Abstract
Foodborne bacteria interconnect food and human health. Despite significant progress in food safety regulation, bacterial contamination is still a serious public health concern and the reason for significant commercial losses. The screening of the microbiome in meals is one of the main aspects of food production safety influencing the health of the end-consumers. Our research provides an overview of proteomics findings in the field of food safety made over the last decade. It was believed that proteomics offered an accurate snapshot of the complex networks of the major biological machines called proteins. The proteomic methods for the detection of pathogens were armed with bioinformatics algorithms, allowing us to map the data onto the genome and transcriptome. The mechanisms of the interaction between bacteria and their environment were elucidated with unprecedented sensitivity, specificity, and depth. Using our web-based tool ScanBious for automated publication analysis, we analyzed over 48,000 scientific articles on antibiotic and disinfectant resistance and highlighted the benefits of proteomics for the food safety field. The most promising approach to studying safety in food production is the combination of classical genomic and metagenomic approaches and the advantages provided by proteomic methods with the use of panoramic and targeted mass spectrometry.
Keywords:
meat packing plant; meat processing; bacteria; food safety; Salmonella; Listeria; Pseudomonas; antibiotics; resistance 1. Introduction
The safety of meat products is one of the most important current and future challenges requiring the attention of the food industry, scientists, and public health authorities.
Despite the significant advancement of sequencing technologies and the giant pools of NGS data accumulated, robust, cost-effective, and reliable techniques, allowing the annotation of the microbial status of food, remain a challenge for the scientific community. Today, MS-based technologies to detect foodborne pathogens complement the classical methods and NGS. We would like to highlight two directions in the trends in pathogen proteomics. The first direction, which has obvious practical value, is supported by the efforts of the entire proteomic community to ensure in-process control and reduce batch-to-batch variations, especially in label-free techniques.
Taking into account the short duration of the “life” of certain meat products, there is a need for robust and cost-effective methods for pathogen detection, applicable both in the laboratory and in marketable products [1]. We expect the classic “from farm to fork” path to become “from farm through mass spec to fork” soon, especially if the community develops reliable identification criteria, such as standardized lists of proteotypic peptides that could be used as reliable proof of protein presence.
The second hot topic in the field of foodborne pathogens is of high fundamental importance in the context of host–pathogen interactions covering all stages of infection from invasion to dissemination. It would not be an exaggeration to say that the interactions between molecules enable life. To understand the molecular mechanisms behind the life cycle of common foodborne infections, a complete map of the interactions between the proteins and metabolites of pathogens, food products, and, in fact, the end consumer must be formed. We are seeing similar interactive initiatives in the fight against seed-borne diseases in wheat, which lead to the reduced quality and quantity of the crop, and expect similar initiatives from the research community focused on meat products [2,3].
To date, genomic and transcriptomic methods have a clear advantage over proteomic methods in the field of food production state monitoring due to several reasons. Genomic and transcriptomic methods are more mature and have a longer history of use, the instruments are cheaper and, accordingly, more common. If we compare the genomic and proteomic research intensities for food production safety monitoring, there are 6-times more publications that correspond to genomic research (3122 articles according to PubMed, December 2022) than to proteomic research (500 articles according to PubMed, December 2022). In addition, some mechanisms of bacterial resistance are based on genome changes. Furthermore, for each technology, we can observe a gap between the fundamental research and the applied application in food production safety monitoring for both genomics and proteomics. The turning point is, among other things, the emergence of regulatory documents that allow the use of technology to assess the safety of production. WGS (whole-genome sequencing) gained acceptance as a prospective surveillance tool for foodborne illness in 2016 [4,5,6], and now it is increasingly replacing traditional microbial typing and characterization techniques, providing faster and more precise answers [7]. As for proteomics, there are no regulatory documents for its use for food safety monitoring; the normative and methodical bases are still under construction [8].
At the same time, proteomic methods give an opportunity to detect rapid changes in the molecular responses of bacterial cells to aggressive stimuli such as antibiotic damage. Therefore, the usefulness of these methods will provide more information about the mechanisms of bacterial resistance and the strains’ life cycles in food production.
Traditionally, meat spoilage pathogens are identified with culture-based methods focused on the biochemical, physiological, and morphological properties of the microorganisms under study. These classical methods typically require several days to obtain the results. Moreover, these methods do not provide valuable information on the dead pathogen cells or toxins. Despite the tremendous democratization of NGS methods, high-throughput mass spectrometry-based proteomics has been recognized as an essential tool to reliably identify and quantify thousands of proteins in complex biological matrices.
Foodomics renders mass spectrometry as a major “working horse” for its “direct” nature of qualitative and quantitative data on the molecules of interest with great performance and adaptability.
Technical progress and scientific curiosity are bringing to the market new MS devices with increased robustness, sensitivity, and selectivity, and decreased sample demand. The variety of mass spectrometric methods is expansive; however, in a broad sense, all MS approaches can be divided into panoramic (or “shotgun”) and targeted groups [9,10]. In the context of food safety, the panoramic MS is used for the exploration of a meat proteome from a bird’s eye view to massively identify potential biomarkers of foodborne pathogens. Several commonly used variations of the panoramic strategy are focused on the intact proteins (“top-down”) and individual peptides (“bottom-up”) in which selective proteases digest the proteins into proteotypic peptides that are submitted into the MS system [11]. In turn, the methods of targeted proteomics are tailored to quantify the preselected peptides with high sensitivity and precision.
Obviously, all fundamental and practical questions cannot be incorporated into one analytical device, providing panoramic or targeted snapshots of the sample under study. The range of problems that proteomics solves is wider than just the panoramic or targeted search. Often, a researcher analyzes proteins with specific biochemical properties (charge, size, or hydrophobicity), conformational changes, post-translational modifications, or certain interacting partners [12,13]. To answer all these questions, researchers use panoramic or targeted mass spectrometry but enhance it with a whole arsenal of superior supporting technologies: e.g., sample preparation (antibody depletion or enrichment), fractionation (two-dimensional gel electrophoresis or multidimensional chromatography), and quantification (iTRAQ or label-free) [14,15]. Thus, speaking of panoramic or targeted mass spectrometry, we describe an extensive range of methods that can comprehensively explore a particular sample’s proteome. The current review summarizes the significant achievements in proteomic analysis for food production. We summarized the most up-to-date proteomic achievements to highlight the relevance of this omics discipline in elucidating bacterial lifestyle, developing food safety biomarkers, and innovating food protection strategies.
The first part of the article is devoted to the methods used for microbial strain analysis, focused on food pathogens. Further review is focused on three key strains (Salmonella enterica, Listeria monocytogenes, and Pseudomonas aeruginosa) and the poisoning products that cause infectious diseases. The final part includes data on the markers of disinfectant and antibiotic resistance.
It is important to note that we have chosen the PubMed database as a source of publications on the bacterial resistance specific to food production. Despite the fact that this database is not specialized for the problems of food production, it has a number of advantages in the context of our study. First of all, it is one of the most complete and structured databases in the field of biological research and very well adapted for the automatic analysis of a large number of publications. Secondly, omics technologies are at the crossroads of many natural sciences, including biology, medicine, and agriculture, and the peculiarities of their use in the context of bacterial research can be published in various thematic journals. At the same time, proteomic methods are still quite expensive and highly specialized; therefore, they are more often used in fundamental research than in commercial food production control.
