Abstract
We aimed to investigate neutralizing antibody titers (NtAbT) to the P.1 and B.1 SARS-CoV-2 variants in a cohort of healthy health care workers (HCW), including 20 previously infected individuals tested at baseline (BLinf, after a median of 298 days from diagnosis) and 21 days after receiving one vaccine dose (D1inf) and 15 uninfected subjects tested 21 days after the second-dose vaccination (D2uninf). All the subjects received BNT162b2 vaccination. D1inf NtAbT increased significantly with respect to BLinf against both B.1 and P.1 variants, with a fold-change significantly higher for P.1. D1inf NtAbT were significantly higher than D2uninf NtAbT, against B.1 and P.1. NtAbT against the two strains were highly correlated. P.1 NtAbT were significantly higher than B.1 NtAbT. This difference was significant for post-vaccination sera in infected and uninfected subjects. A single-dose BNT162b2 vaccination substantially boosted the NtAb response to both variants in the previously infected subjects. NtAb titers to B.1 and P.1 lineages were highly correlated, suggesting substantial cross-neutralization. Higher titers to the P.1 than to the B.1 strain were driven by the post-vaccination titers, highlighting that cross-neutralization can be enhanced by vaccination.
1. Introduction
Administration of a variety of SARS-CoV-2 vaccines is proceeding at different paces in diverse geographical areas. The key question on the rate and durability of protection with the different vaccines has been further complicated by the emergence of fast-spreading variants, including the United Kingdom (B.1.1.7, alpha), South Africa (B.1.351, beta), and Brazil (P.1, gamma) lineages [1]. Indeed, SARS-CoV-2 variants escaping vaccine-induced protection have the potential to re-ignite virus transmission even in regions with optimal vaccine coverage. The first approved SARS-CoV-2 vaccines BNT162b2 and mRNA1273 have been administered to millions of people, and both are based on mRNA coding for the full-length prefusion spike glycoprotein derived from the original SARS-CoV-2 strain (lineage B). A series of small studies based on different methods for assessment of virus-neutralizing antibody (NtAb) have shown that the B.1.1.7 (alpha) variant is effectively neutralized by convalescent sera from individuals recovering from first-wave wild-type SARS-CoV-2 infection as well as by sera from BNT162b2 or mRNA1273 vaccine recipients, while neutralization of the B.1.351 (beta) variant appears to be weaker, although generally still robust [2,3,4,5,6].
Fewer and less consistent data have been obtained with the P.1 (gamma) variant. The P.1 (gamma) variant was identified for the first time in the city of Manaus, North Brazil, in December 2020 and further in passengers from Brazil tested at an airport nearby Tokyo [7]. Variant spike protein substitutions are L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, and T1027I [8]. P.1 (gamma) was defined a variant of concern because it shared mutations with other variants of concern and because of its transmissibility: a two-category dynamical model integrating genomic and mortality data estimated that P.1 (gamma) could be between 1.7 and 2.4 times more transmissible in Manaus than non-P.1 lineages [9]; moreover P.1 (gamma) led to re-infection after a previous SARS-CoV-2 infection [10]. In another cohort of individuals who received two doses of BNT162b2 or mRNA1273 vaccines, NtAb responses were also significantly decreased to the P.1 (gamma) strain, particularly in BNT162b2 vaccine recipients [2]. Interestingly, the six vaccinees reporting prior COVID-19 had high NtAb titers to most variants and exhibited substantial cross-neutralization even to the distantly related 2002–2003 SARS-CoV and pre-emergent bat-derived WIV1-CoV. This suggests that vaccination following prior infection substantially increases the breadth of cross-reactive NtAb. Similarly, Lustig et al. showed that one BNT162b2 vaccine dose increased neutralizing activity by 2 orders of magnitude against all variants tested (B.1.1.7 or alpha, B.1.351 or beta, and P.1 or gamma) in all of six previously infected health care workers (HCW) analyzed before and after vaccination [11].
