Abstract
Nephrolithiasis is a medical condition characterized by the existence or development of calculi, commonly referred to as stones within the renal system, and poses significant health challenges. Calcium phosphate and calcium oxalate are the predominant constituents of renal calculi and are introduced into the human body primarily via dietary sources. The presence of oxalates can become particularly problematic when the delicate balance of the normal flora residing within the gastrointestinal tract is disrupted. Within the human gut, species of Oxalobacter, Lactobacillus, and Bifidobacterium coexist in a symbiotic relationship. They play a pivotal role in mitigating the risk of stone formation by modulating certain biochemical pathways and producing specific enzymes that can facilitate the breakdown and degradation of oxalate salts. The probiotic potential exhibited by these bacteria is noteworthy, as it underscores their possible utility in the prevention of nephrolithiasis. Investigating the mechanisms by which these beneficial microorganisms exert their effects could lead to novel therapeutic strategies aimed at reducing the incidence of kidney stones. The implications of utilizing probiotics as a preventive measure against kidney stone formation represent an intriguing frontier in both nephrology and microbiome research, meriting further investigation to unlock their full potential.
1. Introduction
The condition known as nephrolithiasis is characterized by solid deposits in the urinary system, commonly called stones, which hinder normal urine flow. These deposits form due to elevated levels of calcium, oxalic acid, phosphate, urate, and cystine in the urine. Regrettably, the prevalence of urolithiasis has significantly risen in the last three decades, possibly attributed to environmental shifts such as suboptimal dietary habits and reduced physical exertion. Presently, nephrolithiasis holds the third position among the most prevalent urological disorders. Urolithiasis varies from 7 to 13% in North America, 5 to 9% in Europe, and 1 to 5% in Asia. Nephrolithiasis primarily affects males aged 40 to 50 and females aged 50 to 70 [1]. Deposits found in the urinary system exhibit variability in their chemical makeup; the majority of stones, approximately 80%, consist of calcium, while 9% are composed of uric acid, 10% of struvite, and 1% of cysteine. Within the category of calcium stones, half are calcium oxalate, 5% are calcium phosphate, and the remaining 45% are a combination of both types. The process of kidney stone formation is intricate and influenced by various factors, including intrinsic ones such as age, gender, and genetic predisposition, as well as extrinsic factors like geographical location, climate, dietary habits, mineral content, and water consumption patterns [2]. Oxalic acid (pKa = 1.2), a potent organic acid, is abundantly present in nature and can be found in various plants and animals. Within plants, it typically builds up as a metabolic byproduct in the shape of free acid, serving various functions within the organism. Interestingly, oxalic acid also exhibits chelating properties towards cations and is frequently encountered in the form of soluble sodium or potassium oxalate. Moreover, it tends to precipitate as insoluble calcium oxalate, further showcasing its diverse chemical behavior in biological systems [3,4].
The primary objective of the present review was to categorize the various biological techniques that can potentially decrease the levels of oxalates found in food products while also examining the methodologies employed for the quantification of oxalates in these food items. We have performed a literature search for oxalate-degrading bacteria using the internet and authenticated research articles, especially focusing on the role of lactic acid bacteria and bifidobacteria. We also tried to analyze the major outcomes of the different studies and compile them in tabular form for ease of understanding and discuss them comprehensively in the text.
Oxalic acid and its oxalate salts are present in the blood (plasma) and urine of both animals and humans [1,4]. It is noteworthy that a portion of the oxalate observed in the human body is obtained via the consumption of oxalate-rich plant material, primarily through the intake of strawberries, spinach, rhubarb, beets, nuts, wheat bran, chocolate, tea, and coffee, as outlined in Table 1. The concentration of oxalate in plant foods can vary for several reasons, such as cultivar, harvesting time, processing, etc. It is also reported that free calcium can reduce soluble oxalate in food by forming insoluble oxalate [5].
It has been observed that the processing method can significantly impact oxalate levels in different plant-derived foods. For instance, boiling plant leaves can leach out soluble oxalates; hence, it reduces oxalate levels. Similarly, hot-air drying can also reduce oxalate levels in rice paddy herbs [5]. Oxalate-degrading microorganisms also reduce their levels via biochemical reactions mediated by the production of enzymes, and, therefore, fermentation may also observe a similar effect.
Simultaneously, a certain quantity of oxalate is internally generated in the liver as a byproduct of the metabolic processes involving glycine, glyoxylate, and ascorbic acid [6]. Despite this understanding, the precise ratios at which exogenous and endogenous origins contribute to the overall oxalate levels in the gastrointestinal tract remain a topic of ongoing discussion and investigation (Figure 1). This debate underscores the complexity of oxalate metabolism and the importance of understanding the sources and pathways through which oxalate is introduced into the human system, thereby emphasizing the need for further research in this area [7].
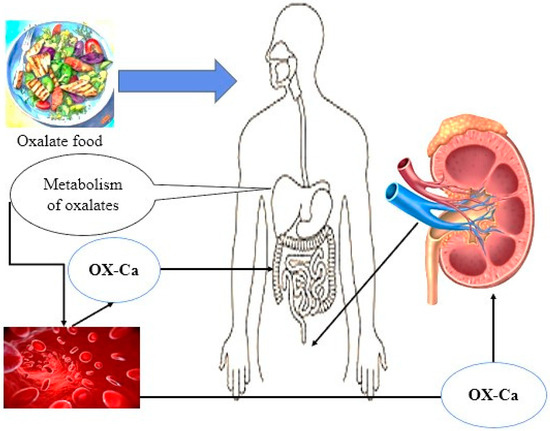
Figure 1.
Food and internal sources (liver metabolism) are the two main sources of oxalate in the body.
The intricate interplay between dietary intake, internal production, and physiological mechanisms governing oxalate levels represents the multifaceted nature of oxalate homeostasis and the necessity for a holistic approach to exploring the factors influencing oxalate concentrations in the body [8]. In light of the potential implications for health and disease, elucidating the nuanced dynamics of oxalate sources and their impact on bodily processes is crucial for advancing our knowledge in this field and informing strategies for managing oxalate-related conditions. Oxalobacter formigenes is recognized as a bacterium that degrades oxalate, utilizing intestinal oxalate as its exclusive source of carbon to regulate oxalate homeostasis within the biological system. Nevertheless, the utilization of this bacterium as a probiotic has been constrained primarily due to its demanding nutritional requirements, limited capacity for colonization, and highly specialized oxalotrophic characteristics [9]. Conversely, lactic acid bacteria (LAB) constitute essential components of the human intestinal environment and have been widely employed as probiotics owing to their advantageous effects on the health of the host organism [10,11]. Various research studies have validated the association between the oral intake of Lactobacillus or Bifidobacterium species and their significant involvement in reducing luminal oxalate levels, consequently leading to a decline in the probability of urinary oxalate excretion in both human subjects and animals [12,13]. Furthermore, this review aimed to compile and showcase the diverse strategies utilized by different researchers across the world for the reduction of oxalates and to ascertain the different approaches for determining the presence of oxalates in food samples.