2. Materials and Methods
The research was performed using a combination of automatic text-analysis algorithms, ScanBious [16,17] and biocuration [18]. The automatic text-analysis algorithms used in this work have previously shown their effectiveness for reconstructing metabolic pathways [19], assessing the current state of the selected subject area [20], and investigating trends in scientific research in the field of biomedicine [21]. The PubMed/MEDLINE library of scientific publications in biology and medicine was used as a data source [22]. Each article was associated with a set of keywords (MeSH (Medical Subject Heading) terms) assigned by the curators of the resource for indexing and briefly characterizing the content of each publication. ScanBious is a free-ware Web-system for highlighting key concepts revealed from PubMed abstracts and related MeSH terms; it provides an interface to the transparent algorithm, which relies on the co-occurrence of two terms in one abstract [20]. One can make a search query to ScanBious and get a list of relevant articles similar to the PubMed/MEDLINE system. Unlike PubMed/MEDLINE, the user can process the obtained set of publications: select the most relevant to the search query articles and their abstracts, extract their MeSH terms, select target MeSH terms, and visualize them as a semantic network based on the measure of interrelation (Jaccard index). The nodes of the semantic network are the objects (MeSH terms) selected by the user and the links of which are a list of publications in which both objects are described [20]. The size of the nodes reflects the number of publications; the thickness and the color of the edge–Jaccard’s index.
Using ScanBious, a semantic map of the subject area was built. Visualization was carried out using the frequency characteristics of the MeSH terms of the publications corresponding to the search query [“meat production” OR “meat products” OR “meat processing” OR “meat industry” OR “meat processing products”, “meat” OR “food production” OR “food industry”].
About 48,000 relevant publications were processed. The nodes of the map are the MeSH terms, and the edges between the nodes reflect the joint mention of the terms in the same study. The ScanBious system provides an opportunity to work with the publications associated with the nodes and the edges of the semantic map. Thus, after a short dive into the subject area using the semantic map, detailed information can be obtained from the texts of the relevant articles.
The hierarchical structure of the organization of the MeSH dictionary of terms also allows for the limiting of the display, on the semantic map of the subject area, of terms related to specific categories. Thus, one can set the visualization of the MeSH terms related to the subject area and characterize the methods of study (MeSH category “Analytical, Diagnostic and Therapeutic Techniques, and Equipment”) or, for example, diseases (“Diseases”).
3. Results and Discussion
3.1. Subject Area Semantic Network Description
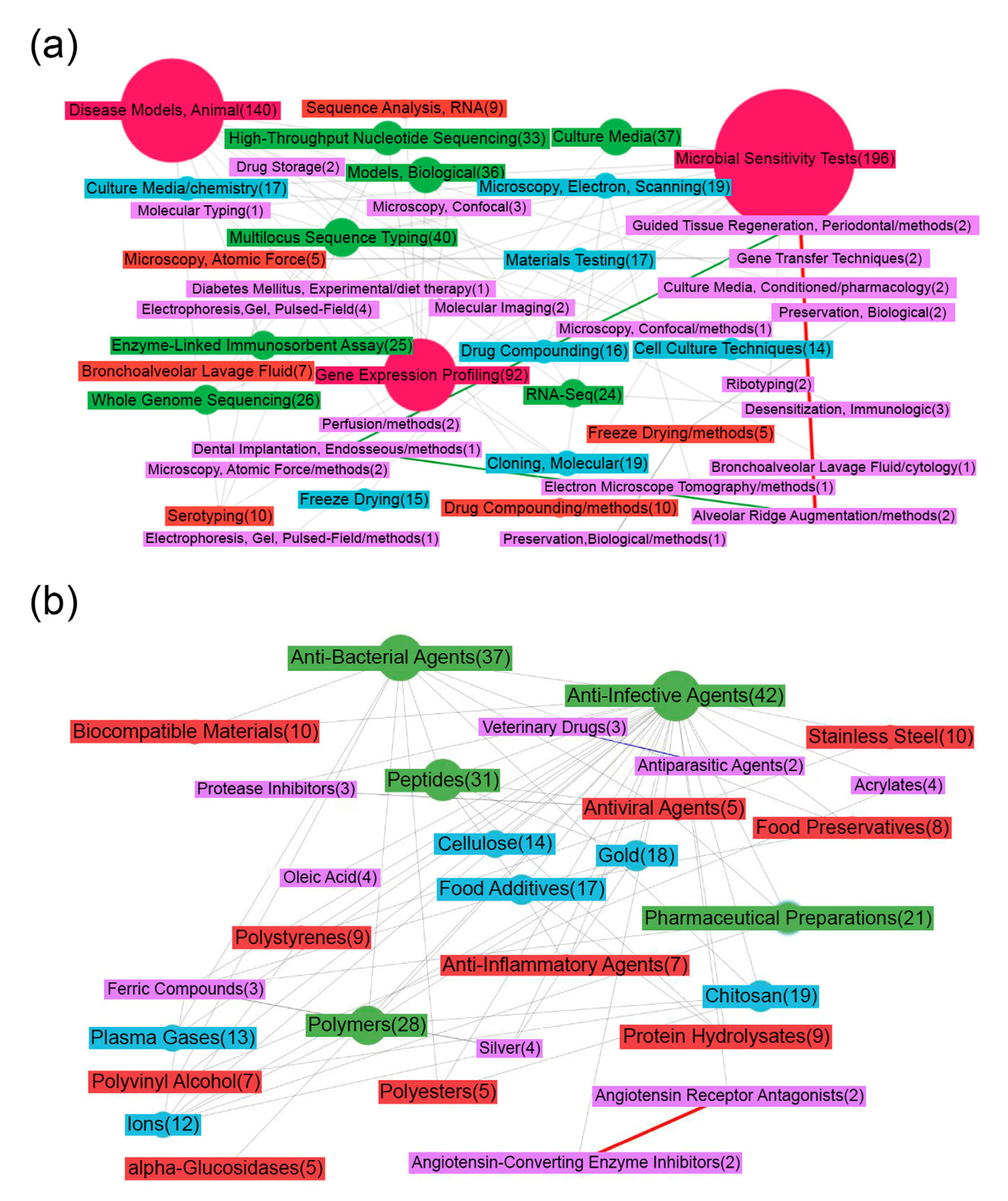
Figure 1a shows a map of the MeSH terms that characterize the publications downloaded on demand (see the Materials and Methods section). It draws attention to the fact that the most researched issue related to food production is the creation of microbiological control tests (“Microbial Tests”). To profile and assess the composition of microbial communities, genomic and transcriptome sequencing is mainly used; the semantic map contains nodes that confirm this statement (for example, “High-Throughput Nucleotide Sequencing”, “RNA sequence Analysis”, “Multilocus Sequence Typing”, “Gene Expression Profiling”, “Whole genome sequencing”) and various microscopy methods (electron-scanning, atomic force, confocal). Proteomic approaches for the analysis of microbial communities (“Gel Electrophoresis”, “Enzyme-Linked Immunosorbent Assay”) are much less widely used.
Figure 1.
Map of MeSH terms resulting from ScanBious processing of publications in the PubMed/MEDLINE system associated with food (meat) production research. The map displays the MeSH terms related to the category “Analytical, Diagnostic and Therapeutic Techniques, and Equipment” (a) and “Chemical and Drug” (b). The number in parentheses indicates the number of articles in which the term was found. The color and thickness of the connecting lines and nodes reflects the degree of interaction between the two terms: the thicker the line, the more articles included both terms simultaneously. The gray color corresponds to Jaccard index indices with values of 0.2 and below, the blue color corresponds to values from 0.2–0.4, the green corresponds to values from 0.4–0.6, the yellow corresponds to values from 0.6–0.8, and the red corresponds to values of 0.8 and above.