2. Materials and Methods
To further investigate the role of BNT162b2 vaccination in NtAb titers to the P.1 variant, we studied a cohort of healthy HCW, including 20 previously infected individuals tested at baseline (BLinf) and 21 days after receiving one vaccine dose (D1inf) and 15 uninfected subjects tested 21 days after the second-dose vaccination (D2uninf). The previously infected group had a median age of 46 (37–52) years, included 70% females, had a laboratory diagnosis of SARS-CoV-2 infection in the Veneto region during the first outbreak in March–April 2020 when the original B.1 lineage was highly prevalent, and received a single-dose vaccination after a median of 298 (283–304) days from diagnosis. All the previously infected subjects enrolled were asymptomatic or had mild disease [12]: the diagnosis was made because of clinical suspicion or in the context of the hospital surveillance program. Eight of the twenty patients had mild disease, all with fever. A detailed description is reported in Table 1.

Table 1.
Description of signs and/or symptoms of the health care workers with mild COVID-19.
The uninfected group included HCW from the same region, had a median age of 49 (30–58) years with 67% females, and completed the two-dose vaccination schedule in January–February 2021. NtAb to the live virus belonging to the B.1 and P.1 lineages, as assessed by NGS (submitted to GISAID, accession number EPI_ISL_2472918 for P.1 and EPI_ISL_2472896 for B.1), were titrated in duplicate by testing two-fold serial dilutions of sera with 100 TCID50 of the corresponding SARS-CoV-2 strain in Vero E6 cells with automated measurement of cell viability by the Cell-titer Glo 2.0 system in a GloMax Discover luciferase plate reader (Promega, Madison, WI, USA) [13,14,15]. The NtAb titer (ID50) was defined as the reciprocal value of the sample dilution that showed a 50% protection from the virus-induced cytopathic effect. Each run included an uninfected control, an infected control, and the virus back-titration to confirm the virus inoculum. NtAb titers were expressed as median (IQR), and the non-parametric Wilcoxon signed-rank sum test and Mann–Whitney test were used to analyze changes in paired and unpaired data, respectively. Spearman analysis was used to measure the correlation between NtAb titers to the two viral strains. Analyses were run in IBM SPSS Statistics, version 20 (IBM Corp., Armonk, NY, USA) and all p-values were 2-sided.
Written informed consent was obtained from all the HCW to be enrolled in a neutralizing antibody (NtAb) follow-up study, as approved by the comitato per la sperimentazione clinica di Treviso e Belluno (protocol code 812/2020).
3. Results
In the previously infected patient group, D1inf NtAb titers increased significantly with respect to BLinf against both the B.1 (median values 1706 (993–3186) vs. 38 (16–67); p < 0.001) and the P.1 (median values 4087 (3324–5053) vs. 66 (30–129); p < 0.001) variant, with a fold-change significantly higher for the P.1 variant vs. the B.1 wild type (94 (29–134) vs. 44 (15–70); p = 0.03).
At 20 days post-vaccination, D1inf NtAb titers were significantly higher than D2uninf NtAb titers against both the B.1 (median values 1706 (993–3186) vs. 190 (119–302); p < 0.0001) and the P.1 (median values 4087 (3324–5053) vs. 288 (147–904) p < 0.0001) variant. Overall, NtAb titers against the two strains were highly correlated (rho = 0.924, p < 0.001). However, P.1 NtAb titers were significantly higher with respect to B.1 NtAb titers (median 394 (73–368) vs. 225 (75–1227), p < 0.001). Interestingly, this difference was significant for post-vaccination data in infected and uninfected subjects (D1inf: 4087 (3227–5125) vs. 1706 (993–3186), p < 0.001; D2uninf 288 (147–904) vs. 190 (119–302), p = 0.02) but not for BLinf (66 (29–130) vs. 38 (16–167), p = 0.13) (Figure 1).
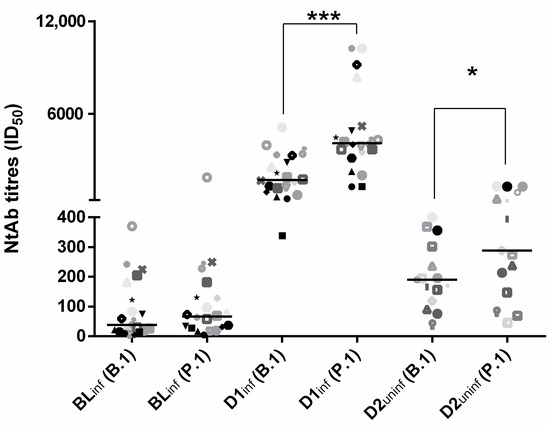
Figure 1.