Table 1.
Percentage of oxalate concentration in some types of food and beverages.
Table 1.
Percentage of oxalate concentration in some types of food and beverages.
| Food and Beverage Types | Concentration of Oxalate (mg/100 g or mL) | References |
|---|---|---|
| Citrus aurantium | 2.07–10.64 | [14] |
| Musa acuminata Musa balbisiana | 0.00–9.90 | |
| Spinacia oleracea | 364.44–1145.00 | |
| Beta vulgaris | 36.90–794.12 | |
| Arachis hypogaea | 64.57–348.58 | |
| Glycyrrhiza glabra | 3343.20–3795.40 | [15] |
| Phaseolus vulgaris | 500.60–593.20 | |
| Oryza sativa | <10 | |
| Ipomoea batatas | 467.30–523.90 | |
| Abelmoschus esculentus | 56.30–317.20 | |
| Glycine max | 33.40–42.50 | |
| Pisum sativum | 244.70–294.00 | [16] |
| Lens culinaris | 168.60–289.10 | |
| Vicia faba | 241.50–291.40 | |
| Cicer arietinum | 92.20–214.00 | |
| Black Tea | 78.00–112.00 | [17] |
| Green Tea | 40.00–50.00 | |
| Red wine and beer | <1 | |
| Carrot juice | 5.81–6.20 | |
| Tomato juice | 1.43–4.43 | |
| Milk | <10 |
2. Oxalate Utilization by Flora Intestinal Bacteria
The use of oxalate by anaerobic and facultative anaerobic intestinal bacteria is an important process that plays a key role in the gut microbiome [18]. The ability of these bacteria to utilize oxalate contributes to maintaining gut health and metabolism by regulating the concentration of this molecule and preventing potential health-damaging effects. The human body harbors a diverse and constantly evolving community consisting of hundreds of different types of microorganisms, with the majority residing in the digestive system [19]. The composition of the microbiota in the intestines of adults is unique to each person. It tends to remain relatively consistent, although it may undergo fluctuations in response to dietary changes or the consumption of antibiotics. These variations can occur over time within an individual, highlighting the complex and dynamic nature of the interactions between the host and its gut microbiome [20].
Intestinal bacteria are responsible for carrying out a variety of biochemical reactions that have the potential to impact human health and nutrition significantly. Certain bacterial genera, such as Lactobacillus and Bifidobacterium, have gained extensive importance as probiotics in both food products and pharmaceuticals due to their natural occurrence and beneficial contributions to human health. These probiotics are known for their positive effects on the human body [21,22,23]. Intestinal bacteria play a crucial role in breaking down various dietary substances that are indigestible by humans, including oxalate, which is a compound that can be degraded by these bacteria. Oxalic acid, a simple dicarboxylic acid, is a substance that can be harmful in large quantities and is generally not the main source of energy for most bacteria because it yields a low amount of energy during the metabolic process. Despite its potential toxicity, oxalic acid can be metabolized by intestinal bacteria, showcasing the complex interactions between these microorganisms and the substances present in the human digestive system [24]. There are two main categories of oxalotrophs known as the “generalist oxalotrophs”, which exhibit versatility in utilizing various substrates for fermentation apart from oxalate, and the “specialist oxalotrophs”, which rely predominantly on oxalate as their primary carbon and energy source. These distinct classifications reflect the diverse metabolic capabilities and adaptations of microorganisms in response to their environmental niches [25].
2.1. Oxalobacter formigenes
Oxalobacter formigenes is a Gram-negative, obligatory anaerobic, rod or curve-shaped, non-motile, and non-spore-forming bacterium, and it has been classified within the following taxonomic groups: Bacteria (domain); Proteobacteria (phylum); Betaproteobacteria (class); Burkholderiales (order); Oxalobacteraceae (family); Oxalobacter (genus); formigenes (species). This bacterium plays a crucial role in various biological processes and ecological systems [26]. The optimal growth conditions for O. formigenes in culture are achieved in anaerobic settings with a pH ranging from 6 to 7, utilizing a CO2-bicarbonate buffered undefined medium that includes minerals, oxalate, acetate, and a small portion of yeast extract. Although yeast extract is not mandatory, it has been observed to enhance the growth of certain strains of O. formigenes, particularly during the initial stages of isolation from the gastrointestinal tract. Irrespective of the presence or absence of yeast extract, acetate’s minimal concentration (ranging from 0.5 to 2 mM) is incorporated since O. formigenes utilizes small quantities of carbon from acetate, alongside carbon dioxide, to synthesize cellular biomass. It is important to note that acetate by itself is incapable of sustaining the growth of this microorganism; indeed, no more than 60 different compounds that have been experimented with to date have exhibited growth-promoting properties for O. formigenes [26]. Acetate is crucial for cell synthesis and is essential for some, if not all, strains.
The breakdown of oxalate leads to medium alkalization, where formate is generated in approximately equal amounts to the oxalate metabolized. Isolates of these strains have been recovered from diverse sources such as cattle and sheep rumens and cecal and fecal samples from various animals, including humans, guinea pigs, swine, and domestic and wild rats. Moreover, they have been found in freshwater lakes and marine sediments. These bacteria likely inhabit numerous other anaerobic environments [27]. O. formigenes can colonize the gastrointestinal tract and diminish the oxalate concentration in the urine after an oxalate challenge, thereby mitigating the probability of developing calcium oxalate kidney stones (Table 2). Current research indicates that anaerobic bacteria present in the colon, such as O. formigenes, can metabolize harmful substances within the intestinal environment. This highlights the pivotal role of O. formigenes in modulating oxalate levels and potentially preventing the formation of calcium oxalate kidney stones through its unique biochemical mechanisms [28].

Table 2.
An overview of earlier research on the application of Oxalobacter formigenes in urolithiasis prophylaxis.
