Applications of Proteomic Tools to Study Insect Vector–Plant Virus Interactions
Abstract
1. Introduction
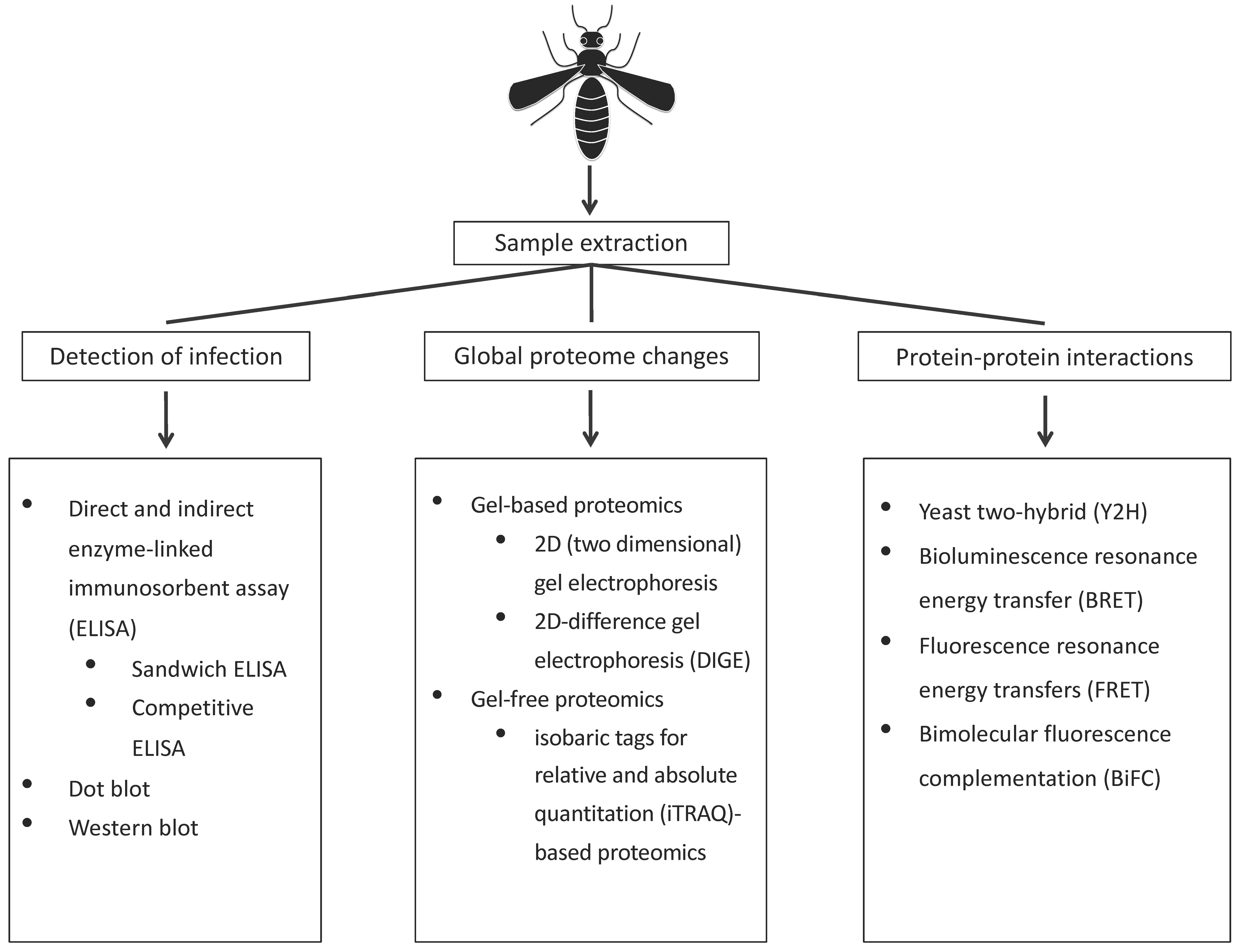
2. Proteomic Approaches to Study of Vector–Virus Interactions
2.1. Detection of Infection
2.2. Techniques to Identify Global Changes to the Vector Proteome upon Infection
2.2.1. Mass Spectrometry for Global Proteomics
2.2.2. Sample Collection and Protein Isolation for Global Proteome
2.2.3. Gel-Based Proteomics
2.2.4. Gel-Free Proteomics
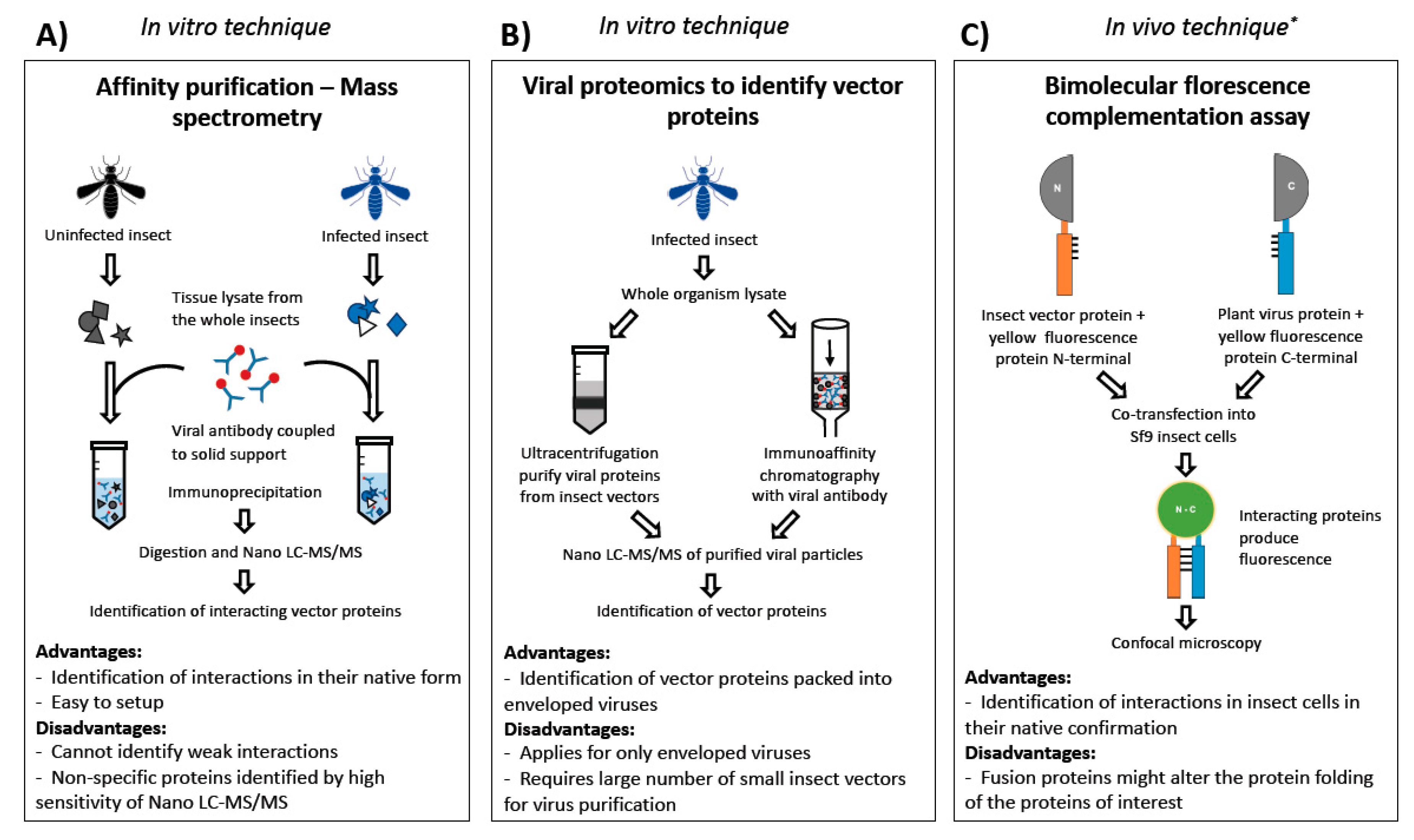
2.3. Techniques to Identify Vector Proteins Interacting with Virus Proteins
3. Combining Proteomics with Multiple ‘-Omics’-Based Techniques
4. Bioinformatic Analysis of Proteomics Data
5. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dietzgen, R.; Mann, K.; Johnson, K. Plant Virus–Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.A.; Roberts, F.M.; Russell, E.J. A comparative study of the transmission of Hyoscyamus virus 3, potato virus Y and cucumber virus 1 by the vectors Myzus persicae (Sulz), M. circumflexus (Buckton), and Macrosiphum gei (Koch). Proc. R. Soc. Lond. Ser. B-Biol. Sci. 1939, 127, 543–576. [Google Scholar] [CrossRef]
- Sylvester, E.S. Beet Yellows Virus Transmission by the Green Peach Aphid. J. Econ. Entomol. 1956, 49, 789–800. [Google Scholar] [CrossRef]
- Pirone, T.P.; Megahed, E.-S. Aphid transmissibility of some purified viruses and viral RNA’s. Virology 1966, 30, 631–637. [Google Scholar] [CrossRef]
- Plisson, C.; Uzest, M.; Drucker, M.; Froissart, R.; Dumas, C.; Conway, J.; Thomas, D.; Blanc, S.; Bron, P. Structure of the Mature P3-virus Particle Complex of Cauliflower Mosaic Virus Revealed by Cryo-electron Microscopy. J. Mol. Biol. 2005, 346, 267–277. [Google Scholar] [CrossRef]
- Blanc, S.; Drucker, M.; Uzest, M. Localizing Viruses in Their Insect Vectors. Annu. Rev. Phytopathol. 2014, 52, 403–425. [Google Scholar] [CrossRef]
- Ng, J.C.K.; Falk, B.W. Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annu. Rev. Phytopathol. 2006, 44, 183–212. [Google Scholar] [CrossRef]
