (48.5, 44,5, and 40) for males, the countries Chile, Malawi, and Canada report the highest bladder cancer rates (9.8, 9.2, and 9.1) in females. Kidney cancer in males is reportedly the highest in the Czech Republic, Lithuania, and Germany (22.1, 17.6, and 16.9), while females report their highest rates in Argentina, the Czech Republic, and the United States (10.4, 9.9, and 8.9). Lung cancer rates in males rank the highest in Turkey, the United States, and Poland (90.1, 84.9, and 67.9), whereas female rates are highest in the United States, Canada, and Denmark (53.6, 48.4, and 35.2). Meanwhile, prostate cancer rates are highest in Ecuador, Brazil, and Austria (177.8, 157.4, and 140.4). Finally, cervical cancer rates are highest in Zimbabwe, Malawi, and China (86.7, 76.3, and 71.8).
When comparing study locations of epigenetic biomarkers isolated from urine and feces, Figure 5 shows the misalignment of the study location with regions reporting high incidence rates of specific cancers. For colorectal cancer, Japan, the Czech Republic, Slovakia, and Germany are notably absent from the study locations. Bladder cancer studies do not include participants from Malawi or Chile. While studies have been conducted within the United States for kidney and lung cancers, other countries with top ranking incidence rates for kidney cancer that are not represented in these studies include the Czech Republic, Lithuania, Germany, and Argentina. Similarly, countries with high incidence rates for lung cancer without participation in these studies include Turkey, Poland, Canada, and Denmark. For prostate cancer, a study in Austria reported findings, but other countries with high incidence rates were not represented (e.g., Ecuador and Brazil). Meanwhile, the top three countries reporting the highest incidence rates for cervical cancer did not include participants in these studies (i.e., Zimbabwe, Malawi, and China).
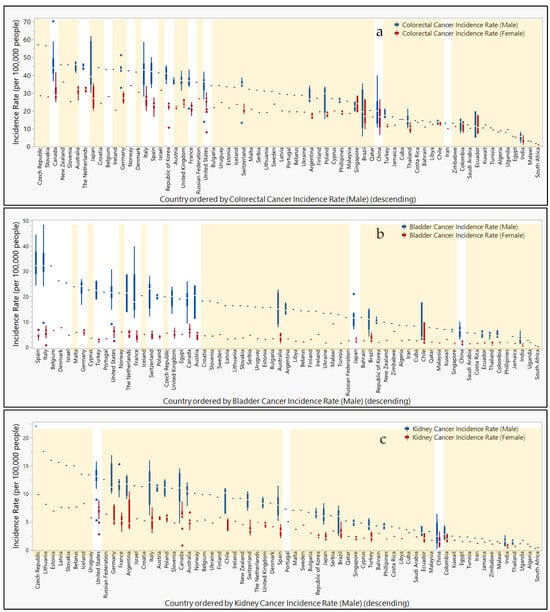
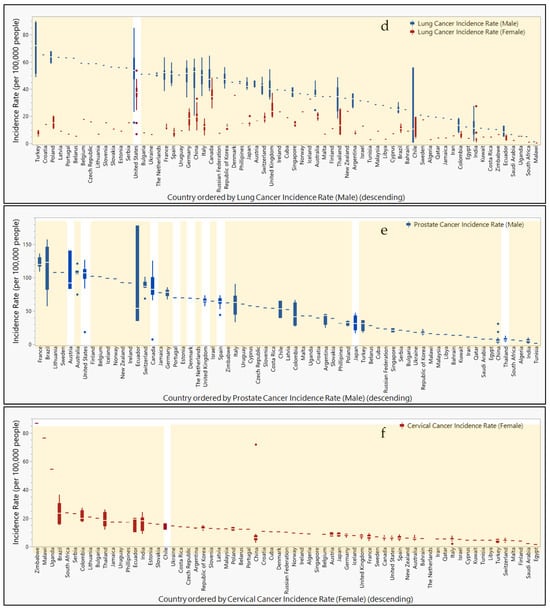
Figure 5.
Countries ranked by cancer incidence compared to urine or fecal epigenetic biomarker panel study locations. Box and whisker plots ranked in descending order by cancer incidence rates (per 100,000 people) for (a) colorectal, (b) bladder, (c), kidney, (d) lung, (e) prostate, and (f) cervical cancers. Male (blue) and female (red) incidence rates are separated and color-coded. Orange highlight bars indicate countries that are lacking representation in urine or fecal epigenetic biomarker panel studies for each disease. Countries not highlighted are represented in urine or fecal epigenetic biomarker panel studies identified in this review.
4. Discussion
This review aimed to assess epigenetic marks found in non-invasive matrices (e.g., urine or feces) as potentially reliable biomarkers of disease. Prior research has shown a relationship between global hypermethylation and the likelihood of receiving a cancer diagnosis [100]. More recently, methylation status of cfDNA in urine has been associated with the prevalence and development of cancer. This paper identified several epigenetic biomarkers and biomarker panels that achieved a sensitivity and specificity above the calculated accuracy threshold (1.5) for several cancers, particularly gastric and urinary cancers. Meanwhile, a global geographic analysis of cancer incidence indicates that many locations with high or the highest incidence of disease are currently underrepresented in studies utilizing epigenetic biomarker panels for disease screening.
4.1. Performance of Epigenetic Biomarker Panels
While it is unlikely that the epigenetic status of cfDNA is identical to DNA within the cells of a tumor or tissue of origin, sensitivity and specificity tests indicate high correlations (Figure 4) between cfDNA methylation profiles and the presence of disease validated from traditional diagnosis methods. Not only have DNA fragmentation sizes and end alterations been associated with cancer pathologies [5], but the results of this review show a significant relationship between cfDNA methylation levels and specific diseases.
The recent literature showed that epigenetic panels tested on urine or feces performed with similarly wide ranges of sensitivity and specificity as panels tested on other liquid biopsy (i.e., plasma) [101,102,103,104,105]. Sensitivity ranged from 60% to 92%, and specificity ranged from 55% to 100% for screening a variety of cancers (i.e., kidney cancer, prostate cancer, and CRC) [101,102,103,104,105]. Meanwhile, more invasive biopsies have shown a higher sensitivity and specificity. For instance, the sensitivity and specificity of the same epigenetic panel to diagnose CRC was notably lower in plasma (81.1% and 96.9%, respectively) compared to tissue (93.8% and 99.9%) [104]. Nevertheless, the authors recognize the importance of testing the utility of the least invasive samples for the benefit of the patient.
The highest values for sensitivity and specificity of epigenetic biomarker panels were obtained from studies which differed markedly in sample size (Supplemental Figure S5). While it should be anticipated that the likelihood of false positive or false negative results would be higher in studies with more subjects, doubts emerge as to the scalability of results reported in studies featuring small sample sizes.
More studies have been undertaken with more promising (more sensitive and selective) biomarkers, whereas 22% of studies focused on biomarkers that were found to be underperforming. Moreover, Figure 2 and Figure 3 reveal little to no standardization in the selection and testing of epigenetic biomarkers in urinary or gastric screening panel studies. The large number of assays and small number of replicating studies suggest the need to focus research efforts to coalesce on utilizing biomarker panels of both high sensitivity and high specificity.