2.2. Lactic Acid Bacteria and Bifidobacteria
Lactic acid bacteria, which are classified as Gram-positive bacteria, are characterized by their inability to produce spores except for the genus Sporolactobacillus, thus setting them apart from other bacterial groups. While some LAB thrive in aerobic environments, others prefer anaerobic conditions; the majority of species exhibit the ability to adapt to both aerobic and anaerobic conditions, classifying them as facultative anaerobes. Through the process of fermentation, these bacteria effectively convert carbohydrates into lactic acid, which is the major fermentation product. This metabolic pathway not only yields lactic acid but also results in the formation of various organic acids and other metabolites that significantly influence the overall sensory profile of the end product, contributing to its flavor, texture, and aroma, thereby enhancing its desirability [38,39].
The groundwork for the systematic classification of LAB was established in 1919, marking a pivotal moment in the understanding of these microorganisms. It is a type of classification framework revolving around specific criteria, including the assessment of glucose fermentation capabilities, cell morphology, the capacity to utilize sugars as carbon sources, and growth patterns under diverse temperature conditions. Initially, the classification encompassed four distinct genera within this bacterial group: Lactobacillus, Pediococcus, Leuconostoc, and Streptococcus. However, with the rapid advancements in genetic methodologies, numerous additional genera have been incorporated into this taxonomic group, broadening the spectrum of lactic acid bacteria. Among the newly identified genera are Aerococcus, Alloiococcus, Carnobacterium, Dolosigranulum, Enterococcus, Lactosphaera, Melissococcus, Oenococcus, Sporolactobacillus, Tetragenococcus, Vagococcus, and Weissella, reflecting the expanding diversity and complexity within this bacterial family [40,41].
In 2020, the taxonomic classification of the genus Lactobacillus underwent a significant reorganization due to the remarkable progress in genetic engineering and genetic diagnosis technologies. This reorganization divided the genus Lactobacillus into 25 genera, each representing a distinct cluster of bacterial species. These newly classified genera include Acetilactobacillus, Agrilactobacillus, Amylolactobacillus, Apilactobacillus, Bombilactobacillus, Companilactobacillus, Dellaglioa, Fructilactobacillus, Furfurilactobacillus, Holzapfelia, Lacticaseibacillus, Latilactobacillus, Lactiplantibacillus, Lapidilactobacillus, Lentilactobacillus, Levilactobacillus, Ligilactobacillus, Limosilactobacillus, Liquorilactobacillus, Loigolactobacilus, Paralactobacillus, Paucilactobacillus, Schleiferilactobacillus, and Secundilactobacillus, as detailed by Zheng et al. [42]. This taxonomic revision reflects the increasing precision and depth of our understanding regarding bacterial diversity and evolution, highlighting the intricate interplay between technological advancements and biological classification systems in the field of microbiology.
Bifidobacterium was first identified in 1924. Gram-positive bacteria that do not retain acid-fast stains, do not form spores, and are non-motile. Typically, cells exhibit irregular staining patterns when subjected to methylene blue. These organisms thrive in anaerobic environments; however, certain species show a tolerance towards oxygen only in the presence of carbon dioxide. Recently identified species like B. psychraerophilum, B. scardovii, and B. tsurumiense have demonstrated the ability to flourish under aerobic conditions. The most favorable temperature range for growth is reported to be between 37 and 41 °C, except B. mongoliense, which shows optimal growth at 30 °C. The lowest temperature suitable for growth ranges from 25 to 28 °C, except for B. mongoliense and B. psychraerophilum, which can thrive at 15 °C and 8 °C, respectively. On the other hand, the maximum temperature conducive to growth spans from 43 to 45 °C, except for B. thermacidophilum, which can thrive up to 49.5 °C. Notably, the ability to grow at 45 °C is a distinguishing factor between strains isolated from animals and those from humans, as most animal strains can survive at this temperature while human strains cannot. The ideal pH for initial growth falls within the range of 6.5 to 7.0; these bacteria do not increase at pH levels ranging from 4.5 to 5.0, except B. thermacidophilum, which can still grow at pH 4.5 or pH 8.0 to 8.5 [43].
Several recent research studies have provided substantial evidence regarding the capacity of lactic acid bacteria, specifically the genera Bifidobacterium and Lactobacillus, to degrade oxalates and transform them into formate and CO2 as highlighted in the works of Murru et al. [44] and Chamberlain et al. [45]. In the study conducted by Sadaf et al. [46], it was mentioned that they isolated four distinct types of Lactobacillus bacteria, which are Lactobacillus acidophilus, Lactiplantibacillus plantarum, Lacticaseibacillus casei, and Lacticaseibacillus zeae, and thoroughly investigated their oxalate breakdown capacity. The research findings revealed that Lactobacillus acidophilus exhibited a superior efficiency in oxalate degradation compared to the other bacterial isolates. Additionally, the research by Karamad et al. [47] reported that Lactobacillus acidophilus bacteria demonstrated the ability to break down oxalates present in artificially prepared gastric fluid by an impressive 48% efficiency rate. These studies collectively shed light on the promising potential of lactic acid bacteria, particularly Lactobacillus strains, in effectively metabolizing oxalates into more benign byproducts, which could have significant implications in various fields such as nutrition, medicine, and biotechnology (Table 3). Furthermore, the research outcomes underscore the importance of exploring the diverse capabilities of probiotic bacteria in promoting gastrointestinal health and potentially mitigating the risks associated with oxalate-related disorders. The findings presented in these studies contribute valuable insights to the existing body of knowledge on the metabolic activities of lactic acid bacteria and their role in oxalate degradation mechanisms. Overall, the research outcomes suggest a promising avenue for future investigations focusing on harnessing the oxalate-degrading potential of lactic acid bacteria for therapeutic or dietary interventions aimed at improving human health and well-being.

Table 3.
A summary of outcomes obtained from previous studies on the application of lactic acid bacteria and bifidobacteria in kidney stone disease.
