Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes
Abstract
1. Introduction
2. Amyloidogenic Proteins
2.1. Amyloid-β Peptide
2.2. IAPP
2.3. Synuclein
2.4. Prion
3. Amyloidogenic Proteins and Model Membranes
3.1. Interaction with Model Membranes
3.2. Interaction with Small Molecules and Ions
4. Lipid-Assisted Protein Transport
5. Symmetry-Breaking Transitions of Oligomers Self-Assembling
6. Common Features, Apparent Irreproducibility and Paradoxes, A Brief Overview
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Scalisi, S.; Sciacca, M.F.M.; Zhavnerko, G.; Grasso, D.M.; Marletta, G.; La Rosa, C. Self-Assembling Pathway of HiApp Fibrils within Lipid Bilayers. ChemBioChem 2010, 11, 1856–1859. [Google Scholar] [CrossRef] [PubMed]
- Quist, A.; Doudevski, I.; Lin, H.; Azimova, R.; Ng, D.; Frangione, B.; Kagan, B.; Ghiso, J.; Lal, R. Amyloid ion channels: A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA 2005, 102, 10427–10432. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Kotler, S.A.; Brender, J.R.; Chen, J.; Lee, D.; Ramamoorthy, A. Two-Step Mechanism of Membrane Disruption by Aβ through Membrane Fragmentation and Pore Formation. Biophys. J. 2012, 103, 702–710. [Google Scholar] [CrossRef]
- Finder, V.H.; Glockshuber, R. Amyloid-β Aggregation. Neurodegener. Dis. 2007, 4, 13–27. [Google Scholar] [CrossRef]
- Thinakaran, G.; Koo, E.H. Amyloid Precursor Protein Trafficking, Processing, and Function. J. Biol. Chem. 2008, 283, 29615–29619. [Google Scholar] [CrossRef]
- Goldgaber, D.; Lerman, M.I.; Mcbride, W.; SAFFIoTrI, U.; Gajdusek, D.C. Characterization and Chromosomal Localimtion of a cDNA Encoding Brain Amyloid of Alzheimer’s Disease. Science 1987, 235, 877–880. [Google Scholar] [CrossRef]
- Cole, S.L.; Vassar, R. The Role of Amyloid Precursor Protein Processing by BACE1, the β-Secretase, in Alzheimer Disease Pathophysiology. J. Biol. Chem. 2008, 283, 29621–29625. [Google Scholar] [CrossRef] [PubMed]
- Buoso, E.; Lanni, C.; Schettini, G.; Govoni, S.; Racchi, M. β-Amyloid precursor protein metabolism: Focus on the functions and degradation of its intracellular domain. Pharmacol. Res. 2010, 62, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Sisodia, S.S. Beta-Amyloid precursor protein cleavage by a membrane-bound protease. Proc. Natl. Acad. Sci. USA 1992, 89, 6075–6079. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Güntert, P.; Döbeli, H.; Wipf, B.; Wüthrich, K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, Aβ(1-40) ox and Aβ(1-42) ox: NMR with Alzheimer peptides in aqueous solution. Eur. J. Biochem. 2001, 268, 5930–5936. [Google Scholar] [CrossRef]
- Jan, A.; Gokce, O.; Luthi-Carter, R.; Lashuel, H.A. The Ratio of Monomeric to Aggregated Forms of Aβ40 and Aβ42 Is an Important Determinant of Amyloid-β Aggregation, Fibrillogenesis, and Toxicity. J. Biol. Chem. 2008, 283, 28176–28189. [Google Scholar] [CrossRef]
- Amaro, M.; Šachl, R.; Aydogan, G.; Mikhalyov, I.I.; Vácha, R.; Hof, M. GM 1 Ganglioside Inhibits β-Amyloid Oligomerization Induced by Sphingomyelin. Angew. Chem. Int. Ed. 2016, 55, 9411–9415. [Google Scholar] [CrossRef]
- Kowall, N.W.; Mckee, A.C. In Vivo Neurotoxicity of Beta-Amyloid [β(1-40)] and the β(25-35) Fragment. Neurobiol. Aging 1992, 13, 537–542. [Google Scholar]
- Sarkar, D.; Chakraborty, I.; Condorelli, M.; Ghosh, B.; Mass, T.; Weingarth, M.; Mandal, A.K.; La Rosa, C.; Subramanian, V.; Bhunia, A. Self-Assembly and Neurotoxicity of β-Amyloid (21–40) Peptide Fragment: The Regulatory Role of GxxxG Motifs. ChemMedChem 2020, 15, 293–301. [Google Scholar] [CrossRef]
- Pannuzzo, M.; Milardi, D.; Raudino, A.; Karttunen, M.; La Rosa, C. Analytical model and multiscale simulations of Aβ peptide aggregation in lipid membranes: Towards a unifying description of conformational transitions, oligomerization and membrane damage. Phys. Chem. Chem. Phys. 2013, 15, 8940–8951. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Monaco, I.; La Rosa, C.; Milardi, D. The active role of Ca 2+ ions in Aβ-mediated membrane damage. Chem. Commun. 2018, 54, 3629–3631. [Google Scholar] [CrossRef]
- Clark, A.; Nilsson, M.R. Islet amyloid: A complication of islet dysfunction or an aetiological factor in Type 2 diabetes? Diabetologia 2004, 47, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.L.; Westermark, G.T.; Westermark, P.; Kahn, S.E. Islet Amyloid: A Critical Entity in the Pathogenesis of Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2004, 89, 3629–3643. [Google Scholar] [CrossRef] [PubMed]