This literature search revealed a lack of convergence toward the unified use of selected biomarkers across multiple studies, suggesting that the use of epigenetic biomarkers of disease in clinical studies relying on non-invasive matrices is still in the early stages of identifying biomarkers’ import to public health. Expanding testing and eventual monitoring to include disease-specific biomarkers in regions with high incidence of disease (the United States, Canada, Japan, and the Czech Republic) presents opportunities for identifying susceptible populations around the world (Supplemental Figure S6). Nevertheless, the selection of regions to be studied needs to be informed both by male and female incidence rates. The sensitivity and specificity of biomarkers for identifying cancer in males should include countries such Italy (colorectal cancer and bladder cancer), Spain and France (bladder cancer), Lithuania and Germany (kidney cancer), Turkey and Poland (lung cancer), as well as Ecuador, Brazil, and Austria (prostate cancer), while studies for identifying cancer in females should be held in New Zealand (colorectal cancer), Chile (bladder cancer), Malawi (bladder cancer and cervical cancer), Argentina (kidney cancer), Denmark (lung cancer), as well as Zimbabwe and China (cervical cancer). The literature revealed that participants from Japan would benefit from studies of colorectal cancer in both males and females (Figure 5). Additionally, studies for bladder cancer in females are absent from the highest-incidence countries, Malawi and Chile. Furthermore, regional selection of participants needs to be informed by the cancers with the highest incidence rates. A study in Chile for cervical cancer could have also included biomarkers for bladder cancer as well to address the high incidence rates of this cancer locally. Additionally, while bladder cancer studies have been performed in several cities in Japan, the high incidence of colorectal cancer in the country suggest studies should also include biomarkers specific to CRC identification. Meanwhile, studies across China for a variety of cancers including colorectal, bladder, kidney, and prostate could have also included biomarkers for cervical cancer to address these extreme rates in females.
To assess the potential individual value of any one biomarker present in multiple panels, panels were disaggregated to allow for each biomarker to be examined individually (Supplemental Figure S3). Various biomarker panels overlapped with others; however, the contribution of each individual marker in the panel could not be evaluated due to a lack of data. Furthermore, the task of individually evaluating each biomarker contained in a biomarker panel was made impossible by their proprietary (i.e., undisclosed) identity.
4.2. Geographic Considerations for Epigenetic Biomarker Panels
Studies of epigenetic markers for the screening of urinary and gastric cancers are scattered in several regions across the world (Figure 4). However, nearly the entire southern hemisphere is lacking studies on epigenetic biomarkers informing the screening for gastric and urinary cancer. Since variation in methylation status in subjects studied across the East Asian countries Taiwan, Hong Kong, and China [43], as well as globally between Siberia, Cambodia, Pakistan, Algeria, and Mexico [106], suggested that geographic location could impact the success of using epigenetic biomarkers as a diagnostic tool, it is necessary to enroll patients and controls in regions not currently represented in the literature. Furthermore, incidence rates reported to the Global Cancer Observatory reveal populations at high risk for cancer in regions not participating in epigenetic biomarker screening studies of urine or feces (Figure 5).
To address the likely heterogeneity in biomarker performance across diverse populations, particularly in geographic regions with relatively few resources, researchers in more affluent nations would improve the efficiency and efficacy of biomarker identification by collaborating with clinics in various regions globally. While several studies included patients from multiple states within the same country or in nearby countries, it was rare to find studies reaching patients across global classifications. Future studies should plan to intentionally recruit patients from countries with high incidence rates of the target cancer, particularly if populations within these countries have traditionally reported low participation in epigenetic panel studies. Of course, funding agencies aiming to reduce global health disparities should also encourage experimental designs incorporating more diverse populations and scopes to include the target cancer type to others of high need in particular countries. Providing funding opportunity announcements (FOAs) targeting epigenetic panel studies for kidney cancer in the Czech Republic, Lithuania, Estonia, Latvia, Slovakia, and Belarus, for lung cancer in Turkey and Croatia, or for cervical cancer in Zimbabwe, Malawi, and Uganda would further encourage research to meet the needs of populations with the highest incidence of these cancers. If research in these regions is not conducted, then the selection of biomarkers for diseases of particular importance to these populations (e.g., high incidence rates, as shown in Figure 5) may not be as effective during screening, leading to reduced diagnosis and treatment of susceptible individuals.
4.3. Translation to Clinic
Several considerations impact the likelihood of a screening test making it to the clinic. For instance, the diagnostic value of a test that is less invasive to the patient needs to be weighed against a more invasive test. A recent systematic review of epigenetic panel studies for CRC compared tissue, plasma, feces, and urine specimens [107]. After calculating threshold scores for each panel, six tests for five single-gene panels (NDRG4 (1.7), SEPT9 (1.9), SFRP2 (1.9; 95% CI: 1.8–1.9), SPG20 (1.9), and TFPI2 (1.9)) were successful when cfDNA was extracted from tissue. Similarly, for fecal specimens, five tests for four genes (ITGA4 (1.8), SFRP2 (1.8; 95% CI: 1.7–1.8), SPG20 (1.8), and TFPI2 (1.8)) were successful. Three tests with cfDNA from plasma for three genes (SEPT9 (1.8), SFRP1 (1.7), and SPG20 (1.8)) were successful. Only one test for one gene (VIM (1.7)) was reported with diagnostic value for CRC. When comparing specimen type, cfDNA extracted from tissue performed slightly better than non-invasive specimens given the relatively higher threshold scores for SPG20; however, cfDNA extracted from fecal and plasma performed comparably. As for panels with multiple biomarkers, two panels reported high threshold scores for tissue (Wif-1 (1.7); RARB2, p16INK4a, MGMT, and APC (1.8)). Two panels were successful for feces (BMP3, NDRG4, VIM, TFPI2, mutant KRAS, B-actin, and Hb (1.8); RARB2, p16INK4a, MGMT, and APC (1.6)), while only one panel reported high sensitivity and specificity for plasma (APC, MGMT, RASSF2A, and Wif-1 (1.8)). The RARB2, p16INK4a, MGMT, and APC panel’s performance was better for the tissue than fecal test, but one could argue that the difference in sensitivity may be acceptable when considering the invasive procedure needed to extract cfDNA from tissue compared to feces.
The possibility of cfDNA degrading in specimens prior to analysis is another concern clinicians need to consider. Storage conditions, such as access to a −80 °C freezer, can prevent further fragmentation and cell lysis [108]. Urine poses a particular challenge due to its high degradation rate when not stabilized with buffers to reduce nuclease activity [109]. Therefore, the development of standards for the development and application of biomarker panels as diagnostic tools will need to include strict storage and sample preparation guidance [110]. Ultimately, the authors acknowledge cfDNA presents limitations and uncertainties (e.g., indirect correlations to disease and degradation over time and in complex matrices) not present in other sample media, such as tissue, where cellular DNA profiles may be analyzed at the source and within the general protection of the nucleus. Nevertheless, cfDNA offers clinicians the opportunity to collect less invasive samples for equally valuable diagnostic potential while inflicting less harm on the patient.
Furthermore, the benefits of translating a viable biomarker into a clinical tool need to outweigh the costs, which can be significant. While many biomarkers have been shown to be viable for gastric and urinary cancers such as bladder and CRC, one study also reported a successful DNA methylation analysis (1.7) of a panel (CDO1, TAC1, HOXA7, HOXA9, SOX17, and ZFP42 promoters) to diagnose lung cancer [20]. Therefore, cfDNA from urine and feces could offer diagnostic value to other highly prevalent cancers. Often, for-profit companies lead the charge in moving a biomarker to clinical practice due to the incentive to sell a product for consumption [110]. Researchers, however, are almost exclusively incentivized to publish independent papers as often as possible. For-profit companies may not consider the viability of a particular biomarker over another, while researchers may not consider methods of detecting biomarkers with clinical infrastructure limitations. Regulatory changes at the national and international levels requiring these two spheres to collaborate rather than simply coexisting could significantly improve the potential for promising biomarkers to make it to patients.
As of this literature search, the U.S. Food and Drug Administration (FDA) has approved several epigenetic screening tests for CRC. The first fecal-based test using epigenetic markers, Cologuard, was approved by the FDA in 2014. However, it was recommended to perform additional screening in clinic [111]. In 2016, the FDA approved the blood-based epigenetic test trademarked Epi proColon as an alternative test to colonoscopy or stool-based fecal immunochemical testing [112]. Therefore, it is reasonable to assert high potential for the FDA to approve a fecal- or urine-based test using epigenetic markers as an alternative to the current more invasive clinical tests upon high sensitivity and specificity reporting in multiple trials.