2.3. Other Gut Bacteria
Many bacterial species inhabit the small intestine of the host (human or animal). They are natural flora and have the ability to decompose oxalate and reduce the concentration of oxalate in the blood and urine. Anaerobic conditions were used to isolate an oxalate-erode degrading Enterococcus faecalis bacterium from human feces. For the bacteria to continue reducing oxalate, they needed a low nutritional environment and frequent sub-culturing. SDS-PAGE analysis revealed that E. faecalis synthesized three proteins (65, 48, and 40 kDa) that were not formed by the non-oxalate-degrading E. faecalis [56]. Another investigation discovered a single Providencia rettgeri bacterial isolate that displayed two proteins (65 kDa and 48 kDa) on SDS-polyacrylamide gel electrophoresis that were absent in P. rettgeri that did not break down oxalate. In Western blotting, antibodies interacted with the P. rettgeri strain’s 65 and 48 kDa proteins. An Oxalobacter formyl-coenzyme from O. formigenes under highly stringent circumstances, a transferase gene probe interacted with P. rettgeri chromosomal DNA on Southern blotting. In contrast, an Oxalobacter formigenes oxalyl-coenzyme with identical circumstances did not cause a decarboxylase gene probe to respond. Oxalate decarboxylase (OXC), a manganese-dependent enzyme, is involved in the catalysis of oxalate oxidation to carbon dioxide with the formation of hydrogen peroxide. The ability of this OXC enzyme to purify from the Pseudomonas strain confirmed that it can be used for the diagnosis of oxalate-related disorders through microtiter plate analysis [57]. A previous study found that Pseudomonas sp. OXDC12 could produce and purify OXC up to 45.3 times, with an overall yield of 7%. The pure OXC may be a hexameric enzyme because it only showed one band, measuring around 40 kDa on SDS-PAGE and 240 kDa on Native PAGE [58].
2.4. Mechanisms of Bacterial Oxalate Degradation
Oxalate is a compound produced by many edible plants. As a terminal metabolite in the liver of mammals, it acts as a toxin that has detrimental effects on human health [59]. Endogenous oxalate-degrading enzymes are absent in humans and other mammals, leading them to depend on the gut microbiota for this crucial metabolic process. Several types of bacteria within the gastrointestinal tract have been recognized for their ability to break down oxalate through various in vitro as well as in vivo studies, with notable examples including Oxalobacter formigenes, various species of Lactobacillus, different Bifidobacterium strains, and members of the Enterobacteriaceae family [60].
Oxalobacter formigenes are particularly remarkable due to their utilization of oxalate as the exclusive source of energy, resulting in the stimulation of host oxalate secretion into the colonic lumen. This bacterium, in conjunction with other microorganisms, harbors crucial oxalate-degrading enzymes (ODEs) such as oxalyl-CoA decarboxylase (OXC) and formyl-CoA transferase (FRC), which play a vital role in oxalate metabolism. These pivotal enzymes are governed by genes that are commonly present in the genetic makeup of oxalate-degrading bacteria. One of the interesting facts is that taxa encoding FRC and OXC are exclusively of bacteria only, whereas those encoding OXDD can be of both fungal and bacterial origin. Through experimental investigations, it has been demonstrated that the colonization of mice with O. formigenes leads to a substantial reduction in urinary oxalate levels, thereby confirming its significance in the degradation of oxalate. Analogously, various other oxalate-degrading bacteria have exhibited the ability to diminish urinary oxalate levels in rodent models of hyperoxaluria, underscoring their potential effectiveness in regulating oxalate concentrations within living organisms [60,61].
In the microbiota of a healthy human, the oxc gene is commonly transcribed by various species, including E. coli, O. formigenes, and members of Muribaculaceae, Bifidobacterium, and Lactobacillus, with E. coli and O. formigenes being the most predominant among them. However, under conditions of diseases such as Inflammatory Bowel Disease (IBD), the transcription of these genes may undergo a significant reduction. It has been observed that despite the presence of frc and oxc genes in patients with IBD, the actual expression levels of these genes are considerably lower in comparison to those in healthy individuals, which is associated with the depletion of O. formigenes, a key oxalate-degrading bacterium. The intricate relationship between the transcription of these genes and the microbial composition in the gut highlights the potential implications of dysbiosis in disease pathogenesis, particularly in the context of IBD [32,62].
The differential transcription observed signifies a significant shift in the functional dynamics of the microbiota, indicating that the abundance of genes may not always align with actual functional activity. Particularly, the transcriptional activity of O. formigenes emerges as a central player in the overall oxalate degradation pathway (ODP), underscoring its crucial role as an oxalate autotroph and the prevalence of FRC and OXC proteins across various growth phases (Figure 2). These findings underscore the paramount importance of relying on transcriptional evidence rather than mere gene abundance when assessing microbiome functions associated with oxalate breakdown. It is crucial to acknowledge that environmental variables, such as dietary constituents and pH levels, have a significant impact on the effectiveness of oxalate-degrading bacteria. The expression levels of oxc and frc genes, which directly influence oxalate degradation efficiency, are notably influenced by these external factors, highlighting the intricate nature of oxalate metabolism within diverse environmental frameworks [59,60].

Figure 2.
Decomposition of oxalate molecules through biological processes by different species of bacteria within the host.
3. Health Implications
The gut microbiome, which consists of many micro-organisms like bacteria, viruses, fungi, and archaea, is fundamentally important in maintaining human health through its influence on processes such as digestion, absorption of nutrients, and regulation of the immune system. The disruption or dysbiosis of the gut microbiome, characterized by an imbalance in its composition, has been associated with the development and progression of various medical conditions, including inflammatory bowel disease (IBD), diabetes, and kidney stone disease. These findings underscore the intricate relationship between the gut microbiome and human health, highlighting the potential therapeutic implications of targeting the microbiome to manage and prevent a wide range of diseases.
3.1. Inflammatory Bowel Disease (IBD)
The gut microbiome composition in patients suffering from inflammatory bowel disease (IBD) shows a significant deviation from that of individuals in good health, characterized by a decrease in bacterial diversity and a decline in beneficial species such as butyrate-producing bacteria while being marked by an increase in opportunistic species referred to as pathobionts [63]. Metabolites generated by the gut microbiota, including short-chain fatty acids, tryptophan metabolites, and bile acids, have been linked to the onset and advancement of IBD, emphasizing the critical role of these compounds in the disease process. The alterations in the gut microbiome and its metabolites highlight the intricate interplay between the microbial community residing in the gut and the pathophysiology of IBD, shedding light on potential therapeutic targets for managing this complex disorder. Alterations in the gut microbiota have been correlated with two prevalent gastrointestinal disorders, namely inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS). In this study, researchers conducted a case-control examination utilizing shotgun metagenomic sequencing of fecal samples from 1792 individuals diagnosed with IBD and IBS in comparison to control subjects from the general population [64]. The findings from the examination of the gut microbiome composition in Saudi individuals diagnosed with inflammatory bowel disease (IBD) revealed the presence of three detrimental bacterial biomarkers: the Paraprevotellaceae, the Muribaculaceae families belonging to the Bacteroidetes phylum, and the Leuconostocaceae family from the Firmicutes phylum, which exhibited a higher relative prevalence in healthy subjects as opposed to those suffering from IBD. Moreover, it was observed that specific microbiota signatures at particular genera and species levels, such as Prevotella copri, Bifidobacterium adolescentis, Ruminococcus callidus, Coprococcus sp., Ruminococcus gnavus, Dorea formicigenerans, Leuconostoc, Dialister, Catenibacterium, Eubacterium biforme, and Lactobacillus mucosae, were virtually absent in nearly all IBD patients. In contrast, Veillonella dispar was completely absent among all healthy individuals [65].