- Haataja, L.; Gurlo, T.; Huang, C.J.; Butler, P.C. Islet Amyloid in Type 2 Diabetes, and the Toxic Oligomer Hypothesis. Endocr. Rev. 2008, 29, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Marzban, L. Islet amyloid polypeptide and type 2 diabetes. Exp. Gerontol. 2003, 38, 347–351. [Google Scholar] [CrossRef]
- Cooper, G.J.; Willis, A.C.; Clark, A.; Turner, R.C.; Sim, R.B.; Reid, K.B. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. USA 1987, 84, 8628–8632. [Google Scholar] [CrossRef]
- Westermark, P.; Wernstedt, C.; Wilander, E.; Hayden, D.W.; O’Brien, T.D.; Johnson, K.H. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. USA 1987, 84, 3881–3885. [Google Scholar] [CrossRef]
- Milardi, D.; Pappalardo, M.; Pannuzzo, M.; Grasso, D.M.; Rosa, C.L. The role of the Cys2-Cys7 disulfide bridge in the early steps of Islet amyloid polypeptide aggregation: A molecular dynamics study. Chem. Phys. Lett. 2008, 463, 396–399. [Google Scholar] [CrossRef]
- Cooper, G.J.; Leighton, B.; Dimitriadis, G.D.; Parry-Billings, M.; Kowalchuk, J.M.; Howland, K.; Rothbard, J.B.; Willis, A.C.; Reid, K.B. Amylin found in amyloid deposits in human type 2 diabetes mellitus may be a hormone that regulates glycogen metabolism in skeletal muscle. Proc. Natl. Acad. Sci. USA 1988, 85, 7763–7766. [Google Scholar] [CrossRef]
- Cluck, M.W.; Chan, C.Y.; Adrian, T.E. The Regulation of Amylin and Insulin Gene Expression and Secretion. Prancreas 2005, 30, 1–14. [Google Scholar]
- Gurlo, T.; Ryazantsev, S.; Huang, C.; Yeh, M.W.; Reber, H.A.; Hines, O.J.; O’Brien, T.D.; Glabe, C.G.; Butler, P.C. Evidence for Proteotoxicity in β Cells in Type 2 Diabetes. Am. J. Pathol. 2010, 176, 861–869. [Google Scholar] [CrossRef]
- Milardi, D.; Sciacca, M.F.M.; Pappalardo, M.; Grasso, D.M.; La Rosa, C. The role of aromatic side-chains in amyloid growth and membrane interaction of the islet amyloid polypeptide fragment LANFLVH. Eur. Biophys. J. 2011, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sciacca, M.F.; Pappalardo, M.; Attanasio, F.; Milardi, D.; La Rosa, C.; Grasso, D.M. Are fibril growth and membrane damage linked processes? An experimental and computational study of IAPP 12–18 and IAPP 21–27 peptides. New J. Chem. 2010, 34, 200–207. [Google Scholar] [CrossRef]
- Pannuzzo, M.; Raudino, A.; Milardi, D.; La Rosa, C.; Karttunen, M. α-helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes. Sci. Rep. 2013, 3, 2781. [Google Scholar] [CrossRef] [PubMed]
- Abedini, A.; Raleigh, D.P. Destabilization of Human IAPP Amyloid Fibrils by Proline Mutations Outside of the Putative Amyloidogenic Domain: Is There a Critical Amyloidogenic Domain in Human IAPP? J. Mol. Biol. 2006, 355, 274–281. [Google Scholar] [CrossRef]
- Pappalardo, G.; Milardi, D.; Magrì, A.; Attanasio, F.; Impellizzeri, G.; La Rosa, C.; Grasso, D.; Rizzarelli, E. Environmental Factors Differently Affect Human and Rat IAPP: Conformational Preferences and Membrane Interactions of IAPP17–29 Peptide Derivatives. Chem. A Eur. J. 2007, 13, 10204–10215. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.C.; Gnutt, D.; Gao, M.; Wärmländer, S.K.T.S.; Jarvet, J.; Gräslund, A.; Winter, R.; Ebbinghaus, S.; Strodel, B. Effects of in vivo conditions on amyloid aggregation. Chem. Soc. Rev. 2019, 48, 3946–3996. [Google Scholar] [CrossRef] [PubMed]
- Susa, A.C.; Wu, C.; Bernstein, S.L.; Dupuis, N.F.; Wang, H.; Raleigh, D.P.; Shea, J.-E.; Bowers, M.T. Defining the Molecular Basis of Amyloid Inhibitors: Human Islet Amyloid Polypeptide–Insulin Interactions. J. Am. Chem. Soc. 2014, 136, 12912–12919. [Google Scholar] [CrossRef]
- Jaikaran, E.T.A.S.; Nilsson, M.R.; Clark, A. Pancreatic β-cell granule peptides form heteromolecular complexes which inhibit islet amyloid polypeptide fibril formation. Biochem. J. 2004, 377, 709–716. [Google Scholar] [CrossRef]
- Knight, J.D.; Williamson, J.A.; Miranker, A.D. Interaction of membrane-bound islet amyloid polypeptide with soluble and crystalline insulin. Protein Sci. 2008, 17, 1850–1856. [Google Scholar] [CrossRef]
- Knight, J.D.; Miranker, A.D. Phospholipid Catalysis of Diabetic Amyloid Assembly. J. Mol. Biol. 2004, 341, 1175–1187. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Crowther, R.A.; Spillantini, M.G. Parkinson’s Disease, Dementia with Lewy Bodies, and Multiple System Atrophy as α-Synucleinopathies. Available online: https://link.springer.com/protocol/10.1385/1-59259-142-6:33 (accessed on 7 August 2020).