4.4. Applications of Population Epigenetics for Public Health
Epigenetic marks that are highly correlated to patients with disease compared to controls have the potential to be observed at the population level. If these marks can be observed from cfDNA in urine or feces, then composited samples in wastewater may offer an efficient, effective, and timely media to observe varying concentrations of epigenetic marks between populations. Wastewater-based epidemiology (WBE) has recently become popularized for its utility in predicting surges in infectious disease rates during the height of the COVID-19 pandemic [113,114]. Since the methodology to detect SARS-CoV-2 requires comparable technology for genomics analysis (e.g., RT-qPCR, Illumina sequencing) and epigenetic marker analysis, it stands to reason that WBE pipelines could be adapted to detect epigenetic marks. If a WBE pipeline was developed for epigenetic marks associated with cancers in this review, populations could be monitored and subsequently supported with targeted clinical screening, treatment, and other healthcare resources.
4.5. Potential Confounding Results
Biomarkers that are not specific to disease represent a challenge for public health interventions. Epigenetic changes may be indicative of one or more diseases, such as global hypermethylation or hypomethylation. Biomarkers that are associated with only one disease would be better suited for monitoring each disease of interest. It is therefore imperative to select genetic biomarkers that are specific to disease.
Additionally, studies with high sensitivity and specificity results may not be viable due to the potential for a Type 1 error (Figure 1 and Figure 4). A study for colorectal cancer with a sample size of n = 10 [76] and another for bladder cancer with a sample size of only n = 4 for a particular region [44] gives the reviewers pause when considering recommendations of biomarkers. Increasing sample sizes would increase confidence in results prior to selection for further study.
Due to the challenges mentioned above and other aspects, some markers that rank highly (Figure 2) ultimately may not be as well suited compared to others for future clinical application. To be of utility, biomarkers tracked by epigenetic panel testing must occur at appreciable concentrations. For a marker to be a reliable indicator of public health, it would ideally be both abundant, i.e., occurring at high concentrations, and highly modulated in concentration as a function of disease status to produce an epigenetic signal that is both detectable and quantitatively informative. If biomarker panel(s) are widely implemented at the clinical setting for diagnostic purposes, epidemiologists tracking disease rates at the population level (e.g., using WBE methodology) will rely on the knowledge that the chosen biomarkers are highly sensitive, specific, and quantifiable. WBE methodology analyzing wastewater samples compositing urine and feces from a significant number of individuals is more likely to detect signals of epigenetic mark differences in regions with high prevalence of disease. The choice to use WBE for quantification of epigenetic marks at the population level would therefore hinge on whether a high intensity signal with a high dynamic range could be expected given a region’s incidence and prevalence of disease.
4.6. Literature Search Window Extension
The authors would also like to note that funding was made available to support this literature review through the year 2021. Since the preparation of the present manuscript, several primary research articles have been published utilizing urine or feces as a diagnostic matrix for epigenetic marker testing, particularly regarding screening for bladder cancer. A recent study evaluating BladMetrix in Norwegian patients, a proprietary epigenetic test for bladder cancer using urine, reported a sensitivity of 96% and a sensitivity of 95% in a sample size of 112 individuals [115]. Another study in China found that a genome-wide DNA methylation profile test for bladder cancer was 100% sensitive to high-grade bladder cancer and 62% sensitive to low-grade bladder cancer, both with 100% specificity [116]. Hypermethylation of individual biomarkers TWIST1, hTERT, NID2, and VIM was detected with a sensitivity of 92%, 97%, 84%, and 83%, respectively, and a specificity of 100% for each using urine sediment samples in Moroccan bladder cancer patients [117]. The performance of a panel of ZNF671, OTX1, and IRF8 developed with a decision tree method attained a sensitivity of 75% and a specificity of 91% in Taiwanese patients [118]. A final study published this year for the detection of CRC in Iranian patients found a sensitivity of 52% and a specificity of 100% using a test for the methylation status of the CDX1 gene in fecal samples [119]. The new study representing bladder cancer patients in Norway is a particularly valuable addition to the literature analyzed in the prior pool collected prior to 2022, as Norway was not previously represented and yet reports a higher incidence of bladder cancer relative to most other nations globally. Nevertheless, the addition of these studies does not change the performance results shared in this literature review, as the best panels remain for both bladder cancer (with 100% sensitivity and specificity for the SOX1, TJP2, MYOD, HOXA9_1, HOXA9_2 panel [12]) and CRC (with a sensitivity of 98% and a specificity of 90% for the BMP3 and NDRG4 panel [18]). We sincerely hope future research continues to narrow biomarker panels by scrutinizing performance for the screening of bladder cancer, CRC, and other diseases.
5. Conclusions
Methylated genes detectable in urine and feces of individuals and human populations have the potential to enhance the early diagnosis of a variety of cancers. Development and broad-scale adoption of epigenetic diagnostic panels may enhance both clinical screening of individuals. Researchers offering expertise or potentially partnering with health officials to decide on the best means to use these methods in systematic resource allocation would open a collaboration to assist in the early detection of disease in local communities. Tracing epigenetic marks in composited urine and feces of large populations could potentially inform healthcare professionals regionally as to where and when to mobilize clinical tools to aid in the diagnosis and treatment of difficult-to-detect major diseases.
The possibility exists of improving health outcomes of cancer with a diagnostic strategy that proved helpful during the COVID-19 pandemic: the population-wide monitoring for threat agents and disease biomarkers in composited urine and stool from human communities, followed by a targeted deployment of clinical interventions to reduce morbidity and mortality. This scenario appears plausible given the large spectrum of epigenetic biomarkers identified in this study. However, there are many significant limitations to utilize these diagnostics most effectively. Whether a transition is possible from monitoring individuals only to surveilling whole populations via WBE for cancer will depend on many factors, including the following: the quantity of biomarkers excreted, the stability of epigenetic markers in wastewater, the dynamic range of marker expression, the ratio of expression in controls versus the diseased, etc. Epigenetic testing applied from the individual to the population level could improve the quality of life of both patients and their caregivers while reducing healthcare costs. Exploring this possibility will require time and further studies, however, as the science of population epigenetics and population diagnostics is still in its infancy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life15030482/s1, Supplemental Figure S1. Publication Trends in Epigenetics in Urine and Feces PRISMA Literature Review Search; Supplemental Figure S2. PRISMA Systematic Literature Review Diagram of Liquid Biopsy (Urine and Feces) Epigenetic Marks; Supplemental Figure S3. Rank-ordered Epigenetic Biomarker Sensitivity Urine and Feces by Biomarker and Disease; Supplemental Figure S4. Epigenetic Biomarker Sensitivity and Specificity in Liquid Biopsy (Urine and Feces) by Publication Count; Supplemental Figure S5. Epigenetic Biomarker Sensitivity and Specificity in Liquid Biopsy (Urine and Feces) scaled by Sample Size; Supplemental Figure S6. Cancer Incidence Hotspots; Supplemental Table S1. Metadata of Epigenetic Biomarker Panel Studies include in this Review.
Author Contributions
M.E.N.: conceptualization; data curation; formal analysis; investigation; methodology; software; visualization; writing—original draft; and writing—review and editing. A.B.: conceptualization; data curation; formal analysis; investigation; methodology; software; validation; visualization; and writing—review and editing. A.A.: conceptualization; formal analysis; investigation; methodology; and writing—review and editing. R.R.: writing—review and editing R.U.H.: conceptualization; funding acquisition; methodology; project administration; supervision; and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Glen Swette Memorial Fund [GF000000002135]. An external funding source other than discretionary funds available to the authors was not involved in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit this paper for publication. Rolf U. Halden is a founding member of OneWaterOneHealth (OWOH), a non-profit project of the ASU Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
We are thankful for the financial support gifted by the Swette family and the Glen Swette Memorial Fund. We acknowledge the institutional support provided from the Biodesign Institute and Arizona State University.