3.2. Kidney Stone Disease
The research carried out by Pisani et al. [63] has demonstrated a connection between the gut microbiome in patients with inflammatory bowel disease (IBD) and increased levels of fecal oxalate, especially in individuals diagnosed with Crohn’s disease (CD) and ulcerative colitis (UC). This association has the potential to raise the likelihood of developing kidney stones. Notably, the increased fecal oxalate concentrations in patients with CD are primarily observed in those with ileocolonic inflammation, indicating that the specific location of IBD within the gastrointestinal tract plays a critical role in assessing the associated risks [63,64].
Kidney stone disease, which is a condition that impacts an estimated 10 to 15% of the populace on a global scale, exhibits a significant correlation with the phenomenon of dysbiosis within the gut microbiota [66]. The predominant classification of kidney stones, specifically calcium oxalate stones, constitutes a substantial 76% of all documented cases of this medical condition [67]. The state of dysbiosis observed in patients afflicted with kidney stones is distinguished by a particular microbial profile within the gut, which may have the potential to significantly affect both the formation of these stones and their subsequent recurrence [68]. The persistence of kidney stones over an extended period can lead to a multitude of severe health complications, which may include but are not limited to urinary tract obstruction, the development of infections, and the potential for irreversible damage to renal structures. Innovative and individualized therapeutic approaches, including but not limited to microbial supplementation, the use of probiotics, the implementation of synbiotics, and dietary modifications informed by the unique characteristics of the gut microbiome, have demonstrated considerable promise in the realms of both the prevention of stone formation as well as the reduction of recurrence rates [69].
4. Future Perspectives and Directions
The significance of the gut microbiome in the manifestation and progression of various pathological conditions underscores the vast potential for the development and implementation of personalized medical interventions that are tailored to individual patient profiles. A comprehensive understanding of the complex and multifaceted interactions that exist between dietary practices, the diverse composition of the microbiota, and the inflammatory responses that occur within the body could ultimately facilitate the formulation of innovative strategies aimed at both the prevention and the treatment of diseases such as Inflammatory Bowel Disease (IBD) and Kidney stones (KSs). The identification and validation of promising biomarkers found within the blood and fecal specimens may prove instrumental in enhancing the accuracy of diagnoses as well as in forecasting the trajectory of disease progression and the efficacy of therapeutic interventions, thereby paving the way for the advent of more customized and effective treatment regimens that are specifically designed to meet the unique needs of each patient.
The persistent quest for innovative microbial strains exhibiting superior fermentative capabilities and probiotic characteristics is anticipated to play a crucial role in forthcoming developments. Scholars advocate for the employment of bacteriocin-producing strains as initial inoculate to effectively combat foodborne pathogens. Nonetheless, the possible adverse effects associated with bacteriocins, particularly their toxicity, require thorough examination. Although evidence suggests that bacteriocins are typically non-immunogenic and display low cytotoxicity, there have been instances documenting considerable cytotoxic repercussions. These effects may fluctuate according to concentration and the type of target cells, thus warranting additional research.
The prospective trajectory of microbiome research is poised to advance significantly through the meticulous amalgamation of metabolomic data with an array of other critical biomarkers, which encompass the proteome as well as circulating antibodies, all of which should be examined across a wide spectrum of diverse and multiethnic cohorts that reflect the complexity of human health. Such a comprehensive and integrative methodological framework has the potential not only to substantially enhance the precision and reliability of diagnostic tests but also to yield profound insights into the intricate nature of disease heterogeneity and its progression over time, thereby informing targeted therapeutic strategies. Forthcoming research endeavors must concentrate on mechanistic inquiries to substantiate the significance of novel metabolites or pathways linked to ailments such as inflammatory bowel disease (IBD) and to prioritize biomolecules for the formulation of innovative therapeutic strategies. Such inquiries will be essential in differentiating metabolites that instigate diseases from the by-products resulting from inflammatory processes. Previous prospective cohort studies have already delineated serum metabolomic markers, including sarcosine, carnitine, propionyl-l-carnitine, and sorbitol, which correlate with clinical relapses in individuals diagnosed with IBD.
Ultimately, forthcoming investigations seek to reassess various methodologies employed in research centered on advantageous microorganisms and bacteriocins as preservative agents. This entails the identification of novel bacterial strains sourced from fermented food products, the examination of bacteriocins produced by these microorganisms, and the evaluation of their viability as probiotics. Scholars advocate for empirical studies to incorporate bacteriocins into particular food items and to analyze their effectiveness in mitigating foodborne pathogens. This domain presents considerable promise for the creation of pioneering strategies aimed at managing oxalate degradation while simultaneously improving food safety and overall quality.
5. Conclusions
It is possible to conclude that the intricate ecologies of various bacterial strains serve a significantly crucial function in enhancing their overall efficiency in the solubilization processes of mineral compounds, which are essential for numerous biological and geological functions. Various strains belonging to the genera Oxalobacter, Lactobacillus, and Bifidobacterium have demonstrated considerable potential as effective probiotic supplements, which can play a pivotal role in meticulously regulating and monitoring the levels of rock-forming salts that accumulate within the kidneys, thereby contributing to the maintenance of renal health.