An essential task in determining the composition of microbial communities is assessing resistance to various types of disinfectants, for example, antibiotics; the effectiveness of measures aimed at the safety of food production directly depends on this. The scientific works in this area characterize the MeSH terms associated with drugs (“Drug Compounding”, “Drug Storage”) and methods for studying antibiotic resistance in animals and cell lines (“Models Biological”, “Cell Culture Techniques”, “Disease Models, Animal”). Figure 1b depicts the MeSH terms from the category “Chemical and Drug”. Most of the publications were indexed using high-level MeSH terms (“Anti-Infective Agents”, “Protease Inhibitors”), not defining exact compounds.
In a study [23] on the metagenetic analysis of food production, it was shown that the leaders among infectious agents are Proteobacteria (found in 71.37% of bacterial communities) and Actinobacteria (79.72%). As biofilms mature, the species composition changes somewhat, but Pseudomonas, Enterobacteriaceae, and Firmicutes are also widely encountered [24].
Pseudomonas are ubiquitous bacteria due to their versatile set of proteins involved in metabolism [25]. Studies conducted over several decades on different continents have shown that Pseudomonas from raw materials or treated surfaces, cooling air, or water can proliferate in a food product, especially under aerobic conditions [26]. At the end of the shelf life, Pseudomonas dominates and spoils the meat through proteolytic, lipolytic, saccharolytic, and biosurfactant processes [27]. Pseudomonas aeruginosa remains one of the most important species in terms of antibiotic resistance due to its high tendency to acquire polyresistance [28,29,30].
Gram-negative Salmonella enterica (family Enterobacteriaceae) is an intracellular parasite that invades small intestinal mucosal cells. Entering the human body with food, this bacterium causes the dangerous disease salmonellosis, which accounts for 25% of all human diarrheal diseases, ranging from mild infections to death [31]. Salmonella strains with resistance to multiple antimicrobials have long spread in developed and developing countries [32]. It is generally accepted that, for the most part, such strains are of zoonotic origin. They acquire resistance due to the use of antimicrobials for prophylactic or therapeutic purposes in the food industries.
Listeria monocytogenes is a member of Firmicutes, a rod-shaped, Gram-positive pathogenic bacterium that is a facultative intracellular parasite. Once in the human body, Listeria monocytogenes causes the dangerous invasive disease listeriosis, an infection that accounts for about 28% of deaths due to foodborne diseases [33]. This pathogen can cross three critical barriers in the human host, namely the intestinal epithelium, the blood–brain barrier, and the placenta, and can subsequently spread to other organs. Listeria monocytogenes can grow at refrigeration temperatures [34], thereby increasing the potential hazard of eating refrigerated and ready-to-eat (RTE) foods.
Bacterial resistance research is complicated due to the bacteria’s ability to form biofilms. A biofilm is a structured community of bacterial cells enclosed in a self-produced matrix that adheres to inert or living surfaces, including tissues, industrial surfaces, and artificial devices, such as intrauterine contraceptive devices, implants and prosthetic medical devices, catheters, dental materials, cardiac valves, and contact lenses. Biofilms form when bacterial colonizers adhere to surfaces in aqueous environments and excrete a slimy, glue-like substance composed of exopolysaccharides (EPS) [35]. It gives them consistency and resistance to antibiotics and disinfectants [36,37]. Biofilm formation and proliferation affects many aspects of public health and industrial processes, including being a source of contamination in the food and beverage industries [38,39,40].
The essence of this problem is that bacteria, organized on any surface as complex communities (biofilms), acquire qualitatively new properties. The biofilm bacteria can increase resistance to immune system effectors, antibiotics, and disinfectants. Food preservatives, disinfectants, and resistance to antiseptics are under-explored [41], and biofilms complicate the research process.
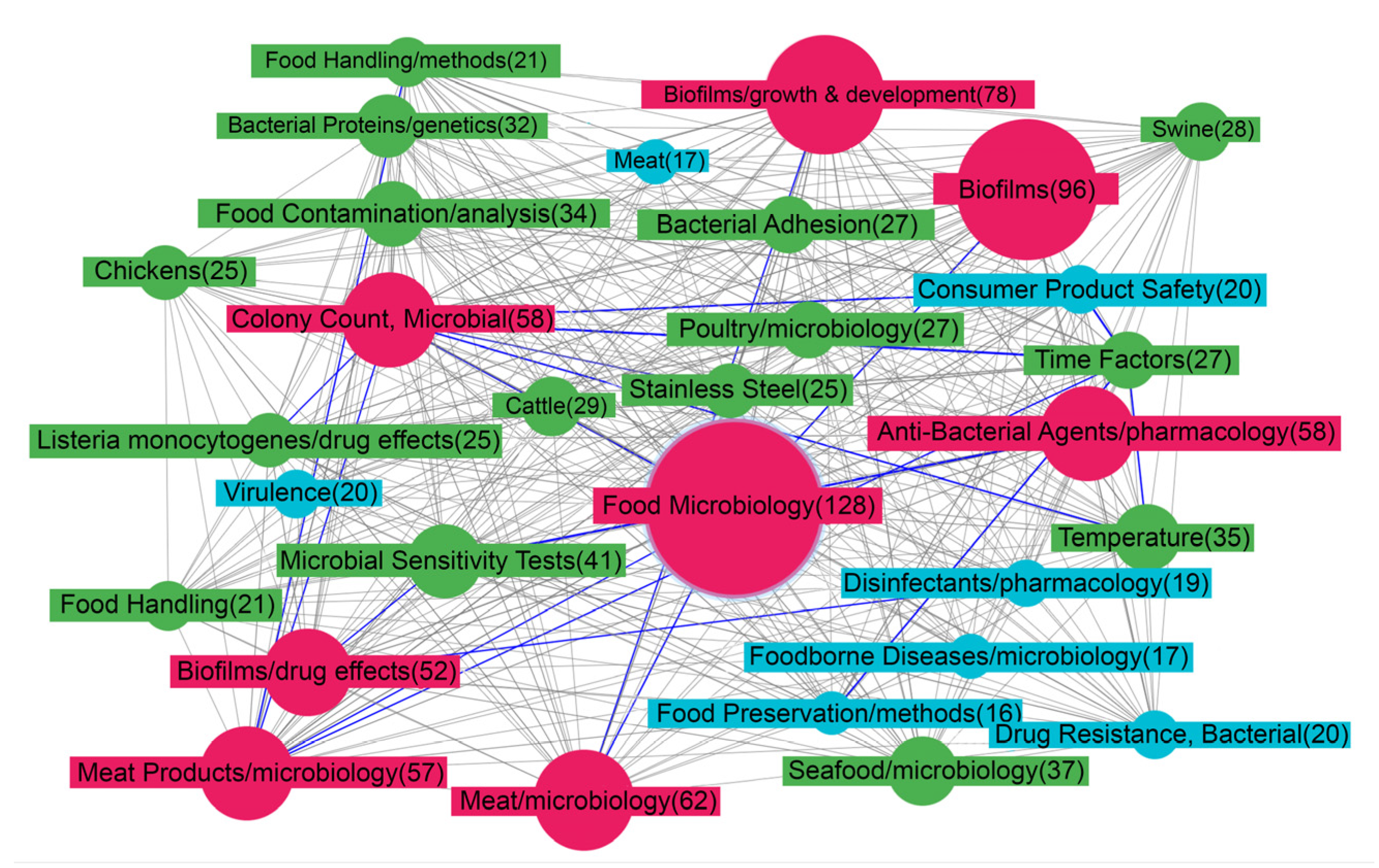
One of the leading positions in Figure 2 belongs to the term “Biofilms”. The closest nodes depict some conditions that affect biofilm development: “Temperature”, “Time-factors”, “Bacterial adhesion”. A given species or strain of bacteria responds to environmental conditions via a finite number of key regulatory factors and pathways, which affect the enzymatic and structural elements that are needed for biofilm formation and dispersal. Among the conditions that affect biofilm development are temperature, pH, O2 levels, hydrodynamics, osmolality, the presence of specific ions, nutrients, and factors derived from the biotic environment. The integration of these influences ultimately determines the pattern of behavior of a given bacterium with respect to biofilm development [42].