- Galvagnion, C. The Role of Lipids Interacting with α-Synuclein in the Pathogenesis of Parkinson’s Disease. JPD 2017, 7, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Shahmoradian, S.H.; Lewis, A.J.; Genoud, C.; Hench, J.; Moors, T.E.; Navarro, P.P.; Castaño-Díez, D.; Schweighauser, G.; Graff-Meyer, A.; Goldie, K.N.; et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 2019, 22, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Lashuel, H.A. Do Lewy bodies contain alpha-synuclein fibrils? and Does it matter? A brief history and critical analysis of recent reports. Neurobiol. Dis. 2020, 141, 104876. [Google Scholar] [CrossRef] [PubMed]
- Maroteaux, L.; Campanelli, J.; Scheller, R. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 1988, 8, 2804–2815. [Google Scholar] [CrossRef]
- Shtilerman, M.D.; Ding, T.T.; Lansbury, P.T. Molecular Crowding Accelerates Fibrillization of α-Synuclein: Could an Increase in the Cytoplasmic Protein Concentration Induce Parkinson’s Disease? Biochemistry 2002, 41, 3855–3860. [Google Scholar] [CrossRef]
- Iwai, A.; Masliah, E.; Yoshimoto, M.; Ge, N.; Fianagan, L.; Kittei, A.; Saitoh, T. The Precursor Protein of Non-Ap Component of Alzheimer’s Disease Amyloid Is a Presynaptic Protein of the Central Nervous System. Neuron 1995, 14, 467–475. [Google Scholar] [CrossRef]
- Cooper, A.A. α-Synuclein Blocks ER-Golgi Traffic and Rab1 Rescues Neuron Loss in Parkinson’s Models. Science 2006, 313, 324–328. [Google Scholar] [CrossRef]
- Burré, J. The Synaptic Function of α-Synuclein. JPD 2015, 5, 699–713. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. α-Synuclein Promotes SNARE-Complex Assembly in Vivo and in Vitro. Science 2010, 329, 5. [Google Scholar] [CrossRef]
- El-Agnaf, O.M.A.; Irvine, G.B. Review: Formation and Properties of Amyloid-like Fibrils Derived from α-Synuclein and Related Proteins. J. Struct. Biol. 2000, 130, 300–309. [Google Scholar] [CrossRef]
- Aarsland, D.; Marsh, L.; Schrag, A. Neuropsychiatric symptoms in Parkinson’s disease. Mov. Disord. 2009, 24, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Aarsland, D.; Larsen, J.P.; Lim, N.G.; Janvin, C.; Karlsen, K.; Tandberg, E.; Cummings, J.L. Range of neuropsychiatric disturbances in patients with Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1999, 67, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Lin Chua, C.E.; Tang, B.L. α—synuclein and Parkinson’s disease: The first roadblock. J. Cell. Mol. Med. 2006, 10, 828–837. [Google Scholar] [CrossRef]
- George, J.M.; Jin, H.; Woods, W.S.; Clayton, D.F. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 1995, 15, 361–372. [Google Scholar] [CrossRef]
- Beyer, K. Mechanistic aspects of Parkinson’s disease: α-synuclein and the biomembrane. Cell Biochem. Biophys. 2007, 47, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Fusco, G.; De Simone, A.; Gopinath, T.; Vostrikov, V.; Vendruscolo, M.; Dobson, C.M.; Veglia, G. Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat. Commun. 2014, 5, 3827. [Google Scholar] [CrossRef]
- Bodner, C.R.; Dobson, C.M.; Bax, A. Multiple Tight Phospholipid-Binding Modes of α-Synuclein Revealed by Solution NMR Spectroscopy. J. Mol. Biol. 2009, 390, 775–790. [Google Scholar] [CrossRef]
- Jang, H.; Connelly, L.; Teran Arce, F.; Ramachandran, S.; Kagan, B.L.; Lal, R.; Nussinov, R. Mechanisms for the Insertion of Toxic, Fibril-like β-Amyloid Oligomers into the Membrane. J. Chem. Theory Comput. 2013, 9, 822–833. [Google Scholar] [CrossRef]
- O’Leary, E.I.; Lee, J.C. Interplay between α-synuclein amyloid formation and membrane structure. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2019, 1867, 483–491. [Google Scholar] [CrossRef]
- Conway, K.A.; Lee, S.-J.; Rochet, J.-C.; Ding, T.T.; Williamson, R.E.; Lansbury, P.T. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 571–576. [Google Scholar] [CrossRef]
- Conway, K.A.; Harper, J.D.; Lansbury, P.T. Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat. Med. 1998, 4, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, R.L.; Polymeropoulos, M.H. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 1997, 6, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H. Ubiquitination of a New Form of alpha-Synuclein by Parkin from Human Brain: Implications for Parkinson’s Disease. Science 2001, 293, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000, 10, 34–39. [Google Scholar] [CrossRef]
- Ellis, R.J. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001, 11, 114–119. [Google Scholar] [CrossRef]
- Hashimoto, M.; Rockenstein, E.; Mante, M.; Mallory, M.; Masliah, E. beta-Synuclein inhibits alpha-synuclein aggregation: A possible role as an anti-parkinsonian factor. Neuron 2001, 32, 213–223. [Google Scholar] [CrossRef]
- Uversky, V.N.; Li, J.; Souillac, P.; Millett, I.S.; Doniach, S.; Jakes, R.; Goedert, M.; Fink, A.L. Biophysical Properties of the Synucleins and Their Propensities to Fibrillate: INHIBITION OF α-SYNUCLEIN ASSEMBLY BY β- AND γ-SYNUCLEINS. J. Biol. Chem. 2002, 277, 11970–11978. [Google Scholar] [CrossRef]