Conflicts of Interest
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: Rolf U. Halden reports financial support was provided by the Glen Swette Memorial Fund. Rolf U. Halden reports a relationship with OneWaterOneHealth that includes board membership. The other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Glossary
| biomarker | biological compound produced in the body indicating disease |
| epigenetic | marks and changes (e.g., methylation, acetylation) to genetic machinery by environmental stimuli; not mutation |
| incidence | the rate of individuals diagnosed with a disease at a given time |
| liquid biopsy | analysis of bodily fluids to detect biomarkers of disease |
| sensitivity | ability for a test to indicate individuals with disease |
| specificity | ability for a test to indicate individuals without a disease |
References
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Francesc, B.; Ajay, G. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; Goldman, L.R.; Brebi-Mieville, P.; Ili-Gangas, C.; LeBron, C.; Hernández-Arroyo, M.; Witter, F.R.; Apelberg, B.J.; Roystacher, M.; Jaffe, A.; et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 2010, 5, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G. A Brief History of Epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6, a018200. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Angeles, A.K.; Janke, F.; Bauer, S.; Christopoulos, P.; Riediger, A.L.; Sültmann, H. Liquid biopsies beyond mutation calling: Genomic and epigenomic features of cell-free dna in cancer. Cancers 2021, 13, 5615. [Google Scholar] [CrossRef]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rodriguez, I.R.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative origins of cell-free DNA in humans: A review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, D.; Xu, J.; Wang, Z.; Chen, Y.; Lei, C.; Li, Y.; Liu, G.; Jiang, Y. The framework for population epigenetic study. Brief Bioinform. 2018, 19, 89–100. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Jatkoe, T.A.; Karnes, R.J.; Freedland, S.J.; Wang, Y.; Le, A.; Baden, J. A urine-based methylation signature for risk stratification within low-risk prostate cancer. Br. J. Cancer 2015, 112, 802–808. [Google Scholar] [CrossRef]
- Battagli, C.; Uzzo, R.G.; Dulaimi, E.; De Caceres, I.I.; Krassenstein, R.; Al-Saleem, T.; Greenberg, R.E.; Cairns, P. Promoter Hypermethylation of Tumor Suppressor Genes in Urine from Kidney Cancer Patients. Cancer Res. 2003, 63, 8695–8699. [Google Scholar]
- Power, M.; Fell, G.; Wright, M. Principles for high-quality, high-value testing. BMJ Evid.-Based Med. 2013, 18, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Chihara, Y.; Kanai, Y.; Fujimoto, H.; Sugano, K.; Kawashima, K.; Liang, G.; Jones, P.A.; Fujimoto, K.; Kuniyasu, H.; Hirao, Y. Diagnostic markers of urothelial cancer based on DNA methylation analysis. BMC Cancer 2013, 13, 275. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.L.; Henrique, R.; Danielsen, S.A.; Duarte-Pereira, S.; Eknaes, M.; Skotheim, R.I.; Rodrigue, Â.; Magalhães, J.S.; Oliveira, J.; Lothe, R.A.; et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin. Cancer Res. 2010, 16, 5842–5851. [Google Scholar] [CrossRef]
- Monteiro-Reis, S.; Leça, L.; Almeida, M.; Antunes, L.; Monteiro, P.; Dias, P.C.; Morais, A.; Oliveira, J.; Henrique, R.; Jerónimo, C. Accurate detection of upper tract urothelial carcinoma in tissue and urine by means of quantitative GDF15, TMEFF2 and VIM promoter methylation. Eur. J. Cancer 2014, 50, 226–233. [Google Scholar] [CrossRef]
- D’Andrea, D.; Soria, F.; Zehetmayer, S.; Gust, K.M.; Korn, S.; Witjes, J.A.; Shariat, S.F. Diagnostic accuracy, clinical utility and influence on decision-making of a methylation urine biomarker test in the surveillance of non-muscle-invasive bladder cancer. BJU Int. 2019, 123, 959–967. [Google Scholar] [CrossRef]
- Trenti, E.; D’Elia, C.; Mian, C.; Schwienbacher, C.; Hanspeter, E.; Pycha, A.; Kafka, M.; Degener, S.; Danuser, H.; Roth, S.; et al. Diagnostic predictive value of the Bladder EpiCheck test in the follow-up of patients with non–muscle-invasive bladder cancer. Cancer Cytopathol. 2019, 127, 465–469. [Google Scholar] [CrossRef]
- Witjes, J.A.; Morote, J.; Cornel, E.B.; Gakis, G.; van Valenberg, F.J.P.; Lozano, F.; Sternberg, I.A.; Willemsen, E.; Hegemann, M.L.; Paitan, Y.; et al. Performance of the Bladder EpiCheck™ Methylation Test for Patients Under Surveillance for Non–muscle-invasive Bladder Cancer: Results of a Multicenter, Prospective, Blinded Clinical Trial. Eur. Urol. Oncol. 2018, 1, 307–313. [Google Scholar] [CrossRef]
- Lidgard, G.P.; Domanico, M.J.; Bruinsma, J.J.; Light, J.; Gagrat, Z.D.; Oldham-Haltom, R.L.; Fourrier, K.D.; Allawi, H.; Yab, T.C.; Taylor, W.R.; et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin. Gastroenterol. Hepatol. 2013, 11, 1313–1318. [Google Scholar] [CrossRef]
- Sun, J.; Fei, F.; Zhang, M.; Li, Y.; Zhang, X.; Zhu, S.; Zhang, S. The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019, 19, 450. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.; Miao, J.; Li, H.; Ma, Y.; Liu, X.; Li, S.; Zhu, Y.; Xiong, S.; Zheng, M.; et al. Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front. Genet. 2020, 11, 324. [Google Scholar] [CrossRef]
- Abern, M.R.; Owusu, R.; Inman, B.A. Clinical performance and utility of a DNA methylation urine test for bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 51.e21–51.e26. [Google Scholar] [CrossRef] [PubMed]
- Renard, I.; Joniau, S.; van Cleynenbreugel, B.; Collette, C.; Naômé, C.; Vlassenbroeck, I.; Nicolas, H.; de Level, J.; Straub, J.; Van Criekinge, W.; et al. Identification and Validation of the Methylated TWIST1 and NID2 Genes through Real-Time Methylation-Specific Polymerase Chain Reaction Assays for the Noninvasive Detection of Primary Bladder Cancer in Urine Samples. Eur. Urol. 2010, 58, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Yegin, Z.; Gunes, S.; Buyukalpelli, R. Hypermethylation of TWIST1 and NID2 in tumor tissues and voided urine in urinary bladder cancer patients. DNA Cell Biol. 2013, 32, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Fantony, J.J.; Abern, M.R.; Gopalakrishna, A.; Owusu, R.; Jack Tay, K.; Lance, R.S.; Inman, B.A. Multi-institutional external validation of urinary TWIST1 and NID2 methylation as a diagnostic test for bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 387.e1–387.e6. [Google Scholar] [CrossRef]