Figure 2.
Map of MeSH terms resulting from ScanBious processing of publications in the PubMed/MEDLINE system associated with food (meat) production research. The map displays the MeSH terms related to the category “Phenomena and Processes”. The color and thickness of the connecting lines and nodes reflects the degree of interaction between the two terms: the thicker the line, the more articles included both terms simultaneously. The gray color corresponds to Jaccard index indices with values of 0.2 and below, the blue color corresponds to values from 0.2–0.4, the green corresponds to values from 0.4–0.6, the yellow corresponds to values from 0.6–0.8, and the red corresponds to values of 0.8 and above.
3.2. Genetic Determinants of Resistance
Bacteria, including the considered Salmonella enterica, Listeria monocytogenes, and Pseudomonas aeruginosa, acquire resistance to antimicrobial agents through the following mechanisms: (1) structure change of the antibiotic target (mutation), (2) enzymatic degradation or restructuring of the antibiotic, (3) active cell release of antibiotics or efflux, (4) reduced permeability to antibiotics, (5) use of alternative metabolic pathways, (6) and formation of biofilms [43,44].
Resistance mechanisms may be driven by acquiring three types of mobile genetic elements: self-transferring plasmids, mobilizable plasmids, and conjugative transposons [45]. Regarding antibiotic and disinfectant resistance in bacteria, researchers often consider genotypic research methods. Isolated bacterial strains from patient material or food are subjected to antibiotic susceptibility tests. The bacterial strains are treated with the antibiotic of interest after which the colony’s inhibition zone’s size is determined. Then, the researcher’s task is reduced to a search for the resistance gene and the donor plasmid.
Determining the resistance genes and assessing their expression at the transcriptomic level do not recreate the complete picture of the systemic rearrangements of cellular processes under the stress caused by antibiotics. Currently, studies are underway to establish the position of small non-coding RNAs in rearranging metabolic networks under external stress, including that caused by antibiotics [46,47]. RNA-binding proteins play an essential role in modulating the interactions of regulatory small non-coding RNAs [48]. In addition to the metabolic rearrangements in response to stress in bacteria, proteins can undergo post-translational modifications, which can be involved in developing resistance [49].
Combining several omics technologies makes it possible to recreate the complete network of molecular interactions. Proteomics is evolving rapidly, and its flexibility and potential have yet to be fully exploited. Sidoli et al. even supposed that, in the next ten years, every biological institute will have at least one mass spectrometer [50]. Mass spectrometry (MS) provides valuable insights into the signaling, post-translational changes, and microbial–host interactions relevant to bacterial pathogenesis. Although transcriptomics still outperforms mass spectrometry in several ways (for example, in terms of the estimated cost of the experiment), proteomic methods provide many undeniable advantages.
The results of the literature analysis devoted mainly to the genotypic methods for studying the resistance in selected strains are presented in Table 1. This table accumulates information on the available resistance determinants: the gene name, the corresponding UniProt ID, and the neutralizable antimicrobial agent.

Table 1.
Accumulated information on the genes and proteins associated with resistance development in three types of microorganisms.
According to Table 1, most of the protein products of resistance genes have the “unreviewed” status according to the UniProt database. This means that computational methods predicted the sequences of these proteins. These facts define proteomics as the missing link in the chain of research to establish resistance mechanisms and search for alternative targets for treating bacterial infections.
3.3. Proteomic Methods for Studying the Microbiome of Food Industries
- (a)
- Panoramic mass spectrometry
Panoramic mass spectrometry, or shotgun proteomics, determines the complete set of proteins in a complex multi-component cell extract. Shotgun proteomics uses a liquid chromatograph with a tandem mass spectrometer. This method is considered panoramic because the mass spectrometer registers all peptide ions of the isolated proteins. Shotgun proteomics for studying the mechanisms of the development of resistance in bacteria can be applied similarly to the genotypic approach to measure protein concentrations in strains pretreated with antimicrobial agents, identify overexpression, and try to establish a relationship.
Coldham et al. [54] studied the physiological response of Salmonella enterica (Typhimurium serovar) to fluoroquinolone antibiotics using proteomic methods. The protein expression was assessed using two-dimensional HPLC-MS. The increased expression of AcrAB/TolC proteins (efflux pump proteins) is associated with the development of antibiotic resistance. Qi et al. also associated the AcrB efflux pump protein with antibiotic resistance, but they also mentioned the BaeSR protein system [57]. They suggested that these proteins play a role in the phosphorylation of other Salmonella enterica proteins, which, in turn, leads to the development of drug resistance.
Ultra-high-performance liquid chromatography with tandem mass spectrometry is often used to detect bacterial aggression factors. For example, the authors of the following work identified enterotoxins of Staphylococcus aureus, which has 15 variants of the toxin [79].
Similarly, using tandem mass spectrometry with electrospray ionization coupled with liquid chromatography (LC–ESI–MS/MS) in a large-scale study [68], 395 proteins with a direct role in the pathogenicity of Listeria were identified. These proteins were virulence factors, toxins, antitoxins, and molecules associated with antibiotic resistance or resistance to toxic substances. In particular, β-lactamases, MarR transcription regulators involved in the transduction of β-lactamase metallopeptidase M56, penicillin-binding proteins, etc., were found among the latter.
As mentioned earlier, the post-translational modifications of proteins also impact bacterial resistance to antibiotics [55]. For example, Li et al. proved that the acetylation status of lysine in the proteins associated with fluoroquinolone resistance varies in resistant and susceptible strains of Salmonella enterica. A high content of acetylated lysine characterizes the proteins of resistant Salmonella enterica.
Similarly, in work [80] devoted to the resistance of Pseudomonas aeruginosa to ofloxacin, post-translational modifications of PheA and SpuC proteins (succinylation and acetylation) were revealed. The study showed that the modifications of pheA and spuC modulate resistance to ofloxacin. The presence of post-translational modifications on these two proteins could direct the protein to a different cellular function and promote the survival of P. aeruginosa.
The changes in the metabolic pathways that cause the development of antibiotic resistance can also be captured using proteomic methods. Erdmann J. and colleagues established the quantitative ratio of the core proteome for 27 clinical isolate samples using the DIA technology (data independent acquisition) on the nanoLC-ESI-MS/MS platform (TripleTOF 5600+) [81]. Genomic and transcriptomic analyses were also performed for each sample. The study of soft-core biofilm proteins showed that, despite significant variability between the strains, there was an increase in the concentration of proteins involved in the iron sequestration and iron metabolism processes (PchDG, FptA), Pseudomonas quinolone signal transduction (PQS) (PqsBCDH), phenazine biosynthesis (PhzB1, PhzB2, PhzE2, PhzF2, PhzM), outer membrane proteins (OprCFG, MexI), and fatty acid biosynthesis (FabG).