- Tsigelny, I.F.; Bar-On, P.; Sharikov, Y.; Crews, L.; Hashimoto, M.; Miller, M.A.; Keller, S.H.; Platoshyn, O.; Yuan, J.X.-J.; Masliah, E. Dynamics of α-synuclein aggregation and inhibition of pore-like oligomer development by β-synuclein: Modeling of α-syn oligomer formation. FEBS J. 2007, 274, 1862–1877. [Google Scholar] [CrossRef]
- Sung, Y.; Eliezer, D. Residual Structure, Backbone Dynamics, and Interactions within the Synuclein Family. J. Mol. Biol. 2007, 372, 689–707. [Google Scholar] [CrossRef]
- Sode, K.; Ochiai, S.; Kobayashi, N.; Usuzaka, E. Effect of Reparation of Repeat Sequences in the Human α-Synuclein on Fibrillation Ability. Int. J. Biol. Sci. 2007, 3, 1–7. [Google Scholar] [CrossRef]
- Prusiner, S.B. Molecular Biology of Prion Diseases. Science 1991, 252, 8. [Google Scholar] [CrossRef] [PubMed]
- Cohen, F.E.; Prusiner, S.B. Pathologic Conformations of Prion Proteins. Annu. Rev. Biochem. 1998, 67, 793–819. [Google Scholar] [CrossRef] [PubMed]
- Colby, D.W.; Prusiner, S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011, 3, a006833. [Google Scholar] [CrossRef] [PubMed]
- Abskharon, R.; Wang, F.; Wohlkonig, A.; Ruan, J.; Soror, S.; Giachin, G.; Pardon, E.; Zou, W.; Legname, G.; Ma, J.; et al. Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion. PLoS Pathog. 2019, 15, e1008139. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.-M.; Baldwin, M.; Nguyen, J.; Gasset, M.; Mehlhorn, I.; Huang, Z.; Fletterick, R.J.; Cohenu, F.E.; Prusiner, S.B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 10962–10966. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y. Molecular dynamics studies on 3D structures of the hydrophobic region PrP (109–136). Acta Biochim. Biophys. Sin. 2013, 45, 509–519. [Google Scholar] [CrossRef][Green Version]
- Garnier, J.; Osguthorpe, D.J.; Robson, B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 1978, 120, 97–120. [Google Scholar] [CrossRef]
- Riek, R.; Wider, G.; Billeter, M.; Hornemann, S.; Glockshuber, R.; Wuthrich, K. Prion protein NMR structure and familial human spongiform encephalopathies. Proc. Natl. Acad. Sci. USA 1998, 95, 11667–11672. [Google Scholar] [CrossRef]
- Donne, D.G.; Viles, J.H.; Groth, D.; Mehlhorn, I.; James, T.L.; Cohen, F.E.; Prusiner, S.B.; Wright, P.E.; Dyson, H.J. Structure of the recombinant full-length hamster prion protein PrP(29-231): The N terminus is highly flexible. Proc. Natl. Acad. Sci. USA 1997, 94, 13452–13457. [Google Scholar] [CrossRef]
- Liu, H.; Farr-Jones, S.; Ulyanov, N.B.; Llinas, M.; Marqusee, S.; Groth, D.; Cohen, F.E.; Prusiner, S.B.; James, T.L. Solution Structure of Syrian Hamster Prion Protein rPrP(90−231). Biochemistry 1999, 38, 5362–5377. [Google Scholar] [CrossRef]
- Riek, R.; Hornemann, S.; Wider, G.; Glockshuber, R.; Wüthrich, K. NMR characterization of the full-length recombinant murine prion protein, m PrP(23-231). FEBS Lett. 1997, 413, 282–288. [Google Scholar] [CrossRef]
- Zahn, R.; Liu, A.; Luhrs, T.; Riek, R.; von Schroetter, C.; Lopez Garcia, F.; Billeter, M.; Calzolai, L.; Wider, G.; Wuthrich, K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. USA 2000, 97, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Huang, Y.; Yu, S.; Yin, S.; Plomp, M.; Qiu, S.R.; Lakshminarayanan, R.; Moradian-Oldak, J.; Sy, M.-S.; De Yoreo, J.J. A Multistage Pathway for Human Prion Protein Aggregation in Vitro: From Multimeric Seeds to β-Oligomers and Nonfibrillar Structures. J. Am. Chem. Soc. 2011, 133, 8586–8593. [Google Scholar] [CrossRef] [PubMed]
- Bocharova, O.V.; Breydo, L.; Parfenov, A.S.; Salnikov, V.V.; Baskakov, I.V. In vitro Conversion of Full-length Mammalian Prion Protein Produces Amyloid Form with Physical Properties of PrPSc. J. Mol. Biol. 2005, 346, 645–659. [Google Scholar] [CrossRef]
- Baskakov, I.V.; Bocharova, O.V. In Vitro Conversion of Mammalian Prion Protein into Amyloid Fibrils Displays Unusual Features. Biochemistry 2005, 44, 2339–2348. [Google Scholar] [CrossRef]
- Safar, J.; Wille, H.; Itri, V.; Groth, D.; Serban, H.; Torchia, M.; Cohen, F.E.; Prusiner, S.B. Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 1998, 4, 1157–1165. [Google Scholar] [CrossRef]
- Prusiner, S.B.; McKinley, M.P.; Bowman, K.A.; Bolton, D.C.; Bendheim, P.E.; Groth, D.F.; Glenner, G.G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983, 35, 349–358. [Google Scholar] [CrossRef]
- Wille, H.; Michelitsch, M.D.; Guénebaut, V.; Supattapone, S.; Serban, A.; Cohen, F.E.; Agard, D.A.; Prusiner, S.B. Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci. USA 2002, 99, 3563–3568. [Google Scholar] [CrossRef]
- Chesebro, B.; Trifilo, M.; Race, R.; Meade-White, K.; Teng, C.; Lacasse, R.; Raymond, L.; Favara, C.; Baron, G.; Priola, S.; et al. Anchorless Prion Protein Results in Infectious Amyloid Disease Without Clinical Scrapie. Science 2005, 308, 1435–1439. [Google Scholar] [CrossRef]