- Fantony, J.J.; Longo, T.A.; Gopalakrishna, A.; Owusu, R.; Lance, R.S.; Foo, W.C.; Inman, B.A.; Abern, M.R. Urinary NID2 and TWIST1 methylation to augment conventional urine cytology for the detection of bladder cancer. Cancer Biomark. 2017, 18, 381–387. [Google Scholar] [CrossRef]
- Hermanns, T.; Savio, A.J.; Olkhov-Mitsel, E.; Mari, A.; Wettstein, M.S.; Saba, K.; Bhindi, B.; Kuk, C.; Poyet, C.; Wild, P.J.; et al. A noninvasive urine-based methylation biomarker panel to detect bladder cancer and discriminate cancer grade. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 603.e1–603.e7. [Google Scholar] [CrossRef]
- Moreira-Barbosa, C.; Barros-Silva, D.; Costa-Pinheiro, P.; Torres-Ferreira, J.; Constâncio, V.; Freitas, R.; Oliveira, J.; Antunes, L.; Henrique, R.; Jerónimo, C. Comparing diagnostic and prognostic performance of two-gene promoter methylation panels in tissue biopsies and urines of prostate cancer patients. Clin. Epigenet. 2018, 10, 132. [Google Scholar] [CrossRef]
- Kandimalla, R.; Masius, R.; Beukers, W.; Bangma, C.H.; Orntoft, T.F.; Dyrskjot, L.; Van Leeuwen, N.; Lingsma, H.; Van Tilborg, A.A.G.; Zwarthoff, E.C. A 3-plex methylation assay combined with the FGFR3 mutation assay sensitively detects recurrent bladder cancer in voided urine. Clin. Cancer Res. 2013, 19, 4760–4769. [Google Scholar] [CrossRef]
- Wang, K.; Tian, Y.; Xu, H. Improved noninvasive bladder cancer diagnosis using urine sediments and novel DNA methylation biomarker panels. Clin. Lab. 2016, 62, 327–336. [Google Scholar] [CrossRef]
- Feber, A.; Dhami, P.; Dong, L.; de Winter, P.; Tan, W.S.; Martínez-Fernández, M.; Paul, D.S.; Hynes-Allen, A.; Rezaee, S.; Gurung, P.; et al. UroMark—A urinary biomarker assay for the detection of bladder cancer. Clin. Epigenetics 2017, 9, 8. [Google Scholar] [CrossRef]
- Roperch, J.P.; Grandchamp, B.; Desgrandchamps, F.; Mongiat-Artus, P.; Ravery, V.; Ouzaid, I.; Ropret, M.; Phe, V.; Ciofu, C.; Tubach, F.; et al. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer 2016, 16, 704. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.M.; Chen, P.C.; Hsieh, H.Y.; Jou, Y.C.; Lin, C.T.; Tsai, M.H.; Huang, H.Y.; Wang, Y.T.; Lin, R.I.; Chen, S.S.; et al. Methylomics analysis identifies ZNF671 as an epigenetically repressed novel tumor suppressor and a potential non-invasive biomarker for the detection of urothelial carcinoma. Oncotarget 2015, 6, 29555–29572. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, A.G.; Mengual, L.; Ingelmo-Torres, M.; Lozano, J.J.; van Rijt-van de Westerlo, C.C.M.; Baixauli, M.; Geavlete, B.; Moldoveanud, C.; Ene, C.; Dinney, C.P.; et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin. Epigenetics 2018, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.H.T.; Jiang, P.; Teoh, J.Y.C.; Heung, M.M.S.; Tam, J.C.W.; Sun, X.; Lee, W.S.; Ni, M.; Chan, R.C.K.; Ng, C.F.; et al. Noninvasive detection of bladder cancer by shallow-depth genome-wide bisulfite sequencing of urinary cell-free DNA for methylation and copy number profiling. Clin. Chem. 2019, 65, 927–936. [Google Scholar] [CrossRef]
- Piatti, P.; Chew, Y.C.; Suwoto, M.; Yamada, T.; Jara, B.; Jia, X.Y.; Guo, W.; Ghodoussipour, S.; Daneshmand, S.; Ahmadi, H.; et al. Clinical evaluation of Bladder CARE, a new epigenetic test for bladder cancer detection in urine samples. Clin. Epigenetics 2021, 13, 84. [Google Scholar] [CrossRef]
- Bosschieter, J.; Nieuwenhuijzen, J.A.; Hentschel, A.; Van Splunter, A.P.; Segerink, L.I.; Vis, A.N.; Wilting, S.M.; Lissenberg-Witte, B.I.; Van Moorselaar, R.J.A.; Steenbergen, R.D.M. A two-gene methylation signature for the diagnosis of bladder cancer in urine. Epigenomics 2019, 11, 337–347. [Google Scholar] [CrossRef]
- Yu, J.; Zhu, T.; Wang, Z.; Zhang, H.; Qian, Z.; Xu, H.; Gao, B.; Wang, W.; Gu, L.; Meng, L.; et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin. Cancer Res. 2007, 13, 7296–7304. [Google Scholar] [CrossRef]
- Guo, R.Q.; Xiong, G.Y.; Yang, K.W.; Zhang, L.; He, S.M.; Gong, Y.Q.; He, Q.; Li, X.Y.; Wang, Z.C.; Bao, Z.Q.; et al. Detection of urothelial carcinoma, upper tract urothelial carcinoma, bladder carcinoma, and urothelial carcinoma with gross hematuria using selected urine-DNA methylation biomarkers: A prospective, single-center study. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 342.e15–342.e23. [Google Scholar] [CrossRef]
- Pietrusiński, M.; Kpczyński, Ł.; Jdrzejczyk, A.; Borkowska, E.; Traczyk-Borszyńska, M.; Constantinou, M.; Kauzewski, B.; Borowiec, M. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark. 2017, 18, 47–59. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Ye, R.; Zhang, D.; Li, Q.; An, D.; Fang, L.; Lin, Y.; Hou, Y.; Xu, A.; et al. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget 2016, 7, 2754–2764. [Google Scholar] [CrossRef]
- Padrão, N.A.; Monteiro-Reis, S.; Torres-Ferreira, J.; Antunes, L.; Leça, L.; Montezuma, D.; Ramalho-Carvalho, J.; Rias, P.C.; Monteiro, P.; Oliveira, J.; et al. MicroRNA promoter methylation: A new tool for accurate detection of urothelial carcinoma. Br. J. Cancer 2017, 116, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Urakami, S.; Shiina, H.; Enokida, H.; Kawakami, T.; Kawamoto, K.; Hirata, H.; Tanaka, Y.; Kikuno, N.; Nakagawa, M.; Igawa, M.; et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin. Cancer Res. 2006, 12, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Tsai, M.H.; Yip, S.K.; Jou, Y.C.; Ng, C.F.; Chen, Y.; Wang, X.; Huang, W.; Tung, C.L.; Chen, G.C.; et al. Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med. Genom. 2011, 4, 45. [Google Scholar] [CrossRef]
- Su, S.F.; De Castro Abreu, A.L.; Chihara, Y.; Tsai, Y.; Andreu-Vieyra, C.; Daneshmand, S.; Skinner, E.C.; Jones, P.A.; Siegmund, K.D.; Liang, G. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin. Cancer Res. 2014, 20, 1978–1989. [Google Scholar] [CrossRef]
- Reinert, T.; Borre, M.; Christiansen, A.; Hermann, G.G.; Ørntoft, T.F.; Dyrskjøt, L. Diagnosis of Bladder Cancer Recurrence Based on Urinary Levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 Hypermethylation. PLoS ONE 2012, 7, e46297. [Google Scholar] [CrossRef]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.P.J. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1483–1491. [Google Scholar] [CrossRef]