- Stewart, L.R.; Medina, V.; Tian, T.; Turina, M.; Falk, B.W.; Ng, J.C.K. A Mutation in the Lettuce Infectious Yellows Virus Minor Coat Protein Disrupts Whitefly Transmission but Not In Planta Systemic Movement. J. Virol. 2010, 84, 12165–12173. [Google Scholar] [CrossRef]
- Chen, A.Y.S.; Walker, G.P.; Carter, D.; Ng, J.C.K. A virus capsid component mediates virion retention and transmission by its insect vector. Proc. Natl. Acad. Sci. USA 2011, 108, 16777–16782. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef]
- Rotenberg, D.; Jacobson, A.L.; Schneweis, D.J.; Whitfield, A.E. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015, 15, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dessau, M.; Rotenberg, D.; Rasmussen, D.A.; Whitfield, A.E. Entry of bunyaviruses into plants and vectors. In Advances in Virus Research; Kielian, M., Mettenleiter, T.C., Roossinck, M.J., Eds.; Virus Entry; Academic Press: Cambridge, MA, USA, 2019; Volume 104, pp. 65–96. [Google Scholar]
- Falk, B.W.; Tsai, J.H. Biology and Molecular Biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998, 36, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Cilia, M.; Ghanim, M. Circulative, “Nonpropagative” Virus Transmission. Adv. Virus. Res. 2014, 89, 141–199. [Google Scholar] [CrossRef] [PubMed]
- Bragard, C.; Caciagli, P.; Lemaire, O.; Lopez-Moya, J.J.; MacFarlane, S.; Peters, D.; Susi, P.; Torrance, L. Status and Prospects of Plant Virus Control Through Interference with Vector Transmission. Annu. Rev. Phytopathol. 2013, 51, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479, 278–289. [Google Scholar] [CrossRef]
- Wilson, J.R.; DeBlasio, S.L.; Alexander, M.M.; Heck, M. Looking Through the Lens of ’Omics Technologies: Insights into the Transmission of Insect Vector-borne Plant Viruses. Curr. Issues Mol. Biol. 2019, 34, 113–144. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the Microplate Method of Enzyme-Linked Immunosorbent Assay for the Detection of Plant Viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Lopez, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Gorris, M.T.; Llop, P.; Penyalver, R.; Cambra, M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef]
- Uehara-Ichiki, T.; Shiba, T.; Matsukura, K.; Ueno, T.; Hirae, M.; Sasaya, T. Detection and diagnosis of rice-infecting viruses. Front. Microbiol. 2013, 4, 289. [Google Scholar] [CrossRef]