- Pappalardo, M.; Milardi, D.; Grasso, D.; La Rosa, C. Steered molecular dynamics studies reveal different unfolding pathways of prions from mammalian and non-mammalian species. New J. Chem. 2007, 31, 901–905. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Lansbury, P.T. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 2006, 39, 167–201. [Google Scholar] [CrossRef] [PubMed]
- Hebda, J.A.; Miranker, A.D. The Interplay of Catalysis and Toxicity by Amyloid Intermediates on Lipid Bilayers: Insights from Type II Diabetes. Annu. Rev. Biophys. 2009, 38, 125–152. [Google Scholar] [CrossRef] [PubMed]
- Hirakura, Y.; Yiu, W.W.; Yamamoto, A.; Kagan, B.L. Amyloid peptide channels: Blockade by zinc and inhibition by Congo red (amyloid channel block). Amyloid 2000, 7, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Sciacca, M.F.M.; Milardi, D.; Messina, G.M.L.; Marletta, G.; Brender, J.R.; Ramamoorthy, A.; La Rosa, C. Cations as Switches of Amyloid-Mediated Membrane Disruption Mechanisms: Calcium and IAPP. Biophys. J. 2013, 104, 173–184. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Brender, J.R.; Lee, D.-K.; Ramamoorthy, A. Phosphatidylethanolamine Enhances Amyloid Fiber-Dependent Membrane Fragmentation. Biochemistry 2012, 51, 7676–7684. [Google Scholar] [CrossRef]
- Engel, M.F.M.; Khemtemourian, L.; Kleijer, C.C.; Meeldijk, H.J.D.; Jacobs, J.; Verkleij, A.J.; de Kruijff, B.; Killian, J.A.; Hoppener, J.W.M. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc. Natl. Acad. Sci. USA 2008, 105, 6033–6038. [Google Scholar] [CrossRef]
- Zhu, M.; Li, J.; Fink, A.L. The Association of α-Synuclein with Membranes Affects Bilayer Structure, Stability, and Fibril Formation. J. Biol. Chem. 2003, 278, 40186–40197. [Google Scholar] [CrossRef]
- Sanghera, N.; Pinheiro, T.J.T. Binding of prion protein to lipid membranes and implications for prion conversion. J. Mol. Biol. 2002, 315, 1241–1256. [Google Scholar] [CrossRef]
- Terzi, E.; Holzemann, G. Alzheimer, & Amyloid Peptide 25-35: Electrostatic Interactions with Phospholipid Membranest. Biochemistry 1994, 33, 7434–7441. [Google Scholar]
- Hane, F.; Drolle, E.; Gaikwad, R.; Faught, E.; Leonenko, Z. Amyloid-β Aggregation on Model Lipid Membranes: An Atomic Force Microscopy Study. JAD 2011, 26, 485–494. [Google Scholar] [CrossRef]
- Terzi, E.; Hölzemann, G.; Seelig, J. Interaction of Alzheimer β-Amyloid Peptide(1−40) with Lipid Membranes. Biochemistry 1997, 36, 14845–14852. [Google Scholar] [CrossRef] [PubMed]
- Sciacca, M.F.; Pappalardo, M.; Milardi, D.; Grasso, D.M.; La Rosa, C. Calcium-activated membrane interaction of the islet amyloid polypeptide: Implications in the pathogenesis of type II diabetes mellitus. Arch. Biochem. Biophys. 2008, 477, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Brender, J.R.; Krishnamoorthy, J.; Messina, G.M.L.; Deb, A.; Vivekanandan, S.; Rosa, C.L.; Penner-Hahn, J.E.; Ramamoorthy, A. Zinc stabilization of prefibrillar oligomers of human islet amyloid polypeptide. Chem. Commun. 2013, 49, 3339–3341. [Google Scholar] [CrossRef] [PubMed]
- Ahyayauch, H.; de la Arada, I.; Masserini, M.E.; Arrondo, J.L.R.; Goñi, F.M.; Alonso, A. The Binding of Aβ42 Peptide Monomers to Sphingomyelin/Cholesterol/Ganglioside Bilayers Assayed by Density Gradient Ultracentrifugation. Int. J. Mol. Sci. 2020, 21, 1674. [Google Scholar] [CrossRef]
- Wakabayashi, M.; Matsuzaki, K. Ganglioside-induced amyloid formation by human islet amyloid polypeptide in lipid rafts. FEBS Lett. 2009, 583, 2854–2858. [Google Scholar] [CrossRef]
- Hoshino, T.; Mahmood, M.I.; Mori, K.; Matsuzaki, K. Binding and Aggregation Mechanism of Amyloid β-Peptides onto the GM1 Ganglioside-Containing Lipid Membrane. J. Phys. Chem. B 2013, 117, 8085–8094. [Google Scholar] [CrossRef]
- Wakabayashi, M.; Okada, T.; Kozutsumi, Y.; Matsuzaki, K. GM1 ganglioside-mediated accumulation of amyloid β-protein on cell membranes. Biochem. Biophys. Res. Commun. 2005, 328, 1019–1023. [Google Scholar] [CrossRef]
- Di Scala, C.; Troadec, J.-D.; Lelièvre, C.; Garmy, N.; Fantini, J.; Chahinian, H. Mechanism of cholesterol-assisted oligomeric channel formation by a short Alzheimer β-amyloid peptide. J. Neurochem. 2014, 128, 186–195. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Lolicato, F.; Di Mauro, G.; Milardi, D.; D’Urso, L.; Satriano, C.; Ramamoorthy, A.; La Rosa, C. The Role of Cholesterol in Driving IAPP-Membrane Interactions. Biophys. J. 2016, 111, 140–151. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, J. Cholesterol Promotes the Interaction of Alzheimer β-Amyloid Monomer with Lipid Bilayer. J. Mol. Biol. 2012, 421, 561–571. [Google Scholar] [CrossRef]
- Choo-Smith, L.-P.; Garzon-Rodriguez, W.; Glabe, C.G.; Surewicz, W.K. Acceleration of Amyloid Fibril Formation by Specific Binding of Aβ-(1–40) Peptide to Ganglioside-containing Membrane Vesicles. J. Biol. Chem. 1997, 272, 22987–22990. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.-R.; Wu, Y.; Sui, S. Cholesterol Is an Important Factor Affecting the Membrane Insertion of β-Amyloid Peptide (Aβ1–40), Which May Potentially Inhibit the Fibril Formation. J. Biol. Chem. 2002, 277, 6273–6279. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.L.; Jalali, S.; Yang, Y.; Cruz, L. Role of Cholesterol on Binding of Amyloid Fibrils to Lipid Bilayers. J. Phys. Chem. B 2020, 124, 3036–3042. [Google Scholar] [CrossRef] [PubMed]