Panoramic proteomics can be successfully incorporated into multi-omics projects to visualize the multi-dimensional nature of foodborne pathogens in the context of genomics, proteomics, metabolomics, and clinical data. The major advantage of panoramic MS over targeted techniques is that it fuels non-hypothesis-driven proteomic research, making it superior for the early stages of biomarker discovery when dealing with complex biological matrices. Another MS benefit is the ability to explore the processes performed by the whole microbial community at the time of sample collection in time- and cost-effective ways.
- (b)
- Targeted mass spectrometry
In the targeted proteome analysis, the mass spectrometer is tuned to detect specific peptide ions related to target proteins. Methods for multiple reactions monitoring and selected reactions monitoring (MRM and SRM, respectively) make it possible to select peptide ions with specified mass-to-charge characteristics at the first and second stages of tandem mass spectrometry. Such methods have several undeniable advantages in comparison with panoramic MS analysis. Bao and colleagues applied MRM with isotopically labeled peptide standards to detect and quantify the staphylococcal enterotoxin SEB in raw chicken [82]. Three peptides were used as a standard, which made it possible to achieve high detection accuracy [83].
The extraction of proteins for LC-MS/MS-based analysis involves using gels or long-term immunoaffinity purification to obtain a pure sample for analysis. Anjelkovich and colleagues presented a combination of LC-MS/MS and in silico, solid-phase extraction via MRM [84], eliminating some shortcomings of the conventional extraction methods. This method was used to identify SEB and SEA in various cultures. The proposed approach allows the detection of toxins in milk samples, while the limits of detection of SEA and SEB were 8 and 4 ng/g, lower than that achieved with many other identification methods (for example, using ultra-high-performance liquid chromatography with tandem mass spectrometry in the article by Muratovic et al. [79]).
The LC-SRM method is considered the most sensitive [85]. Dupre et al. [86] used it for the absolute quantification of eight toxins in food matrices: ricin, ETX, SEA, SEB, SED, Shigella dysenteriae shigatoxin, STX1 and STX2 (Escherichia coli), and CDT (Campylobacter jejuni). The experimental results confirmed the method’s high sensitivity to toxin concentrations corresponding to food poisoning. In the same work, a unique author’s method for detecting enterotoxins was presented: immuno-LC-MS/MS in combination with PRM. Antitoxin monoclonal antibodies were covalently bound to magnetic elements after which, upon contact with isotope-labeled toxins, selective recognition and binding of the latter occurred. Thus, using the quadrupole–Orbitrap instrument, at least seven peptides were detected for each of the three toxins (ricin, SEB, and Clostridia epsilon toxin). The advantage of this method is its rapidity; in the experiment, which took 5 h, the limit of quantitation was reached at the level of 1 ng ml-1 or lower.
Mass spectrometers are very sensitive quantitative instruments with an attomole detection limit; however, determining the threshold between signal and noise in MS signals is often challenging. Although mass spectrometry cannot provide the depth of coverage of high-throughput sequencing, this method is actively used to identify the proteomes of bacterial strains. Sequencing techniques are inferior to proteomics based on mass spectrometry since proteins can be altered post-translationally, and their structure and function will be altered accordingly. This information cannot be read from gene sequences or transcripts.
The systemic analysis of the literature was performed using an automated text-mining tool and manual approaches. It revealed that proteomic methods have the potential to identify differentially regulated bacterial proteins in food samples or in response to drugs and disinfectants as well as to describe the mechanisms of biofilm formation and the host–parasite relationship. The current technologies offer two major scenarios of food production proteomic profiling. The first one involves the comprehensive, rapid, and robust analysis of key markers of food production status to detect bacterial strains and/or markers of their resistance [87]. The second scenario is to develop chromatographic approaches to perform deep proteomic profiling and overcome the current limitations in the frequency of scans and the complexity of biological samples. Quantitative profiling will allow for the solving of the fundamental problems with high applied potential, for example, tracking in detail the process of biofilm formation by microbial communities in food production conditions, its speed, and the best approaches to limit it [88]. The equipment needed to implement the scenarios described above is complex, expensive, and requires capacities for sample preparation and computing power for data analysis. Therefore, only the cooperation of the food production industries with scientific laboratories will provide the best insights for food safety development. The application of proteomics will be helpful to uplift human health and develop the fields of animal production, agriculture, food processing, and storage [89].
4. Conclusions
Based on the above, the proteomics approach serves as a complement to the genomic and transcriptomic approaches. Proteomics is effective in the study of complex mapping of bacterial proteomes, post-translational modifications of proteins, and pathogen–host interactions, as well as in antimicrobial resistance and the discovery of new protein biomarkers. A knowledge base is needed in which bacterial genes are annotated in the context of antibiotic resistance. Such an information resource will allow the creation of more sensitive methods for assessing the state of food production and considering the variety of bacteria and the resistance of specific strains to disinfectants. Proteomic bacterial resistance research is at its early stages but shows great perspectives. However, the most advantageous strategy is the use of a multi-omics approach. It provides cross-validation of the results and gives a complete understanding of the molecular processes characterizing the food production bacterial communities. The multi-omics approach can undercover the mechanisms of biofilm resistance, tracking the most dangerous combinations of bacterial strains and surfaces on which they are attached.
The text-mining approach gives us an opportunity to process large amounts of papers and to extract the most valuable information about the proteomic basics of bacterial resistance. The semantic networks are useful instruments for scientific literature surfing.
Author Contributions
Conceptualization, E.I.; writing—original draft preparation, S.T., A.K. and E.S.; writing—review and editing, O.K.; supervision, E.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation for large scientific projects in priority areas of scientific and technological development, grant number 075-15-2020-775.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank the “Human Proteome” Core Facilities of the Institute of Biomedical Chemistry (Russia) for providing resources for data analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jadhav, S.R.; Shah, R.M.; Karpe, A.V.; Morrison, P.D.; Kouremenos, K.; Beale, D.J.; Palombo, E.A. Detection of Foodborne Pathogens Using Proteomics and Metabolomics-Based Approaches. Front. Microbiol. 2018, 9, 3132. [Google Scholar] [CrossRef] [PubMed]
- Kataria, R.; Kaundal, R. WeCoNET: A Host–Pathogen Interactome Database for Deciphering Crucial Molecular Networks of Wheat-Common Bunt Cross-Talk Mechanisms. Plant Methods 2022, 18, 73. [Google Scholar] [CrossRef]
- Piras, C.; Roncada, P.; Rodrigues, P.M.; Bonizzi, L.; Soggiu, A. Proteomics in Food: Quality, Safety, Microbes, and Allergens. Proteomics 2016, 16, 799–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Identification of Salmonella for Public Health Surveillance Using Whole Genome Sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef]
- Whole Genome Sequencing (WGS) Program|FDA. Available online: https://www.fda.gov/food/science-research-food/whole-genome-sequencing-wgs-program (accessed on 4 January 2023).