- Reinert, T.; Modin, C.; Castano, F.M.; Lamy, P.; Wojdacz, T.K.; Hansen, L.L.; Wiuf, C.; Borre, M.; Dyrskjøt, L.; Ørntoft, T.F. Comprehensive genome methylation analysis in bladder cancer: Identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin. Cancer Res. 2011, 17, 5582–5592. [Google Scholar] [CrossRef]
- Lin, H.H.; Ke, H.L.; Huang, S.P.; Wu, W.J.; Chen, Y.K.; Chang, L.L. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol. Oncol. Semin. Orig. Investig. 2010, 28, 597–602. [Google Scholar] [CrossRef]
- Friedrich, M.G.; Toma, M.I.; Chun, J.K.H.F.; Steuber, T.; Budäus, L.; Isbarn, H.; Humand, H. DNA methylation on urinalysis and as a prognostic marker in urothelial cancer of the bladder. Urol.—Ausg. A 2007, 46, 761–768. [Google Scholar] [CrossRef]
- Shimizu, T.; Suzuki, H.; Nojima, M.; Kitamura, H.; Yamamoto, E.; Maruyama, R.; Ashida, M.; Hatahira, T.; Kai, M.; Masumori, N.; et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur. Urol. 2013, 63, 1091–1100. [Google Scholar] [CrossRef]
- Scher, M.B.; Elbaum, M.B.; Mogilevkin, Y.; Hilbert, D.W.; Mydlo, J.H.; Sidi, A.A.; Adelson, M.E.; Mordechai, E.; Trama, J.P. Detecting DNA methylation of the BCL2, CDKN2A and NID2 genes in urine using a nested methylation specific polymerase chain reaction assay to predict bladder cancer. J. Urol. 2012, 188, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Vinci, S.; Giannarini, G.; Selli, C.; Kuncova, J.; Villari, D.; Valent, F.; Orlando, C. Quantitative methylation analysis of BCL2, hTERT, and DAPK promoters in urine sediment for the detection of non-muscle-invasive urothelial carcinoma of the bladder: A prospective, two-center validation study. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Steven, K.; Guldberg, P. Size-based enrichment of exfoliated tumor cells in urine increases the sensitivity for DNA-based detection of bladder cancer. PLoS ONE 2014, 9, e94023. [Google Scholar] [CrossRef] [PubMed]
- Bayramov, B.; Gunes, S.; Buyukalpelli, R.; Aydın, O.; Henkel, R. Promoter methylation analysis of CDH1 and p14ARF genes in patients with urothelial bladder cancer. OncoTargets Ther. 2018, 11, 4189–4196. [Google Scholar] [CrossRef]
- Antony, P.; Rose, M.; Gaisa, N.T.; Alkaya, S.; Heidenreich, A.; Knüchel, R.; Dahl, E. Characterisation of DNA methylation biomarkers for bladder cancer. Pathologe 2010, 31 (Suppl. 2), 244–250. [Google Scholar] [CrossRef]
- Yates, D.R.; Rehman, I.; Meuth, M.; Cross, S.S.; Hamdy, F.C.; Catto, J.W.F. Methylational urinalysis: A prospective study of bladder cancer patients and age stratified benign controls. Oncogene 2006, 25, 1984–1988. [Google Scholar] [CrossRef]
- Hoque, M.O.; Begum, S.; Topaloglu, O.; Chatterjee, A.; Rosenbaum, E.; Van Criekinge, W.; Westra, W.H.; Schoenberg, M.; Zahurak, M.; Goodman, S.N.; et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J. Natl. Cancer Inst. 2006, 98, 996–1004. [Google Scholar] [CrossRef]
- Maldonado, L.; Brait, M.; Michailidi, C.; Munari, E.; Driscoll, T.; Schultz, L.; Bivalacqua, T.; Schoenberg, M.; Sidransky, D.; Netto, G.J.; et al. An epigenetic marker panel for recurrence risk prediction of low grade papillary urothelial cell carcinoma (LGPUCC) and its potential use for surveillance after transurethral resection using urine. Oncotarget 2014, 5, 5218–5233. [Google Scholar] [CrossRef]
- Eissa, S.; Zohny, S.F.; Shehata, H.H.; Hegazy, M.G.A.; Salem, A.M.; Esmat, M. Urinary retinoic acid receptor-β2 gene promoter methylation and hyaluronidase activity as noninvasive tests for diagnosis of bladder cancer. Clin. Biochem. 2012, 45, 402–407. [Google Scholar] [CrossRef]
- Serizawa, R.R.; Ralfkiær, U.; Steven, K.; Lam, G.W.; Schmiedel, S.; Schüz, J.; Hansen, A.B.; Horn, T.; Guldberg, P. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int. J. Cancer 2011, 129, 78–87. [Google Scholar] [CrossRef]
- Costa, V.L.; Henrique, R.; Danielsen, S.A.; Eknaes, M.; Patrício, P.; Morais, A.; Oliveira, J.; Lothe, R.A.; Teixeira, M.R.; Lind, G.E.; et al. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 2011, 6, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, D.; Kaufmann, M.; Hippe, J.; Gajda, M.; Grimm, M.O. High Detection Rate for Non–Muscle-Invasive Bladder Cancer Using an Approved DNA Methylation Signature Test. Clin. Genitourin. Cancer 2020, 18, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Mijnes, J.; Veeck, J.; Gaisa, N.T.; Burghardt, E.; de Ruijter, T.C.; Gostek, S.; Dahl, E.; Pfiser, D.; Schmidt, S.C.; Knüchel, R.; et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin. Epigenetics 2018, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Bernert, H.; Kagohara, L.T.; Maldonado, L.; Brait, M.; Schoenberg, M.; Bivalacqua, T.; Netto, G.J.; Koch, W.; Sidransky, D.; et al. Epigenetic inactivation of VGF associated with Urothelial Cell Carcinoma and its potential as a non-invasive biomarker using urine. Oncotarget 2014, 5, 3350–3361. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Z.; Zhu, T.; Yu, J.; Ma, K.; Zhang, H.; He, Y.; Luo, X.; Zhu, J. Hypermethylated SFRP1, but none of other nine genes “informative” for western countries, is valuable for bladder cancer detection in Mainland China. J. Cancer Res. Clin. Oncol. 2009, 135, 1717–1727. [Google Scholar] [CrossRef]
- Hoffstetter, R.; Riquelme, I.; Andana, A.; Ili, C.G.; Buchegger, K.; Vargas, H.; Brebi, P.; Roa, J.C. Evaluation of DNA methylation in promoter regions of SFRP4 and ZAR1 in urine and plasma of women with cervical lesions. Transl. Cancer Res. 2017, 6, 157–168. [Google Scholar] [CrossRef]
- Lu, H.; Huang, S.; Zhang, X.; Wang, D.; Zhang, X.; Yuan, X.; Zhang, Q.; Huang, Z. DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA. Oncol. Lett. 2014, 8, 1751–1756. [Google Scholar] [CrossRef]
- Han, Y.D.; Oh, T.J.; Chung, T.H.; Jang, H.W.; Kim, Y.N.; An, S.; Kim, N.K. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin. Epigenetics 2019, 11, 51. [Google Scholar] [CrossRef]
- Oh, T.J.; Oh, H.I.; Seo, Y.Y.; Jeong, D.; Kim, C.; Kang, H.W.; Han, Y.D.; Chung, H.C.; Kim, N.K.; An, S. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin. Epigenetics 2017, 9, 126. [Google Scholar] [CrossRef]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Orrù, S.; Ziranu, P.; Pretta, A.; Lai, E.; Puzzoni, M.; Ciccone, L.; Casadei-Gardini, A.; et al. Colorectal cancer early detection in stool samples tracing CPG islands methylation alterations affecting gene expression. Int. J. Mol. Sci. 2020, 21, 4494. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Taylor, W.R.; Mahoney, D.W.; Zou, H.; Domanico, M.; Thibodeau, S.N.; Boardman, L.A.; Berger, B.M.; Lidgard, G.P. The Stool DNA Test Is More Accurate Than the Plasma Septin 9 Test in Detecting Colorectal Neoplasia. Clin. Gastroenterol. Hepatol. 2012, 10, 272–277.e1. [Google Scholar] [CrossRef]