- Chi, E.Y.; Ege, C.; Winans, A.; Majewski, J.; Wu, G.; Kjaer, K.; Lee, K.Y.C. Lipid membrane templates the ordering and induces the fibrillogenesis of Alzheimer’s disease amyloid-β peptide: Lipid Membrane Templates Aβ Fibrillogenesis. Proteins 2008, 72, 1–24. [Google Scholar] [CrossRef]
- Soong, R.; Brender, J.R.; Macdonald, P.M.; Ramamoorthy, A. Association of Highly Compact Type II Diabetes Related Islet Amyloid Polypeptide Intermediate Species at Physiological Temperature Revealed by Diffusion NMR Spectroscopy. J. Am. Chem. Soc. 2009, 131, 7079–7085. [Google Scholar] [CrossRef]
- Brender, J.R.; Lee, E.L.; Cavitt, M.A.; Gafni, A.; Steel, D.G.; Ramamoorthy, A. Amyloid Fiber Formation and Membrane Disruption are Separate Processes Localized in Two Distinct Regions of IAPP, the Type-2-Diabetes-Related Peptide. J. Am. Chem. Soc. 2008, 130, 6424–6429. [Google Scholar] [CrossRef]
- Terzi, E. Self-association of β-Amyloid Peptide (1–40) in Solution and Binding to Lipid Membranes. J. Mol. Biol. 1998, 252, 633–642. [Google Scholar] [CrossRef]
- Morillas, M.; Swietnicki, W.; Gambetti, P.; Surewicz, W.K. Membrane Environment Alters the Conformational Structure of the Recombinant Human Prion Protein. J. Biol. Chem. 1999, 274, 36859–36865. [Google Scholar] [CrossRef]
- Domanov, Y.A.; Kinnunen, P.K.J. Islet Amyloid Polypeptide Forms Rigid Lipid–Protein Amyloid Fibrils on Supported Phospholipid Bilayers. J. Mol. Biol. 2008, 376, 42–54. [Google Scholar] [CrossRef]
- Jayasinghe, S.A.; Langen, R. Lipid Membranes Modulate the Structure of Islet Amyloid Polypeptide. Biochemistry 2005, 44, 12113–12119. [Google Scholar] [CrossRef]
- Sparr, E.; Engel, M.F.M.; Sakharov, D.V.; Sprong, M.; Jacobs, J.; de Kruijff, B.; Höppener, J.W.M.; Antoinette Killian, J. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004, 577, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Bucciantini, M.; Cecchi, C. Biological Membranes as Protein Aggregation Matrices and Targets of Amyloid Toxicity. In Protein Misfolding and Cellular Stress in Disease and Aging; Bross, P., Gregersen, N., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 648, pp. 231–243. ISBN 978-1-60761-755-756. [Google Scholar]
- Sciacca, M.F.M.; Tempra, C.; Scollo, F.; Milardi, D.; La Rosa, C. Amyloid growth and membrane damage: Current themes and emerging perspectives from theory and experiments on Aβ and hIAPP. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Bokvist, M.; Lindström, F.; Watts, A.; Gröbner, G. Two Types of Alzheimer’s β-Amyloid (1–40) Peptide Membrane Interactions: Aggregation Preventing Transmembrane Anchoring Versus Accelerated Surface Fibril Formation. J. Mol. Biol. 2004, 335, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. The Aggregation and Fibrillation of α-Synuclein. Acc. Chem. Res. 2006, 39, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Ambadi Thody, S.; Mathew, M.K.; Udgaonkar, J.B. Mechanism of aggregation and membrane interactions of mammalian prion protein. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
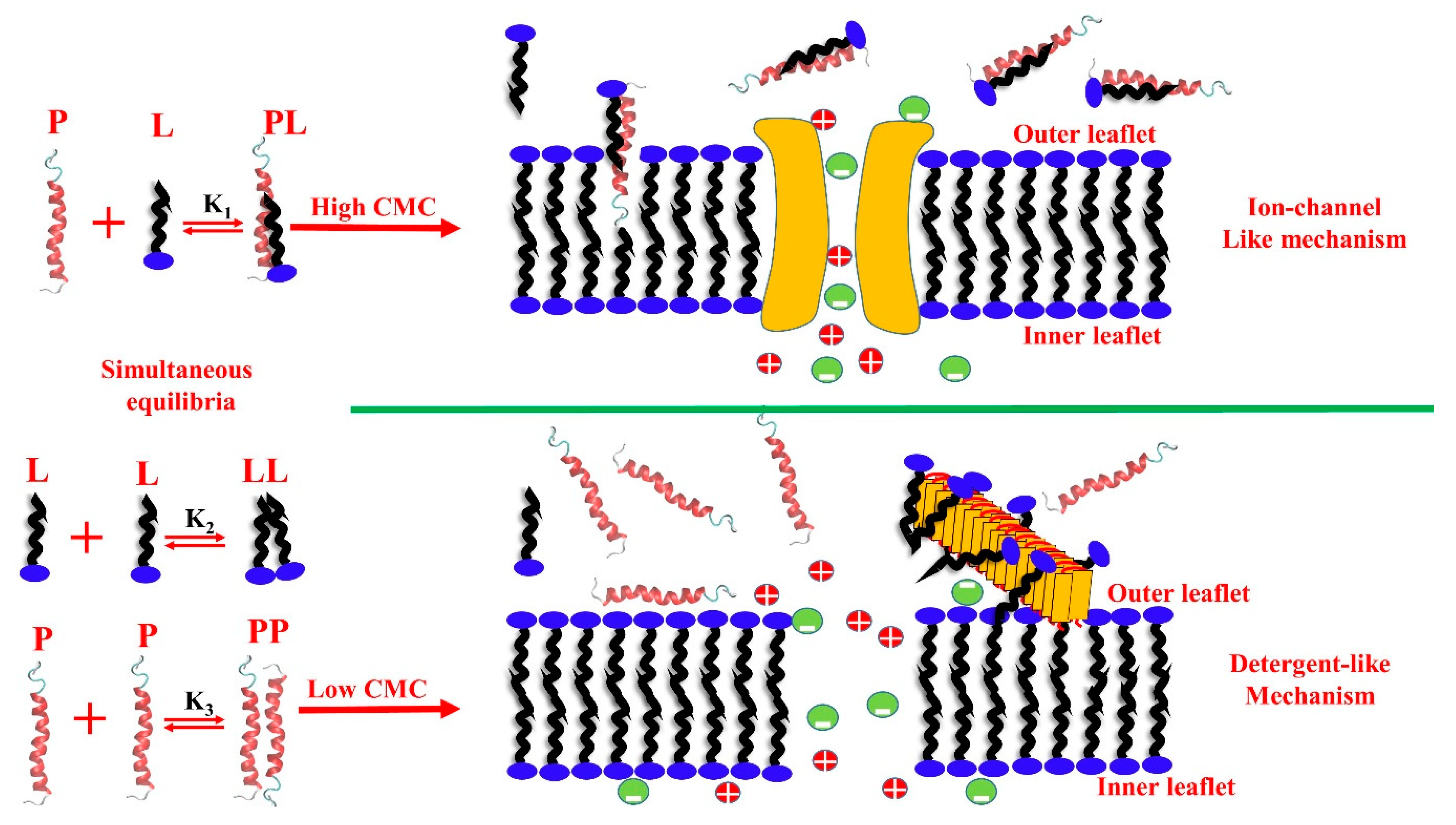
- Scollo, F.; Tempra, C.; Lolicato, F.; Sciacca, M.F.M.; Raudino, A.; Milardi, D.; La Rosa, C. Phospholipids Critical Micellar Concentrations Trigger Different Mechanisms of Intrinsically Disordered Proteins Interaction with Model Membranes. J. Phys. Chem. Lett. 2018, 9, 5125–5129. [Google Scholar] [CrossRef]
- La Rosa, C.; Scalisi, S.; Lolicato, F.; Pannuzzo, M.; Raudino, A. Lipid-assisted protein transport: A diffusion-reaction model supported by kinetic experiments and molecular dynamics simulations. J. Chem. Phys. 2016, 144, 184901. [Google Scholar] [CrossRef]
- Milardi, D.; Sciacca, M.F.M.; Randazzo, L.; Raudino, A.; La Rosa, C. The Role of Calcium, Lipid Membranes and Islet Amyloid Polypeptide in the Onset of Type 2 Diabetes: Innocent Bystanders or Partners in a Crime? Front. Endocrinol. 2014, 5. [Google Scholar] [CrossRef]
- Brender, J.R.; Hartman, K.; Nanga, R.P.R.; Popovych, N.; de la Salud Bea, R.; Vivekanandan, S.; Marsh, E.N.G.; Ramamoorthy, A. Role of Zinc in Human Islet Amyloid Polypeptide Aggregation. J. Am. Chem. Soc. 2010, 132, 8973–8983. [Google Scholar] [CrossRef]
- Hindo, S.S.; Mancino, A.M.; Braymer, J.J.; Liu, Y.; Vivekanandan, S.; Ramamoorthy, A.; Lim, M.H. Small Molecule Modulators of Copper-Induced Aβ Aggregation. J. Am. Chem. Soc. 2009, 131, 16663–16665. [Google Scholar] [CrossRef]
- Ladiwala, A.R.A.; Lin, J.C.; Bale, S.S.; Marcelino-Cruz, A.M.; Bhattacharya, M.; Dordick, J.S.; Tessier, P.M. Resveratrol Selectively Remodels Soluble Oligomers and Fibrils of Amyloid Aβ into Off-pathway Conformers. J. Biol. Chem. 2010, 285, 24228–24237. [Google Scholar] [CrossRef] [PubMed]