Neutralizing antibody titers to the original B.1 lineage and the P.1 SARS-CoV-2 variant in 20 infected subjects tested at baseline (BLinf) and 21 days following single-dose BNT162b2 vaccination (D1inf) and in 15 uninfected subjects tested 21 days following second-dose BNT162b2 vaccination (D2uninf). Data are reported as individual ID50 values and as median value at each study time. The same symbols indicate the same subjects at different time points. Asterisks indicate significance levels: *, p < 0.05, ***, p < 0.001.
Outcomes of studies focusing on P.1 and BNT162b2 are summarized in Table 2.

Table 2.
Description of the studies focusing on the neutralizing antibody response in subjects vaccinated with BNT162b2 mRNA COVID-19 vaccine.
4. Discussion
The NtAb response to SARS-CoV-2 vaccination is being increasingly investigated to determine the impact of virus variability on vaccine efficacy. The P.1 variant is of particular concern due to the resurgence of high infection rates in Brazilian areas with supposedly large prevalence of first-wave infection [26]. Various previous studies have shown that vaccination of uninfected subjects with B lineage mRNA vaccines elicits lower but measurable NtAb titers to the P.1 variant compared to the other variants [2]. In our group of 15 uninfected BNT162b2 vaccinees, the NtAb response to the P.1 variant was not decreased, or even higher, compared to the original B.1 lineage virus. Notably, in the 20 previously infected subjects included in our study, single-dose vaccination substantially boosted the NtAb response to both variants tested, supporting the data reported by Lustig et al. in six analogous patients whose sera had high NtAb titers to the B.1.1.7, B.1.351, and P.1 lineages [11]. This may support the recommendation for vaccinating previously infected patients [27]. However, the increased titer and breadth of NtAb could simply reflect the expected secondary immune response without conferring stronger immunity to secondary infection.
The NtAb titer after a single vaccine dose in previously infected subjects was higher that NtAb titer in uninfected HCW after a complete vaccination schedule: this result is in accord with data published by Mazzoni et al. [28], who had uninfected subjects tested after a shorter (7 days after the second dose) and a longer (50 days from the first vaccine dose) interval than ours (21 days after the second dose). Conversely, comparable titers were reported in the study by Gobbi et al. [29] and in that by Ebinger et al. [30]: possible explanations for the different results may be the low numerosity (15 subjects) for the first one and the racial distribution for the second one (white people were about 50%).
The more intense humoral response in SARS-CoV-2-exposed subjects seemed not related to the vaccine type administered: the anti-receptor-binding domain of SARS-CoV-2 spike protein antibody titers was significantly higher in HCWs with a previous natural infection 4 weeks after the second dose of inactivated SARS-CoV-2 vaccine (CoronaVac) [31]. Of note, we should take into account the risk of an excess of immune activation, followed by switch-off of the response for antigen exhaustion: this mechanism may explain the reduced antibody response after the second vaccine dose in subjects with a previous infection [28,32]. Finally, the extreme heterogeneity of the methods used in the different studies, commercial or in-house methods, using recombinant viruses or authentic viral strains, as in our case, often make this evidence poorly comparable.
NtAb titers to B.1 and P.1 lineages were highly correlated in the whole case file. Intriguingly, paired analysis revealed higher titers to the P.1 than to the B.1 strain. This overall effect was driven by the post-vaccination titers, highlighting that. Interestingly, as recently published, patients infected with the B.1.351.V2 South African variant had higher NtAb titers to the related B.1.351.V3 variant than to the infecting B.1.351.V2 strain [33] and uninfected recipients of the BNT162b2 vaccine responded better to a recombinant virus mimicking the B.1.1.7 variant than to the vaccine strain [6]. These data show that SARS-CoV-2 variants may be well controlled following natural or artificial immunization with a different variant [34]. The characteristics of the antigenicity of the P.1 variant are not completely known [35]: this fact and the different study designs, viral components tested (strain isolated from a clinical sample or generated mutant, as in Chang et al. [36]), and commercial or in-house methods used to evaluate different kinds of antibody titers may explain the multifaceted response to the P.1 variant with respect to other SARS-CoV-2 strains. Gidari et al. [17] described a post-vaccinal NtAb titer to B1 and P.1 specular to ours, and sera from vaccinated individuals showed a significant loss of neutralizing activity against P.1 but the impact was lower than that against B.1.351 [24,25,37]: these data underline the need to include SARS-CoV-2 viral sequencing of PCR-positive COVID-19 subjects in the diagnostic workflow because variant diffusion may reduce vaccine efficacy.