Author Contributions
Investigation, A.K.N.: designing and planning the experiments; A.K.N. supervision, S.H.A.-K. and A.K.N.: writing, reviewing, and editing, S.H.A.-K. and A.K.N.: reviewing and editing an original draft for the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wigner, P.; Bijak, M.; Saluk-Bijak, J. Probiotics in the Prevention of the Calcium Oxalate Urolithiasis. Cells 2022, 11, 284. [Google Scholar] [CrossRef] [PubMed]
- Alelign, T.; Petros, B. Kidney Stone Disease: An Update on Current Concepts. Adv. Urol. 2018, 2018, 3068365. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.E. Oxalic Acid and the Hyperoxaluric Syndromes. Kidney Int. 1978, 13, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Brzica, H.; Breljak, D.; Burckhardt, B.C.; Burckhardt, G.; Sabolić, I. Oxalate: From the Environment to Kidney Stones. Arh. Hig. Rada Toksikol. 2013, 64, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.K.; Nguyen, D.H.; Nguyen, H.V. Effects of processing on oxalate contents in plant foods: A review. J. Food Compos. Anal. 2022, 112, 104685. [Google Scholar] [CrossRef]
- Holmes, R.P.; Knight, J.; Assimos, D.G. Lowering Urinary Oxalate Excretion to Decrease Calcium Oxalate Stone Disease. Urolithiasis 2016, 44, 27–32. [Google Scholar] [CrossRef]
- Kumar, P.; Patel, M.; Oster, R.A.; Yarlagadda, V.; Ambrosetti, A.; Assimos, D.G.; Mitchell, T. Dietary Oxalate Loading Impacts Monocyte Metabolism and Inflammatory Signaling in Humans. Front. Immunol. 2021, 12, 617508. [Google Scholar] [CrossRef]
- Stepanova, N. Oxalate Homeostasis in Non-Stone-Forming Chronic Kidney Disease: A Review of Key Findings and Perspectives. Biomedicines 2023, 11, 1654. [Google Scholar] [CrossRef]
- Cowan, D.A.; Babenko, D.; Bird, R.; Botha, A.; Breecker, D.O.; Clarke, C.E.; Francis, M.L.; Gallagher, T.; Lebre, P.H.; Nel, T.; et al. al. Oxalate and oxalotrophy: An environmental perspective. Sustain. Microbiol. 2024, 1, qvad004. [Google Scholar] [CrossRef]
- Niamah, A.K.; Sahi, A.A.; Al-Sharifi, A.S.N. Effect of Feeding Soy Milk Fermented by Probiotic Bacteria on Some Blood Criteria and Weight of Experimental Animals. Probiotics Antimicrob. Proteins 2017, 9, 284–291. [Google Scholar] [CrossRef]
- Hussein, K.A.; Niamah, A.K.; Majeed, K.R. Strategies and Trends for Application Exopolysaccharides of Lactic Acid Bacteria in the Food and Biomedical. IOP Conf. Ser. Earth Environ. Sci. 2024, 1371, 062017. [Google Scholar] [CrossRef]
- Noonin, C.; Putpim, A.; Thongboonkerd, V. The direct inhibitory effects of Lactobacillus acidophilus, a commensal urinary bacterium, on calcium oxalate stone development. Microbiome 2024, 12, 175. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, S.; Sasikumar, P.; Anbazhagan, K.; Sasikumar, S.; Kavitha, M.; Selvi, M.S.; Selvam, G.S. Screening of Indigenous Oxalate Degrading Lactic Acid Bacteria from Human Faeces and South Indian Fermented Foods: Assessment of Probiotic Potential. Sci. World J. 2014, 2014, 648059. [Google Scholar] [CrossRef] [PubMed]
- Attalla, K.; De, S.; Monga, M. Oxalate Content of Food: A Tangled Web. Urology 2014, 84, 555–560. [Google Scholar] [CrossRef]
- Siener, R.; Seidler, A.; Hönow, R. Oxalate-Rich Foods. Food Sci. Technol. (Braz.) 2021, 41, 169–173. [Google Scholar] [CrossRef]
- Shi, L.; Arntfield, S.D.; Nickerson, M. Changes in Levels of Phytic Acid, Lectins and Oxalates during Soaking and Cooking of Canadian Pulses. Food Res. Int. 2018, 107, 660–668. [Google Scholar] [CrossRef]
- Siener, R.; Seidler, A.; Voss, S.; Hesse, A. The Oxalate Content of Fruit and Vegetable Juices, Nectars and Drinks. J. Food Compos. Anal. 2016, 45, 108–112. [Google Scholar] [CrossRef]
- Sadaf, H.; Raza, S.I.; Hassan, S.W. Role of Gut Microbiota against Calcium Oxalate. Microb. Pathog. 2017, 109, 287–291. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; de Vos, W.M. The First 1000 Cultured Species of the Human Gastrointestinal Microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Kaur, I.P.; Chopra, K.; Saini, A. Probiotics: Potential Pharmaceutical Applications. Eur. J. Pharm. Sci. 2002, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Rodrigues, C.F.; Stojanović-Radić, Z.; Dimitrijević, M.; Aleksić, A.; Neffe-Skocińska, K.; Zielińska, D.; Kołożyn-Krajewska, D.; Salehi, B.; Prabu, S.M.; et al. Probiotics: Versatile Bioactive Components in Promoting Human Health. Medicina 2020, 56, 433. [Google Scholar] [CrossRef] [PubMed]
- Al-Sahlany, S.T.G.; Niamah, A.K. Bacterial Viability, Antioxidant Stability, Antimutagenicity and Sensory Properties of Onion Types Fermentation by Using Probiotic Starter During Storage. Nutr. Food Sci. 2022, 52, 901–916. [Google Scholar] [CrossRef]
- Sotelo, A.; González-Osnaya, L.; Snchez-Chinchillas, A.; Trejo, A. Role of Oxate, Phytate, Tannins and Cooking on Iron Bioavailability from Foods Commonly Consumed in Mexico. Int. J. Food Sci. Nutr. 2010, 61, 29–39. [Google Scholar] [CrossRef]
- Sonke, A.; Trembath-Reichert, E. Expanding the Taxonomic and Environmental Extent of an Underexplored Carbon Metabolism—Oxalotrophy. Front. Microbiol. 2023, 14, 1161937. [Google Scholar] [CrossRef]