- Bieschke, J.; Herbst, M.; Wiglenda, T.; Friedrich, R.P.; Boeddrich, A.; Schiele, F.; Kleckers, D.; Lopez del Amo, J.M.; Grüning, B.A.; Wang, Q.; et al. Small-molecule conversion of toxic oligomers to nontoxic β-sheet–rich amyloid fibrils. Nat. Chem. Biol. 2012, 8, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Wärmländer, S.; Tiiman, A.; Abelein, A.; Luo, J.; Jarvet, J.; Söderberg, K.L.; Danielsson, J.; Gräslund, A. Biophysical Studies of the Amyloid β-Peptide: Interactions with Metal Ions and Small Molecules. ChemBioChem 2013, 14, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.I.; Yang, M.; Brender, J.R.; Subramanian, V.; Sun, K.; Joo, N.E.; Jeong, S.-H.; Ramamoorthy, A.; Kotov, N.A. Inhibition of Amyloid Peptide Fibrillation by Inorganic Nanoparticles: Functional Similarities with Proteins. Angew. Chem. Int. Ed. 2011, 50, 5110–5115. [Google Scholar] [CrossRef]
- D’Urso, L.; Condorelli, M.; Puglisi, O.; Tempra, C.; Lolicato, F.; Compagnini, G.; Rosa, C.L. Detection and characterization at nM concentration of oligomers formed by hIAPP, Aβ(1–40) and their equimolar mixture using SERS and MD simulations. Phys. Chem. Chem. Phys. 2018, 20, 20588–20596. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Romanucci, V.; Zarrelli, A.; Monaco, I.; Lolicato, F.; Spinella, N.; Galati, C.; Grasso, G.; D’Urso, L.; Romeo, M.; et al. Inhibition of Aβ Amyloid Growth and Toxicity by Silybins: The Crucial Role of Stereochemistry. ACS Chem. Neurosci. 2017, 8, 1767–1778. [Google Scholar] [CrossRef]
- Hyung, S.-J.; DeToma, A.S.; Brender, J.R.; Lee, S.; Vivekanandan, S.; Kochi, A.; Choi, J.-S.; Ramamoorthy, A.; Ruotolo, B.T.; Lim, M.H. Insights into antiamyloidogenic properties of the green tea extract (-)-epigallocatechin-3-gallate toward metal-associated amyloid- species. Proc. Natl. Acad. Sci. USA 2013, 110, 3743–3748. [Google Scholar] [CrossRef]
- Sciacca, M.F.; Chillemi, R.; Sciuto, S.; Pappalardo, M.; Rosa, C.L.; Grasso, D.; Milardi, D. Interactions of two O-phosphorylresveratrol derivatives with model membranes. Arch. Biochem. Biophys. 2012, 521, 111–116. [Google Scholar] [CrossRef]
- Sciacca, M.F.M.; Chillemi, R.; Sciuto, S.; Greco, V.; Messineo, C.; Kotler, S.A.; Lee, D.-K.; Brender, J.R.; Ramamoorthy, A.; La Rosa, C.; et al. A blend of two resveratrol derivatives abolishes hIAPP amyloid growth and membrane damage. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 1793–1802. [Google Scholar] [CrossRef]
- Marsh, D. Handbook of Lipid Bilayer, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Korshavn, K.J.; Satriano, C.; Lin, Y.; Zhang, R.; Dulchavsky, M.; Bhunia, A.; Ivanova, M.I.; Lee, Y.-H.; La Rosa, C.; Lim, M.H.; et al. Reduced Lipid Bilayer Thickness Regulates the Aggregation and Cytotoxicity of Amyloid-β. J. Biol. Chem. 2017, 292, 4638–4650. [Google Scholar] [CrossRef]
- Sunde, M.; Blake, C. The Structure of Amyloid Fibrils by Electron Microscopy and X-Ray Diffraction. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 1997; Volume 50, pp. 123–159. ISBN 978-0-12-034250-1. [Google Scholar]
- Fändrich, M. Oligomeric Intermediates in Amyloid Formation: Structure Determination and Mechanisms of Toxicity. J. Mol. Biol. 2012, 421, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, M.D.; Comellas, G.; Nieuwkoop, A.J.; Covell, D.J.; Berthold, D.A.; Kloepper, K.D.; Courtney, J.M.; Kim, J.K.; Barclay, A.M.; Kendall, A.; et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016, 23, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Vilar, M.; Chou, H.-T.; Luhrs, T.; Maji, S.K.; Riek-Loher, D.; Verel, R.; Manning, G.; Stahlberg, H.; Riek, R. The fold of -synuclein fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 8637–8642. [Google Scholar] [CrossRef] [PubMed]
- Luhrs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Dobeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid- (1-42) fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 2019, 10, 4760. [Google Scholar] [CrossRef]
- Guerrero-Ferreira, R.; Taylor, N.M.; Arteni, A.-A.; Kumari, P.; Mona, D.; Ringler, P.; Britschgi, M.; Lauer, M.E.; Makky, A.; Verasdonck, J.; et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 2019, 8, e48907. [Google Scholar] [CrossRef]
- Guerrero-Ferreira, R.; Taylor, N.M.; Mona, D.; Ringler, P.; Lauer, M.E.; Riek, R.; Britschgi, M.; Stahlberg, H. Cryo-EM structure of alpha-synuclein fibrils. eLife 2018, 7, e36402. [Google Scholar] [CrossRef]
- Van den Akker, C.C.; Deckert-Gaudig, T.; Schleeger, M.; Velikov, K.P.; Deckert, V.; Bonn, M.; Koenderink, G.H. Nanoscale Heterogeneity of the Molecular Structure of Individual hIAPP Amyloid Fibrils Revealed with Tip-Enhanced Raman Spectroscopy. Small 2015, 11, 4131–4139. [Google Scholar] [CrossRef]
- La Rosa, C.; Condorelli, M.; Compagnini, G.; Lolicato, F.; Milardi, D.; Do, T.N.; Karttunen, M.; Pannuzzo, M.; Ramamoorthy, A.; Fraternali, F.; et al. Symmetry-breaking transitions in the early steps of protein self-assembly. Eur. Biophys. J. 2020, 49, 1–17. [Google Scholar] [CrossRef]