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The Use of next Generation Sequencing for Improving Food Safety: Translation into Practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, S.A.; Kuvaeva, I.B.; Efimochkina, N.R.; Minaeva, L.P. Microbiological Safety of Food: Development of Normative and Methodical Base. Vopr. Pitan 2020, 89, 125–145. [Google Scholar] [CrossRef]
- Khalilpour, A.; Kilic, T.; Khalilpour, S.; Álvarez, M.M.; Yazdi, I.K. Proteomic-Based Biomarker Discovery for Development of next Generation Diagnostics. Appl. Microbiol. Biotechnol. 2017, 101, 475–491. [Google Scholar] [CrossRef]
- Shevchuk, O.; Begonja, A.J.; Gambaryan, S.; Totzeck, M.; Rassaf, T.; Huber, T.B.; Greinacher, A.; Renne, T.; Sickmann, A. Proteomics: A Tool to Study Platelet Function. Int. J. Mol. Sci. 2021, 22, 4776. [Google Scholar] [CrossRef]
- Fornelli, L.; Toby, T.K.; Schachner, L.F.; Doubleday, P.F.; Srzentić, K.; DeHart, C.J.; Kelleher, N.L. Top-down Proteomics: Where We Are, Where We Are Going? J. Proteom. 2018, 175, 3–4. [Google Scholar] [CrossRef]
- Sitammagari, K.K.; Masood, W. Creutzfeldt Jakob Disease; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar] [PubMed]
- Virág, D.; Dalmadi-Kiss, B.; Vékey, K.; Drahos, L.; Klebovich, I.; Antal, I.; Ludányi, K. Current Trends in the Analysis of Post-Translational Modifications. Chromatographia 2020, 83, 1–10. [Google Scholar] [CrossRef]
- Rodríguez-Vázquez, R.; Mato, A.; López-Pedrouso, M.; Franco, D.; Sentandreu, M.A.; Zapata, C. Measuring Quantitative Proteomic Distance between Spanish Beef Breeds. Food Chem. 2020, 315, 126293. [Google Scholar] [CrossRef]
- Lasch, P.; Schneider, A.; Blumenscheit, C.; Doellinger, J. Identification of Microorganisms by Liquid Chromatography-Mass Spectrometry (LC-MS1) and in Silico Peptide Mass Libraries. Mol. Cell Proteom. 2020, 19, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Ilgisonis, E.; Lisitsa, A.; Kudryavtseva, V.; Ponomarenko, E. Creation of Individual Scientific Concept-Centered Semantic Maps Based on Automated Text-Mining Analysis of PubMed. Adv. Bioinform. 2018, 2018, 4625394. [Google Scholar] [CrossRef]
- Hassani, H.; Beneki, C.; Unger, S.; Mazinani, M.T.; Yeganegi, M.R. Text Mining in Big Data Analytics. Big Data Cogn. Comput. 2020, 4, 1. [Google Scholar] [CrossRef]
- Holinski, A.; Burke, M.L.; Morgan, S.L.; McQuilton, P.; Palagi, P.M. Biocuration-Mapping Resources and Needs. F1000Res 2020, 9, 1094. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Lisitsa, A.V.; Il’gisonis, E.V.; Archakov, A.I. Construction of Protein Semantic Networks Using PubMed/MEDLINE. Mol. Biol. 2010, 44, 152–161. [Google Scholar] [CrossRef]
- Tarbeeva, S.; Lyamtseva, E.; Lisitsa, A.; Kozlova, A.; Ponomarenko, E.; Ilgisonis, E. ScanBious: Survey for Obesity Genes Using PubMed Abstracts and DisGeNET. J. Pers. Med. 2021, 11, 246. [Google Scholar] [CrossRef] [PubMed]
- Ilgisonis, E.V.; Pyatnitskiy, M.A.; Tarbeeva, S.N.; Aldushin, A.A.; Ponomarenko, E.A. How to Catch Trends Using MeSH Terms Analysis? Scientometrics 2022, 127, 1953–1967. [Google Scholar] [CrossRef]
- Yang, H.; Lee, H.J. Research Trend Visualization by MeSH Terms from PubMed. Int. J. Environ. Res. Public Health 2018, 15, 1113. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Cabo, M.L. Tracking Bacteriome Variation over Time in Listeria Monocytogenes-Positive Foci in Food Industry. Int. J. Food Microbiol. 2020, 315, 108439. [Google Scholar] [CrossRef] [PubMed]
- Møretrø, T.; Langsrud, S.; Heir, E.; Møretrø, T.; Langsrud, S.; Heir, E. Bacteria on Meat Abattoir Process Surfaces after Sanitation: Characterisation of Survival Properties of Listeria Monocytogenes and the Commensal Bacterial Flora. Adv. Microbiol. 2013, 3, 255–264. [Google Scholar] [CrossRef]
- Palleroni, N.J. Introduction to the Family Pseudomonadaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 1981; pp. 655–665. [Google Scholar] [CrossRef]
- Hinton, A.; Cason, J.A.; Ingram, K.D. Tracking Spoilage Bacteria in Commercial Poultry Processing and Refrigerated Storage of Poultry Carcasses. Int. J. Food Microbiol. 2004, 91, 155–165. [Google Scholar] [CrossRef]
- Morales, P.A.; Aguirre, J.S.; Troncoso, M.R.; Figueroa, G.O. Phenotypic and Genotypic Characterization of Pseudomonas Spp. Present in Spoiled Poultry Fillets Sold in Retail Settings. LWT 2016, 73, 609–614. [Google Scholar] [CrossRef]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR-Pseudomonas Aeruginosa in Fish Commonly Harbor OprL and ToxA Virulence Genes and BlaTEM, BlaCTX-M, and TetA Antibiotic-Resistance Genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.; Yoon, S.S. Virulence Characteristics and an Action Mode of Antibiotic Resistance in Multidrug-Resistant Pseudomonas Aeruginosa. Sci. Rep. 2019, 9, 487. [Google Scholar] [CrossRef]
- Madaha, E.L.; Mienie, C.; Gonsu, H.K.; Bughe, R.N.; Fonkoua, M.C.; Mbacham, W.F.; Alayande, K.A.; Bezuidenhout, C.C.; Ateba, C.N. Whole-Genome Sequence of Multi-Drug Resistant Pseudomonas Aeruginosa Strains UY1PSABAL and UY1PSABAL2 Isolated from Human Broncho-Alveolar Lavage, Yaoundé, Cameroon. PLoS ONE 2020, 15, e0238390. [Google Scholar] [CrossRef]
- de Mesquita Sousa Saraiva, M.; Benevides, V.P.; da Silva, N.M.V.; de Mello Varani, A.; de Freitas Neto, O.C.; Berchieri, Â.; Delgado-Suárez, E.J.; de Lima Rocha, A.D.; Eguale, T.; Munyalo, J.A.; et al. Genomic and Evolutionary Analysis of Salmonella Enterica Serovar Kentucky Sequence Type 198 Isolated From Livestock In East Africa. Front. Cell. Infect. Microbiol. 2022, 12, 1. [Google Scholar] [CrossRef]
- Threlfall, E.J. Antimicrobial Drug Resistance in Salmonella: Problems and Perspectives in Food- and Water-Borne Infections. FEMS Microbiol. Rev. 2002, 26, 141–148. [Google Scholar] [CrossRef]
- Aureli, P.; Fiorucci, G.C.; Caroli, D.; Marchiaro, G.; Novara, O.; Leone, L.; Salmaso, S. An Outbreak of Febrile Gastroenteritis Associated with Corn Contaminated by Listeria Monocytogenes. N. Engl. J. Med. 2000, 342, 1236–1241. [Google Scholar] [CrossRef]