- Crowther, J.R. The ELISA Guidebook; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar] [CrossRef]
- Hsu, H.T.; Lawson, R.H. Direct tissue blotting for detection of tomato spotted wilt virus in Impatiens. Plant Dis. 1991, 75, 292–295. [Google Scholar] [CrossRef]
- Lister, R.M.; Rochow, W.F. Detection of barley yellow dwarf virus by enzyme-linked immunosorbent assay. Microscopy 1979, 12, 18. [Google Scholar] [CrossRef]
- Olmos, A.; Cambra, M.; Dasi, M.A.; Candresse, T.; Esteban, O.; Gorris, M.T.; Asensio, M. Simultaneous detection and typing of plum pox potyvirus (PPV) isolates by heminested-PCR and PCR-ELISA. J. Virol. Methods 1997, 68, 127–137. [Google Scholar] [CrossRef]
- Stevens, M.; Hull, R.; Smith, H.G. Comparison of ELISA and RT-PCR for the detection of beet yellows closterovirus in plants and aphids. J. Virol. Methods 1997, 68, 9–16. [Google Scholar] [CrossRef]
- Ahoonmanesh, A.; Hajimorad, M.R.; Ingham, B.J.; Francki, R.I. Indirect double antibody sandwich ELISA for detecting alfalfa mosaic virus in aphids after short probes on infected plants. J. Virol. Methods 1990, 30, 271–281. [Google Scholar] [CrossRef]
- Falk, B.W.; Tsai, J.H.; Lommel, S.A. Differences in Levels of Detection for the Maize Stripe Virus Capsid and Major Non-capsid Proteins in Plant and Insect Hosts. J. Gen. Virol. 1987, 68, 1801–1811. [Google Scholar] [CrossRef]
- Gingery, R.E.; Nault, L.R.; Bradfute, O.E. Maize stripe virus: Characteristics of a member of a new virus class. Virology 1981, 112, 99–108. [Google Scholar] [CrossRef]
- Ammar, E.D.; Gingery, R.E.; Madden, L.V. Transmission efficiency of three isolates of maize stripe tenuivirus in relation to virus titre in the planthopper vector. Plant Pathol. 1995, 44, 239–243. [Google Scholar] [CrossRef]
- Mahmoud, A.; Thouvenel, J.-C.; Abol-Ela, S.E.; Sewify, G.H.; Ammar, E.D. Detection of maize yellow stripe tenui-like virus by ELISA and dot-blot tests in host plants and leafhopper vector in Egypt. Phytopathol. Mediterr. 1996, 35, 19–23. [Google Scholar]