- Daniel, S.L.; Moradi, L.; Paiste, H.; Wood, K.D.; Assimos, D.G.; Holmes, R.P.; Nazzal, L.; Hatch, M.; Knight, J. Forty Years of Oxalobacter formigenes, a Gutsy Oxalate Degrading Specialist. Appl. Environ. Microbiol. 2021, 87, e00544-21. [Google Scholar] [CrossRef]
- Garrity, G.M.; Brenner, D.J.; Krieg, N.R.; Staley, J.R. Bergey’s Manual® of Systematic Bacteriology. The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria; Springer: New York, NY, USA, 2005; Volume 2. [Google Scholar]
- Stewart, C.S.; Duncan, S.H.; Cave, D.R. Oxalobacter formigenes and Its Role in Oxalate Metabolism in the Human Gut. FEMS Microbiol. Lett. 2004, 230, 1–7. [Google Scholar] [CrossRef]
- Karamad, D.; Khosravi-Darani, K.; Hosseini, H.; Tavasoli, S.; Miller, A.W. Evaluation of Oxalobacter formigenes DSM 4420 Biodegradation Activity for High Oxalate Media Content: An In Vitro Model. Biocatal. Agric. Biotechnol. 2019, 22, 101378. [Google Scholar] [CrossRef]
- Mogna, L.; Pane, M.; Nicola, S.; Raiteri, E. Screening of Different Probiotic Strains for Their In Vitro Ability to Metabolise Oxalates Any Prospective Use in Humans? J. Clin. Gastroenterol. 2014, 48, S91–S95. [Google Scholar] [CrossRef]
- Giardina, S.; Scilironi, C.; Michelotti, A.; Samuele, A.; Borella, F.; Daglia, M.; Marzatico, F. In Vitro Anti-Inflammatory Activity of Selected Oxalate-Degrading Probiotic Bacteria: Potential Applications in the Prevention and Treatment of Hyperoxaluria. J. Food Sci. 2014, 79, M384–M390. [Google Scholar] [CrossRef]
- Sidhu, H.; Allison, M.J.; Chow, J.M.; Clark, A.; Peck, A.B. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxlobacter formigenes. J. Urol. 2001, 166, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Hatch, M.; Cornelius, J.; Allison, M.; Sidhu, H.; Peck, A.; Freel, R.W. Oxalobacter sp. Reduces Urinary Oxalate Excretion by Promoting Enteric Oxalate Secretion. Kidney Int. 2006, 69, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, D.W.; Kelly, J.P.; Curhan, G.C.; Anderson, T.E.; Dretler, S.P.; Preminger, G.M.; Cave, D.R. Oxalobacter formigenes May Reduce the Risk of Calcium Oxalate Kidney Stones. J. Am. Soc. Nephrol. 2008, 19, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Siener, R.; Bangen, U.; Sidhu, H.; Hönow, R.; Von Unruh, G.; Hesse, A. The Role of Oxalobacter formigenes Colonization in Calcium Oxalate Stone Disease. Kidney Int. 2013, 83, 1144–1149. [Google Scholar] [CrossRef]
- Jiang, J.; Knight, J.; Easter, L.H.; Neiberg, R.; Holmes, R.P.; Assimos, D.G. Impact of Dietary Calcium and Oxalate, and Oxalobacter formigenes Colonization on Urinary Oxalate Excretion. J. Urol. 2011, 186, 135–139. [Google Scholar] [CrossRef]
- Arvans, D.; Jung, Y.C.; Antonopoulos, D.; Koval, J.; Granja, I.; Bashir, M.; Karrar, E.; Roy-Chowdhury, J.; Musch, M.; Asplin, J.; et al. Oxalobacter formigenes-Derived Bioactive Factors Stimulate Oxalate Transport by Intestinal Epithelial Cells. J. Am. Soc. Nephrol. 2017, 28, 876–887. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; da Silva, R.C.; Ibrahim, S.A. Lactic Acid Bacteria: Food Safety and Human Health Applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Al-Sahlany, S.T.G.; Khassaf, W.H.; Niamah, A.K.; Abd Al-Manhel, A.J. Date Juice Addition to Bio-Yogurt: The Effects on Physicochemical and Microbiological Properties during Storage, as well as Blood Parameters In Vivo. J. Saudi Soc. Agric. Sci. 2023, 22, 71–77. [Google Scholar] [CrossRef]
- Parte, A.; Krieg, N.R.; Ludwig, W.; Whitman, W.B.; Hedlund, B.P.; Paster, B.J.; Ward, N.L.; Ludwig, W.; Brown, D. (Eds.) Bergey’s Manual of Systematic Bacteriology: Volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes; Springer Science & Business Media: New York, NY, USA, 2011; Volume 4. [Google Scholar]
- Madigan, M.T.; Imhoff, J.F. Phylum BXIII. Firmicutes. In Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2001; pp. 625–637. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. EvolMicrobiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H.-J.; Trujillo, M.E.; Suzuki, K.-i.; Ludwig, W.; Whitman, W.B. (Eds.) Bergey’s Manual of Systematic Bacteriology. Volume 5: The Actinobacteria; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Murru, N.; Blaiotta, G.; Peruzy, M.F.; Santonicola, S.; Mercogliano, R.; Aponte, M. Screening of Oxalate Degrading Lactic Acid Bacteria of Food Origin. Ital. J. Food Saf. 2017, 6, 6345. [Google Scholar] [CrossRef]
- Chamberlain, C.A.; Hatch, M.; Garrett, T.J. Metabolomic Profiling of Oxalate-Degrading Probiotic Lactobacillus acidophilus and Lactobacillus gasseri. PLoS ONE 2019, 14, e0222393. [Google Scholar] [CrossRef] [PubMed]
- Sadaf, H.; Waqas Hassan, S.; Nawaz, F.; Irfan Raza, S.; Jillani, G. Biodegradation of Calcium Phosphate and Calcium Oxalate by Lactobacillus Strains. Univ. Wah J. Sci. Technol. (UWJST) 2019, 3, 29–34. [Google Scholar]
- Karamad, D.; Khosravi-Darani, K.; Hosseini, H.; Tavasoli, S.; Miller, A.W. Assessment of the Process Variables for Degradation of Oxalate by Lactobacillus acidophilus ATCC 4356 Using Simulated Rumen Fluid Media and Tea. Appl. Food Biotechnol. 2020, 7, 195–204. [Google Scholar] [CrossRef]