- Kathariou, S. Listeria Monocytogenes Virulence and Pathogenicity, a Food Safety Perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Jolivet-Gougeon, A.; Bonnaure-Mallet, M. Biofilms as a Mechanism of Bacterial Resistance. Drug Discov. Today Technol. 2014, 11, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Katsikogianni, M.; Missirlis, Y.F.; Harris, L.; Douglas, J. Concise Review of Mechanisms of Bacterial Adhesion to Biomaterials and of Techniques Used in Estimating Bacteria-Material Interactions. Eur. Cell Mater. 2004, 8, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Johansen, C.; Falholt, P.; Gram, L. Enzymatic Removal and Disinfection of Bacterial Biofilms. Appl. Environ. Microbiol. 1997, 63, 3724–3728. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, 1556–1573. [Google Scholar] [CrossRef] [PubMed]
- Goode, K.R.; Asteriadou, K.; Robbins, P.T.; Fryer, P.J. Fouling and Cleaning Studies in the Food and Beverage Industry Classified by Cleaning Type. Compr. Rev. Food Sci. Food Saf. 2013, 12, 121–143. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53. [Google Scholar] [CrossRef]
- Goller, C.C.; Romeo, T. Environmental Influences on Biofilm Development. Curr. Top Microbiol. Immunol. 2008, 322, 37–66. [Google Scholar] [CrossRef]
- McDermott, P.F.; Walker, R.D.; White, D.G. Antimicrobials: Modes of Action and Mechanisms of Resistance. Int. J. Toxicol. 2003, 22, 135–143. [Google Scholar] [CrossRef]
- Martínez, J.L.; Rojo, F. Metabolic Regulation of Antibiotic Resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A Review of Listeria Monocytogenes from Meat and Meat Products: Epidemiology, Virulence Factors, Antimicrobial Resistance and Diagnosis. Onderstepoort J. Vet. Res. 2020, 87, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pusic, P.; Sonnleitner, E.; Bläsi, U. Specific and Global RNA Regulators in Pseudomonas Aeruginosa. Int. J. Mol. Sci. 2021, 22, 8632. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Li, L.; Girodat, D.; Förstner, K.U.; Said, N.; Corcoran, C.; Śmiga, M.; Papenfort, K.; Reinhardt, R.; Wieden, H.J.; et al. In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-Coding RNA Pathways. Mol. Cell 2017, 65, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Göpel, Y.; Papenfort, K.; Reichenbach, B.; Vogel, J.; Görke, B. Targeted Decay of a Regulatory Small RNA by an Adaptor Protein for RNase E and Counteraction by an Anti-Adaptor RNA. Genes Dev. 2013, 27, 552–564. [Google Scholar] [CrossRef]
- Fritsche, T.R.; Castanheira, M.; Miller, G.H.; Jones, R.N.; Armstrong, E.S. Detection of Methyltransferases Conferring High-Level Resistance to Aminoglycosides in Enterobacteriaceae from Europe, North America, and Latin America. Antimicrob. Agents Chemother. 2008, 52, 1843–1845. [Google Scholar] [CrossRef]
- Sidoli, S.; Kulej, K.; Garcia, B.A. Why Proteomics Is Not the New Genomics and the Future of Mass Spectrometry in Cell Biology. J. Cell Biol. 2017, 216, 21. [Google Scholar] [CrossRef]
- Manafi, L.; Aliakbarlu, J.; Dastmalchi Saei, H. Antibiotic Resistance and Biofilm Formation Ability of Salmonella Serotypes Isolated from Beef, Mutton, and Meat Contact Surfaces at Retail. J. Food Sci. 2020, 85, 2516–2522. [Google Scholar] [CrossRef]
- Hur, J.; Choi, Y.Y.; Park, J.H.; Jeon, B.W.; Lee, H.S.; Kim, A.R.; Lee, J.H. Antimicrobial Resistance, Virulence-Associated Genes, and Pulsed-Field Gel Electrophoresis Profiles of Salmonella Enterica Subsp. Enterica Serovar Typhimurium Isolated from Piglets with Diarrhea in Korea. Can. J. Vet. Res. 2011, 75, 49. [Google Scholar]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin Resistance in Salmonella and Escherichia Coli Isolates from a Pig Farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef]
- Coldham, N.G.; Randall, L.P.; Piddock, L.J.V.; Woodward, M.J. Effect of Fluoroquinolone Exposure on the Proteome of Salmonella Enterica Serovar Typhimurium. J. Antimicrob. Chemother. 2006, 58, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, W.; Zhang, R.; Xu, J.; Wang, R.; Wang, L.; Zhao, X.; Li, J. First Acetyl-Proteome Profiling of Salmonella Typhimurium Revealed Involvement of Lysine Acetylation in Drug Resistance. Vet. Microbiol. 2018, 226, 1–8. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Randall, L.P.; Webber, M.; Piddock, L.J.V.; Humphrey, T.J.; Woodward, M.J.; Coldham, N.G. Phenotypic and Proteomic Characterization of Multiply Antibiotic-Resistant Variants of Salmonella Enterica Serovar Typhimurium Selected Following Exposure to Disinfectants. Appl. Environ. Microbiol. 2008, 74, 1508. [Google Scholar] [CrossRef]
- Qi, C.; Sun, F.; Wei, Q.; Xu, J.; Li, R.; Zhang, L.; Lu, F.; Jiang, X.; Fu, H.; Zhang, C.; et al. Quantitative Phosphoproteomics Reveals the Effect of BaeSR and AcrB Genes on Protein Phosphorylation in Salmonella Enterica Serovar Typhimurium. Res. Microbiol. 2022, 173, 103886. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Omar, M.; Nagaraja, K.V.; Goyal, S.M.; Vidovic, S. Novel Insight into the Effects of Cpxr on Salmonella Enteritidis Cells during the Chlorhexidine Treatment and Non-Stressful Growing Conditions. Int. J. Mol. Sci. 2021, 22, 8938. [Google Scholar] [CrossRef] [PubMed]
- Fillgrove, K.L.; Pakhomova, S.; Schaab, M.R.; Newcomer, M.E.; Armstrong, R.N. Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria Monocytogenes. Biochemistry 2007, 46, 8110–8120. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, T.M.; Tang, S.; Wiedmann, M.; Boor, K.J. Nisin Resistance of Listeria Monocytogenes Is Increased by Exposure to Salt Stress and Is Mediated via LiaR. Appl. Environ. Microbiol. 2013, 79, 5682–5688. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, S.; Bender, J.K.; Klare, I.; Halbedel, S.; Grohmann, E.; Szewzyk, U.; Werner, G. Tigecycline Resistance in Clinical Isolates of Enterococcus Faecium Is Mediated by an Upregulation of Plasmid-Encoded Tetracycline Determinants Tet(L) and Tet(M). J. Antimicrob. Chemother. 2016, 71, 871–881. [Google Scholar] [CrossRef]
- Kode, D.; Nannapaneni, R.; Chang, S. Low-Level Tolerance to Antibiotic Trimethoprim in QAC-Adapted Subpopulations of Listeria Monocytogenes. Foods 2021, 10, 1800. [Google Scholar] [CrossRef]
- Yan, S.; Li, M.; Luque-Sastre, L.; Wang, W.; Hu, Y.; Peng, Z.; Dong, Y.; Gan, X.; Nguyen, S.; Anes, J.; et al. Susceptibility (Re)-Testing of a Large Collection of Listeria Monocytogenes from Foods in China from 2012 to 2015 and WGS Characterization of Resistant Isolates. J. Antimicrob. Chemother. 2019, 74, 1786–1794. [Google Scholar] [CrossRef]