- Ammar, E.-D.; Khlifa, E.A.; Mahmoud, A.; Abol-Ela, S.E.; Peterschmitt, M. Evidence for multiplication of the leafhopper-borne maize yellow stripe virus in its vector using ELISA and dot-blot hybridization. Arch. Virol. 2006, 152, 489–494. [Google Scholar] [CrossRef]
- Reynaud, B.; Peterscmitt, M. A study of the mode of transmission of maize streak virus by Cicadulina mbila using an enzyme-linked immunosorbent assay. Ann. Appl. Biol. 1992, 121, 85–94. [Google Scholar] [CrossRef]
- Margaria, P.; Bosco, L.; Vallino, M.; Ciuffo, M.; Mautino, G.C.; Tavella, L.; Turina, M. The NSs Protein of Tomato spotted wilt virus is Required for Persistent Infection and Transmission by Frankliniella occidentalis. J. Virol. 2014, 88, 5788–5802. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zheng, K.; Dong, J.; Fang, Q.; Wu, S.; Wang, L.; Zhang, Z. Identification of a new tospovirus causing necrotic ringspot on tomato in China. Virol. J. 2014, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Seepiban, C.; Charoenvilaisiri, S.; Warin, N.; Bhunchoth, A.; Phironrit, N.; Phuangrat, B.; Chatchawankanphanich, O.; Attathom, S.; Gajanandana, O. Development and application of triple antibody sandwich enzyme-linked immunosorbent assays for begomovirus detection using monoclonal antibodies against tomato yellow leaf curl Thailand virus. Virol. J. 2017, 14, 99. [Google Scholar] [CrossRef] [PubMed]
- Conrads, T.P.; Issaq, H.J.; Hoang, V.M. Current Strategies for Quantitative Proteomics. In Advances in Protein Chemistry; Smith, R.D., Veenstra, T.D., Eds.; Proteome Characterization and Proteomics; Academic Press: Cambridge, MA, USA, 2003; Volume 65, pp. 133–159. [Google Scholar]
- Saraswathy, N.; Ramalingam, P. 10-Introduction to proteomics. In Concepts and Techniques in Genomics and Proteomics; Saraswathy, N., Ramalingam, P., Eds.; Woodhead Publishing Series in Biomedicine; Woodhead Publishing: Sawston/Cambridge, UK, 2011; pp. 147–158. ISBN 978-1-907568-10-7. [Google Scholar]
- Büyükköroğlu, G.; Dora, D.D.; Özdemir, F.; Hızel, C. Chapter 15-Techniques for Protein Analysis. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 317–351. ISBN 978-0-12-804659-3. [Google Scholar]