The strength of our work lies in the homogeneity of the cohort, in the history of mild or asymptomatic infection, in the long interval from infection to pre-vaccine evaluation, and in the use of two authentic viral isolates into a classical neutralization assay. It must be noted that a number of factors can complicate the interpretation of NtAb data, including ethnicity, use of different test virus strains, and the NtAb titration technology, particularly the live virus vs. pseudovirus or spike recombinant format.
While most studies appear to support a clinically valuable breadth of the NtAb response irrespective of the infecting or vaccine virus variant, surveillance as well as development of reference isolates and technical standards remain mandatory to manage the impact of ongoing SARS-CoV-2 variability on the control of infection.
Author Contributions
Conceptualization, S.G.P.; methodology, M.Z., I.V., A.B. and F.D.; validation, I.V., S.G.P. and M.Z.; formal analysis, M.Z. and I.V.; resources, F.G. and R.S.; data curation, S.G.P.; writing—original draft preparation, M.Z. and S.G.P.; writing—review and editing, M.B., M.Z. and S.G.P.; visualization, I.V.; supervision, S.G.P.; project administration, D.Z. and M.B.; funding acquisition, S.G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Padova (grant nos. DOR-2019 and DOR-2020 to S.G.P.).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the comitato per la sperimentazione clinica di Treviso e Belluno (protocol code 812/2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Raw data can be made available by the corresponding author upon reasonable request.
Acknowledgments
We thank Alessia Lai (University of Milan) and Daniela Francisci (University of Perugia) for providing the anonymized B.1 and P.1 isolates used in the study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- McCormick, K.D.; Jacobs, J.L.; Mellors, J.W. The emerging plasticity of SARS-CoV-2. Science 2021, 371, 1306–1308. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383. [Google Scholar] [CrossRef] [PubMed]
- Kuzmina, A.; Khalaila, Y.; Voloshin, O.; Keren-Naus, A.; Boehm-Cohen, L.; Raviv, Y.; Shemer-Avni, Y.; Rosenberg, E.; Taube, R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host. Microbe 2021, 29, 522–528.e2. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Cai, H.; Sarkar, R.; Chen, W.; Cutler, M.; et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021, 384, 1466–1468. [Google Scholar] [CrossRef]
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Liu, Y.; Liu, J.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D.; et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621. [Google Scholar] [CrossRef]
- Vasireddy, D.; Vanaparthy, R.; Mohan, G.; Malayala, S.V.; Atluri, P. Review of COVID-19 Variants and COVID-19 Vaccine Efficacy: What the Clinician Should Know? J. Clin. Med. Res. 2021, 13, 317–325. [Google Scholar] [CrossRef]
- US COVID-19 Cases Caused by Variants. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html (accessed on 14 August 2021).
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Romano, C.M.; Felix, A.C.; Paula, A.V.; Jesus, J.G.; Andrade, P.S.; Cândido, D.; Oliveira, F.M.; Ribeiro, A.C.; Silva, F.C.D.; Inemami, M.; et al. SARS-CoV-2 reinfection caused by the P.1 lineage in Araraquara city, Sao Paulo State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e36. [Google Scholar] [CrossRef]
- Lustig, Y.; Nemet, I.; Kliker, L.; Zuckerman, N.; Yishai, R.; Alroy-Preis, S.; Mendelson, E.; Mandelboim, M. Neutralizing Response against Variants after SARS-CoV-2 Infection and One Dose of BNT162b2. N. Engl. J. Med. 2021, 384, 2453–2454. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/ (accessed on 14 August 2021).