- Kovacevic, J.; Ziegler, J.; Walecka-Zacharska, E.; Reimer, A.; Kitts, D.D.; Gilmour, M.W. Tolerance of Listeria Monocytogenes to Quaternary Ammonium Sanitizers Is Mediated by a Novel Efflux Pump Encoded by EmrE. Appl. Environ. Microbiol. 2016, 82, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Olsen, R.H.; Shi, L.; Ye, L.; He, J.; Meng, H. Characterization of a Plasmid Carrying Cat, ErmB and TetS Genes in a Foodborne Listeria Monocytogenes Strain and Uptake of the Plasmid by Cariogenic Streptococcus Mutans. Int. J. Food Microbiol. 2016, 238, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Lanza, V.F.; Duval, M.; Coque, T.M. Ecogenetics of Antibiotic Resistance in Listeria Monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk-Balska, A.; Markiewicz, Z. The Intrinsic Cephalosporin Resistome of Listeria Monocytogenes in the Context of Stress Response, Gene Regulation, Pathogenesis and Therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic Characterization of Antibiotic Resistance in Listeria and Production of Antimicrobial and Virulence Factors. Int. J. Mol. Sci. 2021, 22, 8141. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Suárez, J.V.; Ortiz, S.; López-Alonso, V. Potential Impact of the Resistance to Quaternary Ammonium Disinfectants on the Persistence of Listeria Monocytogenes in Food Processing Environments. Front. Microbiol. 2016, 7, 638. [Google Scholar] [CrossRef] [PubMed]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and Antibiotic Resistance in Pseudomonas Spp. from Drinking Water. Sci. Total Environ. 2012, 426, 366–374. [Google Scholar] [CrossRef]
- Ranjan, V.K.; Mukherjee, S.; Basak, C.; Chakraborty, R. Abundance of New Delhi Metallo-β-Lactamase-Producing Acinetobacter, Escherichia, Proteus, and Pseudomonas Spp. in Mahananda and Karala Rivers of India. Microb. Drug Resist. 2021, 27, 1603–1615. [Google Scholar] [CrossRef]
- Ghosh, A.; Saran, N.; Saha, S. Survey of Drug Resistance Associated Gene Mutations in Mycobacterium Tuberculosis, ESKAPE and Other Bacterial Species. Sci. Rep. 2020, 10, 8957. [Google Scholar] [CrossRef]
- Meng, L.; Liu, H.; Lan, T.; Dong, L.; Hu, H.; Zhao, S.; Zhang, Y.; Zheng, N.; Wang, J. Antibiotic Resistance Patterns of Pseudomonas Spp. Isolated From Raw Milk Revealed by Whole Genome Sequencing. Front. Microbiol. 2020, 11, 1005. [Google Scholar] [CrossRef]
- Aoike, N.; Saga, T.; Sakata, R.; Yoshizumi, A.; Kimura, S.; Iwata, M.; Yoshizawa, S.; Sugasawa, Y.; Ishii, Y.; Yamaguchi, K.; et al. Molecular Characterization of Extraintestinal Escherichia Coli Isolates in Japan: Relationship between Sequence Types and Mutation Patterns of Quinolone Resistance-Determining Regions Analyzed by Pyrosequencing. J. Clin. Microbiol. 2013, 51, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Gatzeva-Topalova, P.Z.; May, A.P.; Sousa, M.C. Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance. Structure 2005, 13, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.A.; Skurnik, M. Temperature-Regulated Efflux Pump/Potassium Antiporter System Mediates Resistance to Cationic Antimicrobial Peptides in Yersinia. Mol. Microbiol. 2000, 37, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The Tetracycline Resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Mima, T.; Joshi, S.; Gomez-Escalada, M.; Schweizer, H.P. Identification and Characterization of TriABC-OpmH, a Triclosan Efflux Pump of Pseudomonas Aeruginosa Requiring Two Membrane Fusion Proteins. J. Bacteriol. 2007, 189, 7600–7609. [Google Scholar] [CrossRef]
- Muratovic, A.Z.; Hagstöm, T.; Rosén, J.; Granelli, K.; Hellenäs, K.E. Quantitative Analysis of Staphylococcal Enterotoxins A and B in Food Matrices Using Ultra High-Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MS/MS). Toxins 2015, 7, 3637. [Google Scholar] [CrossRef]
- De Groote, V.N.; Verstraeten, N.; Fauvart, M.; Kint, C.I.; Verbeeck, A.M.; Beullens, S.; Cornelis, P.; Michiels, J. Novel Persistence Genes in Pseudomonas Aeruginosa Identified by High-Throughput Screening. FEMS Microbiol. Lett. 2009, 297, 73–79. [Google Scholar] [CrossRef]
- Erdmann, J.; Thöming, J.G.; Pohl, S.; Pich, A.; Lenz, C.; Häussler, S. The Core Proteome of Biofilm-Grown Clinical Pseudomonas Aeruginosa Isolates. Cells 2019, 8, 1129. [Google Scholar] [CrossRef]
- Bao, K.D.; Letellier, A.; Beaudry, F. Analysis of Staphylococcus Enterotoxin B Using Differential Isotopic Tags and Liquid Chromatography Quadrupole Ion Trap Mass Spectrometry. Biomed. Chromatogr. 2012, 26, 1049–1057. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Zolla, L. We Are What We Eat: Food Safety and Proteomics. J. Proteome Res. 2012, 11, 26–36. [Google Scholar] [CrossRef]
- Andjelkovic, M.; Tsilia, V.; Rajkovic, A.; de Cremer, K.; Van Loco, J. Application of LC-MS/MS MRM to Determine Staphylococcal Enterotoxins (SEB and SEA) in Milk. Toxins 2016, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Ilgisonis, E.V.; Kopylov, A.T.; Ponomarenko, E.A.; Poverennaya, E.V.; Tikhonova, O.V.; Farafonova, T.E.; Novikova, S.; Lisitsa, A.V.; Zgoda, V.G.; Archakov, A.I. Increased Sensitivity of Mass Spectrometry by Alkaline Two-Dimensional Liquid Chromatography: Deep Cover of the Human Proteome in Gene-Centric Mode. J. Proteome Res. 2018, 17, 4258–4266. [Google Scholar] [CrossRef] [PubMed]
- Dupré, M.; Gilquin, B.; Fenaille, F.; Feraudet-Tarisse, C.; Dano, J.; Ferro, M.; Simon, S.; Junot, C.; Brun, V.; Becher, F. Multiplex Quantification of Protein Toxins in Human Biofluids and Food Matrices Using Immunoextraction and High-Resolution Targeted Mass Spectrometry. Anal. Chem. 2015, 87, 8473–8480. [Google Scholar] [CrossRef] [PubMed]
- Afzaal, M.; Saeed, F.; Hussain, M.; Shahid, F.; Siddeeg, A.; Al-Farga, A. Proteomics as a Promising Biomarker in Food Authentication, Quality and Safety: A Review. Food Sci. Nutr. 2022, 10, 2333–2346. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesh, D.S.; Kannegundla, U.; Reddy, R.K. Application of Proteomic Tools in Food Quality and Safety. Adv. Anim. Vet. Sci. 2017, 5, 213–225. [Google Scholar]
- Stryiński, R.; Łopieńska-Biernat, E.; Carrera, M. Proteomic Insights into the Biology of the Most Important Foodborne Parasites in Europe. Foods 2020, 9, 1403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).