- Wang, D.R.; Tang, D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J. Gastroenterol. 2008, 14, 524–531. [Google Scholar] [CrossRef]
- Itzkowitz, S.; Brand, R.; Jandorf, L.; Durkee, K.; Millholland, J.; Rabeneck, L.; Schroy III, P.C.; Sontag, S.; Johnson, D.; Markowitz, S.; et al. A simplified, noninvasive Stool DNA test for colorectal cancer detection. Am. J. Gastroenterol. 2008, 103, 2862–2870. [Google Scholar] [CrossRef]
- Xiao, W.; Zhao, H.; Dong, W.; Li, Q.; Zhu, J.; Li, G.; Zhang, S.; Ye, M. Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol. Lett. 2015, 9, 1383–1387. [Google Scholar] [CrossRef]
- Glöckner, S.C.; Dhir, M.; Joo, M.Y.; McGarvey, K.E.; Van Neste, L.; Louwagie, J.; Chan, T.A.; Kleeberger, W.; De Bruïne, A.P.; Smits, K.M.; et al. Methylation of TFPI2 in stool DNA: A potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009, 69, 4691–4699. [Google Scholar] [CrossRef]
- Leung, W.K.; To, K.F.; Man, E.P.S.; Chan, M.W.Y.; Hui, A.J.; Ng, S.S.M.; Lau, J.Y.W.; Sung, J.J.Y. Detection of hypermethylated DNA or cyclooxygenase-2 messenger rna in fecal samples of patients with colorectal cancer or polyps. Am. J. Gastroenterol. 2007, 102, 1070–1076. [Google Scholar] [CrossRef]
- Itzkowitz, S.H.; Jandorf, L.; Brand, R.; Rabeneck, L.; Schroy III, P.C.; Sontag, S.; Johnson, D.; Skoletsky, J.; Durkee, K.; Markowitz, S.; et al. Improved Fecal DNA Test for Colorectal Cancer Screening. Clin. Gastroenterol. Hepatol. 2007, 5, 111–117. [Google Scholar] [CrossRef]
- Pakbaz, B.; Jabinin, R.; Soltani, N.; Ayatollahi, H.; Farzanehfar, M. Quantitative study of vimentin gene methylation in stool samples for colorectal cancer screening. J. Adv. Pharm. Technol. Res. 2019, 10, 121–125. [Google Scholar]
- Hellebekers, D.M.E.I.; Lentjes, M.H.F.M.; Van Den Bosch, S.M.; Melotte, V.; Wouters, K.A.D.; Daenen, K.L.J.; Smits, K.M.; Akiyama, Y.; Yuasa, Y.; Sanduleanu, S.; et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 2009, 15, 3990–3997. [Google Scholar] [CrossRef]
- Chen, W.D.; Han, Z.J.; Skoletsky, J.; Olson, J.; Sah, J.; Myeroff, L.; Platzer, P.; Lu, S.; Dawson, D.; Willis, J.; et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl. Cancer Inst. 2005, 97, 1124–1132. [Google Scholar] [CrossRef]
- Mayor, R.; Casadomé, L.; Azuara, D.; Moreno, V.; Clark, S.J.; Capellà, G.; Peinado, M.A. Long-range epigenetic silencing at 2q14.2 affects most human colorectal cancers and may have application as a non-invasive biomarker of disease. Br. J. Cancer 2009, 100, 1534–1539. [Google Scholar] [CrossRef]
- Kim, M.S.; Louwagie, J.; Carvalho, B.; Terhaar sive Droste, J.S.; Park, H.L.; Chae, Y.K.; Yamashita, K.; Liu, J.; Ostrow, K.L.; Ling, S.; et al. Promoter DNA methylation of Oncostatin M receptor-β as a novel diagnostic and therapeutic marker in colon cancer. PLoS ONE 2009, 4, e6555. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; de’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Bach, S.; Paulis, I.; Sluiter, N.R.; Tibbesma, M.; Martin, I.; van de Wiel, M.A.; Tuynman, J.B.; Bahce, I.; Kazemier, G.; Steenbergen, R.D.M. Detection of colorectal cancer in urine using DNA methylation analysis. Sci. Rep. 2021, 11, 2363. [Google Scholar] [CrossRef]
- Van Kessel, K.E.M.; Van Neste, L.; Lurkin, I.; Zwarthoff, E.C.; Van Criekinge, W. Evaluation of an Epigenetic Profile for the Detection of Bladder Cancer in Patients with Hematuria. J. Urol. 2016, 195, 601–607. [Google Scholar] [CrossRef]
- Van Kessel, K.E.M.; Beukers, W.; Lurkin, I.; Ziel-van der Made, A.; van der Keur, K.A.; Boormans, J.L.; Dyrskjøt, L.; Márquez, M.; Ørntoft, T.F.; Real, F.X.; et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J. Urol. 2017, 197, 590–595. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, G.; Zhang, N.; Liu, S.; Lin, X.; Perschon, C.; Zheng, S.L.; Ding, Q.; Wang, X.; Na, R.; et al. HOXA9, PCDH17, POU4F2, and ONECUT2 as a Urinary Biomarker Combination for the Detection of Bladder Cancer in Chinese Patients with Hematuria. Eur. Urol. Focus 2020, 6, 284–291. [Google Scholar] [CrossRef]
- Xin, J.; Xu, R.; Lin, S.; Xin, M.; Cai, W.; Zhou, J.; Fu, C.; Zhen, G.; Lai, J.; Li, Y.; et al. Clinical potential of TCF21 methylation in the diagnosis of renal cell carcinoma. Oncol. Lett. 2016, 12, 1265–1270. [Google Scholar] [CrossRef]
- Hoque, M.O.; Begum, S.; Topaloglu, O.; Jeronimo, C.; Mambo, E.; Westra, W.H.; Califano, J.A.; Sidransky, D. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004, 64, 5511–5517. [Google Scholar] [CrossRef]
- Outeiro-Pinho, G.; Barros-Silva, D.; Aznar, E.; Sousa, A.I.; Vieira-Coimbra, M.; Oliveira, J.; Gonçalves, C.S.; Costa, B.M.; Junker, K.; Henrique, R.; et al. MicroRNA-30a-5pme: A novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J. Exp. Clin. Cancer Res. 2020, 39, 98. [Google Scholar] [CrossRef]
- Liu, B.; Filho, J.R.; Mallisetty, A.; Villani, C.; Kottorou, A.; Rodgers, K.; Chen, C.; Ito, T.; Holmes, K.; Gastala, N.; et al. Detection of Promoter DNA Methylation in Urine and Plasma Aids the Detection of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 4339–4348. [Google Scholar] [CrossRef]
- Jarrard, W.E.; Schultz, A.; Etheridge, T.; Damodaran, S.; Allen, G.O.; Jarrard, D.; Yang, B. Screening of urine identifies PLA2G16 as a field defect methylation biomarker for prostate cancer detection. PLoS ONE 2019, 14, e0218950. [Google Scholar] [CrossRef]
- Connell, S.P.; O’Reilly, E.; Tuzova, A.; Webb, M.; Hurst, R.; Mills, R.; Zhao, F.; Bapat, B.; Cooper, C.S.; Perry, A.S.; et al. Development of a multivariable risk model integrating urinary cell DNA methylation and cell-free RNA data for the detection of significant prostate cancer. Prostate 2020, 80, 547–558. [Google Scholar] [CrossRef]
- Ramalho-Carvalho, J.; Martins, J.B.; Cekaite, L.; Sveen, A.; Torres-Ferreira, J.; Graça, I.; Costa-Pinheiro, P.; Eilertsen, I.A.; Antunes, L.; Oliveira, J.; et al. Epigenetic disruption of miR-130a promotes prostate cancer by targeting SEC23B and DEPDC1. Cancer Lett. 2017, 385, 150–159. [Google Scholar] [CrossRef]
- Zhao, F.; Vesprini, D.; Liu, R.S.C.; Olkhov-Mitsel, E.; Klotz, L.H.; Loblaw, A.; Liu, S.K.; Bapat, B. Combining urinary DNA methylation and cell-free microRNA biomarkers for improved monitoring of prostate cancer patients on active surveillance. Urol. Oncol. Semin. Orig. Investig. 2019, 37, 297.e9–297.e17. [Google Scholar] [CrossRef]