- Vicenti, I.; Gatti, F.; Scaggiante, R.; Boccuto, A.; Zago, D.; Basso, M.; Dragoni, F.; Zazzi, M.; Parisi, S.G. Single-dose BNT162b2 mRNA COVID-19 vaccine significantly boosts neutralizing antibody response in health care workers recovering from asymptomatic or mild natural SARS-CoV-2 infection. Int. J. Infect. Dis. 2021, 108, 176–178. [Google Scholar] [CrossRef]
- Vicenti, I.; Gatti, F.; Scaggiante, R.; Boccuto, A.; Zago, D.; Basso, M.; Dragoni, F.; Zazzi, M.; Parisi, S.G. Time course of neutralizing antibody in healthcare workers with mild or asymptomatic COVID-19 infection. Open. Forum. Infect. Dis. 2021, in press. [Google Scholar] [CrossRef]
- Vicenti, I.; Gatti, F.; Scaggiante, R.; Boccuto, A.; Zago, D.; Basso, M.; Dragoni, F.; Parisi, S.G.; Zazzi, M. The second dose of the BNT162b2 mRNA vaccine does not boost SARS-CoV-2 neutralizing antibody response in previously infected subjects. Infection 2021, 1–3. [Google Scholar] [CrossRef]
- Zani, A.; Caccuri, F.; Messali, S.; Bonfanti, C.; Caruso, A. Serosurvey in BNT162b2 vaccine-elicited neutralizing antibodies against authentic B.1, B.1.1.7, B.1.351, B.1.525 and P.1 SARS-CoV-2 variants. Emerg. Microbes Infect. 2021, 10, 1241–1243. [Google Scholar] [CrossRef]
- Gidari, A.; Sabbatini, S.; Bastianelli, S.; Pierucci, S.; Busti, C.; Monari, C.; Luciani Pasqua, B.; Dragoni, F.; Schiaroli, E.; Zazzi, M.; et al. Cross-neutralization of SARS-CoV-2 B.1.1.7 and P.1 variants in vaccinated, convalescent and P.1 infected. J. Infect. 2021. [Google Scholar] [CrossRef]
- Barros-Martins, J.; Hammerschmidt, S.I.; Cossmann, A.; Odak, I.; Stankov, M.V.; Morillas Ramos, G.; Dopfer-Jablonka, A.; Heidemann, A.; Ritter, C.; Friedrichsen, M.; et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat. Med. 2021. [Google Scholar] [CrossRef]
- Lustig, Y.; Zuckerman, N.; Nemet, I.; Atari, N.; Kliker, L.; Regev-Yochay, G.; Sapir, E.; Mor, O.; Alroy-Preis, S.; Mendelson, E.; et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Eurosurveillance 2021, 26, 2100557. [Google Scholar] [CrossRef]
- Collier, D.A.; Ferreira, I.A.T.M.; Kotagiri, P.; Datir, R.P.; Lim, E.Y.; Touizer, E.; Meng, B.; Abdullahi, A. CITIID-NIHR BioResource COVID-19 Collaboration.; Elmer A.; et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature 2021, 596, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Stankov, M.V.; Cossmann, A.; Bonifacius, A.; Dopfer-Jablonka, A.; Ramos, G.M.; Gödecke, N.; Scharff, A.Z.; Happle, C.; Boeck, A.L.; Tran, A.T.; et al. Humoral and cellular immune responses against SARS-CoV-2 variants and human coronaviruses after single BNT162b2 vaccination. Clin. Infect. Dis. 2021, ciab555. [Google Scholar] [CrossRef] [PubMed]
- Leier, H.C.; Bates, T.A.; Lyski, Z.L.; McBride, S.K.; Lee, D.X.; Coulter, F.J.; Goodman, J.R.; Lu, Z.; Curlin, M.E.; Messer, W.B.; et al. Previously infected vaccinees broadly neutralize SARS-CoV-2 variants. medRxiv 2021. [Google Scholar] [CrossRef]
- Anichini, G.; Terrosi, C.; Gori Savellini, G.; Gandolfo, C.; Franchi, F.; Cusi, M.G. Neutralizing antibody response of vaccinees to SARS-CoV-2 variants. Vaccines 2021, 9, 517. [Google Scholar] [CrossRef] [PubMed]