- Iljina, M.; Garcia, G.A.; Dear, A.J.; Flint, J.; Narayan, P.; Michaels, T.C.T.; Dobson, C.M.; Frenkel, D.; Knowles, T.P.J.; Klenerman, D. Quantitative analysis of co-oligomer formation by amyloid-beta peptide isoforms. Sci. Rep. 2016, 6, 28658. [Google Scholar] [CrossRef]
- Milardi, D.; La Rosa, C.; Grasso, D.; Guzzi, R.; Sportelli, L.; Fini, C. Thermodynamics and kinetics of the thermal unfolding of plastocyanin. Eur. Biophys. J. 1998, 27, 273–282. [Google Scholar] [CrossRef]
- La Rosa, C.; Milardi, D.; Grasso, D.M.; Verbeet, M.P.; Canters, G.W.; Sportelli, L.; Guzzi, R. A model for the thermal unfolding of amicyanin. Eur. Biophys. J. 2002, 30, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Manetto, G.D.; Grasso, D.M.; Milardi, D.; Pappalardo, M.; Guzzi, R.; Sportelli, L.; Verbeet, M.P.; Canters, G.W.; La Rosa, C. The Role Played by the α-Helix in the Unfolding Pathway and Stability of Azurin: Switching Between Hierarchic and Nonhierarchic Folding. ChemBioChem 2007, 8, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, M.; Sciacca, M.; Milardi, D.; Grasso, D.; La Rosa, C. Thermodynamics of azurin folding: The role of copper ion. J. Therm. Anal. Calorim. 2008, 93, 575–581. [Google Scholar] [CrossRef]
- La Rosa, C.; Milardi, D.; Amato, E.; Pappalardo, M.; Grasso, D. Molecular mechanism of the inhibition of cytochrome c aggregation by Phe-Gly. Arch. Biochem. Biophys. 2005, 435, 182–189. [Google Scholar] [CrossRef]
- Sciacca, M.; Milardi, D.; Pappalardo, M.; La Rosa, C.; Grasso, D. Role of electrostatics in the thermal stability of ubiquitin: A combined DSC and MM study. J. Therm. Anal. Calorim. 2006, 86, 311–314. [Google Scholar] [CrossRef]
- Manetto, G.; La Rosa, C.; Grasso, D.; Milardi, D. Evaluation of thermodynamic properties of irreversible protein thermal unfolding measured by DSC. J. Therm. Anal. Calorim. 2005, 80, 263–270. [Google Scholar] [CrossRef]
- Romanucci, V.; Milardi, D.; Campagna, T.; Gaglione, M.; Messere, A.; D’Urso, A.; Crisafi, E.; La Rosa, C.; Zarrelli, A.; Balzarini, J.; et al. Synthesis, biophysical characterization and anti-HIV activity of d (TG 3 AG) quadruplexes bearing hydrophobic tails at the 5′-end. Bioorg. Med. Chem. 2014, 22, 960–966. [Google Scholar] [CrossRef]
- Khare, S.D.; Caplow, M.; Dokholyan, N.V. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2004, 101, 15094–15099. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Rumfeldt, J.A.O.; Karbassi, F.; Siddall, C.A.; Lepock, J.R.; Meiering, E.M. Calorimetric Analysis of Thermodynamic Stability and Aggregation for Apo and Holo Amyotrophic Lateral Sclerosis-associated Gly-93 Mutants of Superoxide Dismutase. J. Biol. Chem. 2006, 281, 6184–6193. [Google Scholar] [CrossRef]
- Milardi, D.; Pappalardo, M.; Grasso, D.M.; La Rosa, C. Unveiling the unfolding pathway of FALS associated G37R SOD1 mutant: A computational study. Mol. BioSyst. 2010, 6, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Lella, M.; Mahalakshmi, R. Metamorphic Proteins: Emergence of Dual Protein Folds from One Primary Sequence. Biochemistry 2017, 56, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Brender, J.R.; Krishnamoorthy, J.; Sciacca, M.F.M.; Vivekanandan, S.; D’Urso, L.; Chen, J.; La Rosa, C.; Ramamoorthy, A. Probing the Sources of the Apparent Irreproducibility of Amyloid Formation: Drastic Changes in Kinetics and a Switch in Mechanism Due to Micellelike Oligomer Formation at Critical Concentrations of IAPP. J. Phys. Chem. B 2015, 119, 2886–2896. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Abedini, A.; Wang, H.; Tu, L.-H.; Zhang, X.; Schmidt, A.M.; Raleigh, D.P. Islet amyloid polypeptide toxicity and membrane interactions. Proc. Natl. Acad. Sci. USA 2013, 110, 19279–19284. [Google Scholar] [CrossRef]
- Tomasello, M.F.; Sinopoli, A.; Attanasio, F.; Giuffrida, M.L.; Campagna, T.; Milardi, D.; Pappalardo, G. Molecular and cytotoxic properties of hIAPP17–29 and rIAPP17–29 fragments: A comparative study with the respective full-length parent polypeptides. Eur. J. Med. Chem. 2014, 81, 442–455. [Google Scholar] [CrossRef]
- La Rosa, C. Intrinsically Disordered Proteins Share a Common Molecular Mechanism in Membranes Damages: Lipid-Chaperone Hypothesis. Available online: https://www.morressier.com/article/intrinsically-disordered-proteins-share-common-molecular-mechanism-membranes-damages-lipidchaperone-hypothesis/5e736588cde2b641284ab645 (accessed on 7 August 2020).


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scollo, F.; La Rosa, C. Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes. Life 2020, 10, 144. https://doi.org/10.3390/life10080144
Scollo F, La Rosa C. Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes. Life. 2020; 10(8):144. https://doi.org/10.3390/life10080144
Chicago/Turabian StyleScollo, Federica, and Carmelo La Rosa. 2020. "Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes" Life 10, no. 8: 144. https://doi.org/10.3390/life10080144
APA StyleScollo, F., & La Rosa, C. (2020). Amyloidogenic Intrinsically Disordered Proteins: New Insights into Their Self-Assembly and Their Interaction with Membranes. Life, 10(8), 144. https://doi.org/10.3390/life10080144