- Larsen, L.K.; Jakobsen, J.S.; Abdul-Al, A.; Guldberg, P. Noninvasive Detection of High Grade Prostate Cancer by DNA Methylation Analysis of Urine Cells Captured by Microfiltration. J. Urol. 2018, 200, 749–757. [Google Scholar] [CrossRef]
- O’Reily, E.; Tuzova, A.V.; Walsh, A.L.; Russell, N.M.; O’Brien, O.; Kelly, S.; Dhomhnallain, O.N.; DeBarra, L.; Dale, C.M.; Brugman, R.; et al. epiCaPture: A Urine DNA methylation test for early detection of aggressive prostate cancer. JCO Precis. Oncol. 2019, 3, 1–18. [Google Scholar] [CrossRef]
- Vener, T.; Derecho, C.; Baden, J.; Wang, H.; Rajpurohit, Y.; Skelton, J.; Mehrotra, J.; Varde, S.; Chodary, D.; Stallings, W.; et al. Development of a multiplexed urine assay for prostate cancer diagnosis. Clin. Chem. 2008, 54, 874–882. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Q.; Li, L.L.; Wang, Z.; Ying, J.; Fan, Y.; He, O.; LV, T.; Han, W.; Li, J.; et al. DLEC1, a 3p tumor suppressor, represses NF-κB signaling and is methylated in prostate cancer. J. Mol. Med. 2015, 93, 691–701. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA hypermethylation in disease: Mechanisms and clinical relevance. Epigenetics 2019, 14, 1141–1163. [Google Scholar] [CrossRef]
- Sequeira, J.P.; Constâncio, V.; Salta, S.; Lobo, J.; Barros-Silva, D.; Carvalho-Maia, C.; Rodrigues, J.; Braga, I.; Henrique, R.; Jerónimo, C. LiKidMiRs: A ddPCR-Based Panel of 4 Circulating miRNAs for Detection of Renal Cell Carcinoma. Cancers 2022, 14, 858. [Google Scholar] [CrossRef]
- Bryzgunova, O.; Bondar, A.; Ruzankin, P.; Laktionov, P.; Tarasenko, A.; Kurilshikov, A.; Epifanov, R.; Zaripov, M.; Kabilov, M.; Laktionov, P. Locus-Specific Methylation of GSTP1, RNF219, and KIAA1539 Genes with Single Molecule Resolution in Cell-Free DNA from Healthy Donors and Prostate Tumor Patients: Application in Diagnostics. Cancers 2021, 13, 6234. [Google Scholar] [CrossRef]
- Tänzer, M.; Balluff, B.; Distler, J.; Hale, K.; Leodolter, A.; Röcken, C.; Molnar, B.; Schmidt, R.; Lofton-Day, C.; Schuster, T.; et al. Performance of Epigenetic Markers SEPT9 and ALX4 in Plasma for Detection of Colorectal Precancerous Lesions. PLoS ONE 2010, 5, e9061. [Google Scholar] [CrossRef]
- Rezvani, N.; Alibakhshi, R.; Vaisi-Raygani, A.; Bashiri, H.; Saidijam, M. Detection of SPG20 gene promoter-methylated DNA, as a novel epigenetic biomarker, in plasma for colorectal cancer diagnosis using the MethyLight method. Oncol. Lett. 2017, 13, 3277–3284. [Google Scholar] [CrossRef]
- Fedyuk, V.; Erez, N.; Furth, N.; Beresh, O.; Andreishcheva, E.; Shinde, A.; Jones, D.; Zakai, B.B.; Mavor, Y.; Peretz, T.; et al. Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics. Nat. Biotechnol. 2023, 41, 212–221. [Google Scholar] [CrossRef]
- Carja, O.; MacIsaac, J.L.; Mah, S.M.; Henn, B.M.; Kobor, M.S.; Feldman, M.W.; Fraser, H.B. Worldwide patterns of human epigenetic variation. Nat. Ecol. Evol. 2017, 1, 1577–1583. [Google Scholar] [CrossRef]
- Laugsand, E.A.; Brenne, S.S.; Skorpen, F. DNA methylation markers detected in blood, stool, urine, and tissue in colorectal cancer: A systematic review of paired samples. Int. J. Color. Dis. 2021, 36, 239–251. [Google Scholar] [CrossRef]
- Zandvakili, I.; Lazaridis, K.N. Cell-free DNA testing: Future applications in gastroenterology and hepatology. Ther. Adv. Gastroenterol. 2019, 12, 1756284819841896. [Google Scholar] [CrossRef]
- Nel, I.; Münch, C.; Shamkeeva, S.; Heinemann, M.L.; Isermann, B.; Aktas, B. The Challenge to Stabilize, Extract and Analyze Urinary Cell-Free DNA (ucfDNA) during Clinical Routine. Diagnostics 2023, 13, 3670. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A.; Bossuyt, P.M.M. Waste, Leaks, and Failures in the Biomarker Pipeline. Clin. Chem. 2017, 63, 963–972. [Google Scholar] [CrossRef]
- ESMO. FDA Approves First Non-Invasive DNA Screening Test for Colorectal Cancer. Available online: https://www.esmo.org/oncology-news/archive/fda-approves-first-non-invasive-dna-screening-test-for-colorectal-cancer (accessed on 23 December 2024).
- Honey, K. FDA Approves Blood-Based Colorectal Cancer Screening Test [Internet]. American Association for Cancer Research (AACR). 2016. Available online: https://www.aacr.org/blog/2016/04/26/fda-approval-epi-pro-colon-colorectal-cancer/ (accessed on 23 December 2024).
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Bowes, D.A.; Driver, E.M.; Kraberger, S.; Fontenele, R.S.; Holland, L.A.; Wright, J.; Johnson, B.; Savic, S.; Engstrom Newell, M.; Adhikari, S.; et al. Leveraging an Established Neighbourhood-Level, Open Access Wastewater Monitoring Network to Address Public Health Priorities: A Population-Based Study. The Lancet Microbe [Internet]. Available online: https://www.sciencedirect.com/science/article/pii/S2666524722002890 (accessed on 6 December 2022).
- Pharo, H.D.; Jeanmougin, M.; Ager-Wick, E.; Vedeld, H.M.; Sørbø, A.K.; Dahl, C.; Larsen, L.K.; Honne, H.; Brandt-Winge, S.; Five, M.-B.; et al. BladMetrix: A novel urine DNA methylation test with high accuracy for detection of bladder cancer in hematuria patients. Clin. Epigenetics 2022, 14, 1–115. [Google Scholar] [CrossRef]
- Xiao, Y.; Ju, L.; Qian, K.; Jin, W.; Wang, G.; Zhao, Y.; Jiang, W.; Liu, N.; Wu, K.; Peng, M.; et al. Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNA methylation in a prospective cohort study. Clin. Transl. Med. 2022, 12, e1008. [Google Scholar] [CrossRef]
- El Azzouzi, M.; El Ahanidi, H.; Hafidi Alaoui, C.; Chaoui, I.; Benbacer, L.; Tetou, M.; Hassan, I.; Bensaid, M.; Oukabli, M.; Ameur, A.; et al. Exploring urine sediments as a non-invasive method for DNA methylation detection in bladder cancer. Afr. J. Urol. 2022, 28, 31. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Liu, Y.S.; Wei, Y.C.; Jhang, J.F.; Kuo, H.C.; Huang, H.H.; Chan, M.W.Y.; Lin, G.-L.; Cheng, W.-C.; Lin, S.-C.; et al. Hypermethylation Loci of ZNF671, IRF8, and OTX1 as Potential Urine-Based Predictive Biomarkers for Bladder Cancer. Diagnostics 2024, 14, 468. [Google Scholar] [CrossRef]
- Almasi, S.; Haghnazari, L.; Hosseini, S.O.; Rezvani, N. Detection of caudal type homeobox 1 (CDX1) gene methylated DNA, as a stool-based diagnostic biomarker in colorectal cancer. J. Genet. 2024, 103, 23. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