- Anbazhagan, K.; Sasikumar, P.; Gomathi, S.; Priya, H.P.; Selvam, G.S. In Vitro Degradation of Oxalate by Recombinant Lactobacillus plantarum Expressing Heterologous Oxalate Decarboxylase. J. Appl. Microbiol. 2013, 115, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Kargar, M.; Afkari, R.; Sadegh Ghorbani-Dalinip, P. Oxalate-Degrading Capacities of Gastrointestinal Lactic Acid Bacteria and Urinary Tract Stone Formation. Zahedan J. Res. Med. Sci. 2013, 15, e92834. [Google Scholar]
- Mehra, Y.; Viswanathan, P. High-Quality Whole-Genome Sequence Analysis of Lactobacillus paragasseri UBLG-36 Reveals Oxalate-Degrading Potential of the Strain. PLoS ONE 2021, 16, e0260116. [Google Scholar] [CrossRef]
- Al Othaim, A. Probiotic Lactobacillus Strains Isolated from Date Waste and Wastewater for the Kidney Stone, Intestinal Oxalate-Degradation and Antioxidant Activity. J. King Saud. Univ. Sci. 2023, 35, 102888. [Google Scholar] [CrossRef]
- Soliman, N.R.; Elbakry, H.F.H.; Effat, B.A.M.; Mehanna, N.S.; Tawfik, N.F.; Ibrahim, M.K. The Oxalate-Lowering Effect of Functional Stirred Yogurt in a Rat Model. Biocatal. Agric. Biotechnol. 2024, 58, 103196. [Google Scholar] [CrossRef]
- Turroni, S.; Bendazzoli, C.; Dipalo, S.C.F.; Candela, M.; Vitali, B.; Gotti, R.; Brigidi, P. Oxalate-Degrading Activity in Bifidobacterium animalis Subsp. lactis: Impact of Acidic Conditions on the Transcriptional Levels of the Oxalyl Coenzyme A (CoA) Decarboxylase and Formyl-CoA Transferase Oxalate-Degrading Activity in. Appl. Environ. Microbiol. 2010, 76, 5609–5620. [Google Scholar] [CrossRef]
- Klimesova, K.; Whittamore, J.M.; Hatch, M. Bifidobacterium animalis Subsp. lactis Decreases Urinary Oxalate Excretion in a Mouse Model of Primary Hyperoxaluria. Urolithiasis 2015, 43, 107–117. [Google Scholar] [CrossRef]
- Ferraz, R.R.N.; Marques, N.C.; Froeder, L.; Menon, V.B.; Siliano, P.R.; Baxmann, A.C.; Heilberg, I.P. Effects of Lactobacillus casei and Bifidobacterium breve on Urinary Oxalate Excretion in Nephrolithiasis Patients. Urol. Res. 2009, 37, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Hokama, S.; Honma, Y.; Toma, C.; Ogawa, Y. Oxalate-Degrading Enterococcus faecalis. Microbiol. Immunol. 2000, 44, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Paikrao, H.; Sheikh, R.; Dhakate, L. Isolation, Screening, and Partial Characterization of Oxalate Oxidase Enzyme from Pseudomonas Strain Under Induced Oxalate Stress Condition. J. Microbiol. Biotechnol. Food Sci. 2022, 11, e4174. [Google Scholar] [CrossRef]
- Gupta, S.; Kanwar, S.S. Molecular Characterization and in Silico Analysis of Oxalate Decarboxylase of Pseudomonas sp. OXDC12. J. Biomol. Struct. Dyn. 2023, 41, 1495–1509. [Google Scholar] [CrossRef] [PubMed]
- Karamad, D.; Khosravi-Darani, K.; Khaneghah, A.M.; Miller, A.W. Probiotic Oxalate-Degrading Bacteria: New Insight of Environmental Variables and Expression of the Oxc and Frc Genes on Oxalate Degradation Activity. Foods 2022, 11, 2876. [Google Scholar] [CrossRef]
- Youssef, H.A.I.A.E. Detection of Oxalyl-CoA Decarboxylase (Oxc) and Formyl-CoA Transferase (Frc) Genes in Novel Probiotic Isolates Capable of Oxalate Degradation in Vitro. Folia Microbiol. (Praha) 2024, 69, 423–432. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Bruno-Bárcena, J.M.; Hassan, H.M.; Klaenhammer, T.R. Transcriptional and Functional Analysis of Oxalyl-Coenzyme A (CoA) Decarboxylase and Formyl-CoA Transferase Genes from Lactobacillus acidophilus. Appl. Environ. Microbiol. 2006, 72, 1891–1899. [Google Scholar] [CrossRef]
- Sidhu, H.; Ogden, S.D.; Lung, H.-Y.; Luttge, B.G.; Baetz, A.L.; Peck, A.B. DNA Sequencing and Expression of the Formyl Coenzyme A Transferase Gene, Frc, from Oxalobacter formigenes. J. Bacteriol. 1997, 179, 3378–3381. [Google Scholar] [CrossRef][Green Version]
- Pisani, A.; Rausch, P.; Bang, C.; Ellul, S.; Tabone, T.; Marantidis Cordina, C.; Zahra, G.; Franke, A.; Ellul, P. Dysbiosis in the gut microbiota in patients with inflammatory bowel disease during remission. Microbiol. Spectr. 2022, 10, e00616-22. [Google Scholar] [CrossRef]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef]
- Al-Amrah, H.; Saadah, O.I.; Mosli, M.; Annese, V.; Al-Hindi, R.; Edris, S.; Alshehri, D.; Alatawi, H.; Alatawy, M.; Bahieldin, A. Composition of the gut microbiota in patients with inflammatory bowel disease in Saudi Arabia: A pilot study. Saudi J. Gastroenterol. 2023, 29, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, I.; Mamoulakis, C.; Miyazawa, K.; Rodgers, A.; Talati, J.; Lotan, Y. Epidemiology of stone disease across the world. World J. Urol. 2017, 35, 1301–1320. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.R.; Pearle, M.S.; Robertson, W.G.; Gambaro, G.; Canales, B.K.; Doizi, S.; Traxer, O.; Tiselius, H.-G. Kidney stones. Nat. Rev. Dis. Primers 2016, 2, 16008. [Google Scholar] [CrossRef] [PubMed]
- Lemberger, U.; Pjevac, P.; Hausmann, B.; Berry, D.; Moser, D.; Jahrreis, V.; Özsoy, M.; Shariat, S.F.; Veser, J. The microbiome of kidney stones and urine of patients with nephrolithiasis. Urolithiasis 2023, 51, 27. [Google Scholar] [CrossRef]
- Shastri, S.; Patel, J.; Sambandam, K.K.; Lederer, E.D. Kidney stone pathophysiology, evaluation and management: Core curriculum. Am. J. Kidney Dis. 2023, 82, 617–634. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).