- Caniels, T.G.; Bontjer, I.; van der Straten, K.; Poniman, M.; Burger, J.A.; Appelman, B.; Lavell, A.H.A.; Oomen, M.; Godeke, G.; Valle, C.; et al. Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and 1 vaccination. medRxiv 2021. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the Brazilian P.1 strain of SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Marot, S.; Malet, I.; Leducq, V.; Abdi, B.; Teyssou, E.; Soulie, C.; Wirden, M.; Rodriguez, C.; Fourati, S.; Pawlotsky, J.M.; et al. Neutralization heterogeneity of United Kingdom and South-African SARS-CoV-2 variants in BNT162b2-vaccinated or convalescent COVID-19 healthcare workers. Clin. Infect. Dis 2021, in press. [Google Scholar]
- Mazzoni, A.; Di Lauria, N.; Maggi, L.; Salvati, L.; Vanni, A.; Capone, M.; Lamacchia, G.; Mantengoli, E.; Spinicci, M.; Zammarchi, L.; et al. First-dose mRNA vaccination is sufficient to reactivate immunological memory to SARS-CoV-2 in subjects who have recovered from COVID-19. J. Clin. Investig. 2021, 131, e149150. [Google Scholar] [CrossRef]
- Gobbi, F.; Buonfrate, D.; Moro, L.; Rodari, P.; Piubelli, C.; Caldrer, S.; Riccetti, S.; Sinigaglia, A.; Barzon, L. Antibody Response to the BNT162b2 mRNA COVID-19 Vaccine in Subjects with Prior SARS-CoV-2 Infection. Viruses 2021, 13, 422. [Google Scholar] [CrossRef]
- Ebinger, J.E.; Fert-Bober, J.; Printsev, I.; Wu, M.; Sun, N.; Prostko, J.C.; Frias, E.C.; Stewart, J.L.; Van Eyk, J.E.; Braun, J.G.; et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021, 27, 981–984. [Google Scholar] [CrossRef]
- Soysal, A.; Gönüllü, E.; Karabayır, N.; Alan, S.; Atıcı, S.; Yıldız, İ.; Engin, H.; Çivilibal, M.; Karaböcüoğlu, M. Comparison of immunogenicity and reactogenicity of inactivated SARS-CoV-2 vaccine (CoronaVac) in previously SARS-CoV-2 infected and uninfected health care workers. Hum. Vaccin. Immunother. 2021, 1–5. [Google Scholar] [CrossRef]
- Levi, R.; Azzolini, E.; Pozzi, C.; Ubaldi, L.; Lagioia, M.; Mantovani, A.; Rescigno, M. One dose of SARS-CoV-2 vaccine exponentially increases antibodies in individuals who have recovered from symptomatic COVID-19. J. Clin. Investig. 2021, 131, e149154. [Google Scholar] [CrossRef]
- Moyo-Gwete, T.; Madzivhandila, M.; Makhado, Z.; Ayres, F.; Mhlanga, D.; Oosthuysen, B.; Lambson, B.E.; Kgagudi, P.; Tegally, H.; Iranzadeh, A.; et al. Cross-Reactive Neutralizing Antibody Responses Elicited by SARS-CoV-2 501Y.V2 (B.1.351). N. Engl. J. Med. 2021, 384, 2161–2163. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Xia, H.; Zou, J.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Cutler, M.; Cooper, D.; Muik, A.; et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 2021, in press. [Google Scholar] [CrossRef]
- Imai, M.; Halfmann, P.J.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Chiba, S.; Watanabe, T.; Nakajima, N.; Ito, M.; Kuroda, M.; Kiso, M.; et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc. Natl. Acad. Sci. USA 2021, 118, e2106535118. [Google Scholar] [CrossRef]
- Chang, X.; Augusto, G.S.; Liu, X.; Kündig, T.M.; Vogel, M.; Mohsen, M.O.; Bachmann, M.F. BNT162b2 mRNA COVID-19 vaccine induces antibodies of broader cross-reactivity than natural infection, but recognition of mutant viruses is up to 10-fold reduced. Allergy 2021. [Google Scholar] [CrossRef]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv 2021. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).