- O’Neill, J.R. An Overview of Mass Spectrometry-Based Methods for Functional Proteomics. In Functional Proteomics: Methods and Protocols; Wang, X., Kuruc, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 179–196. ISBN 978-1-4939-8814-3. [Google Scholar]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Badillo-Vargas, I.E.; Rotenberg, D.; Schneweis, D.J.; Hiromasa, Y.; Tomich, J.M.; Whitfield, A.E. Proteomic Analysis of Frankliniella occidentalis and Differentially Expressed Proteins in Response to Tomato Spotted Wilt Virus Infection. J. Virol. 2012, 86, 8793–8809. [Google Scholar] [CrossRef]
- Wang, H.; Wu, K.; Liu, Y.; Wu, Y.; Wang, X. Integrative proteomics to understand the transmission mechanism of Barley yellow dwarf virus-GPV by its insect vector Rhopalosiphum padi. Sci. Rep. 2015, 5, 10971. [Google Scholar] [CrossRef]
- Liu, B.; Qin, F.; Liu, W.; Wang, X. Differential proteomics profiling of the ova between healthy and Rice stripe virus-infected female insects of Laodelphax striatellus. Sci. Rep. 2016, 6, 27216. [Google Scholar] [CrossRef]
- Huang, H.-J.; Lu, J.-B.; Li, Q.; Bao, Y.-Y.; Zhang, C.-X. Combined transcriptomic/proteomic analysis of salivary gland and secreted saliva in three planthopper species. J. Proteom. 2018, 172, 25–35. [Google Scholar] [CrossRef]
- Rao, S.A.K.; Carolan, J.C.; Wilkinson, T.L. Proteomic Profiling of Cereal Aphid Saliva Reveals Both Ubiquitous and Adaptive Secreted Proteins. PLoS ONE 2013, 8, e57413. [Google Scholar] [CrossRef]
- Lomate, P.R.; Bonning, B.C. Distinct properties of proteases and nucleases in the gut, salivary gland and saliva of southern green stink bug, Nezara viridula. Sci. Rep. 2016, 6, 27587. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Vega, L.J.; Stanley, B.A.; Stanley, A.; Felton, G.W. Proteomic analysis of labial saliva of the generalist cabbage looper (Trichoplusia ni) and its role in interactions with host plants. J. Insect Physiol. 2018, 107, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Serteyn, L.; Francis, F. Insight into Salivary Gland Proteomes of Two Polyphagous Stink Bugs: Nezara viridula L. and Halyomorpha halys Stål. Proteomics 2019, 19, 1800436. [Google Scholar] [CrossRef] [PubMed]
- Carolan, J.C.; Caragea, D.; Reardon, K.T.; Mutti, N.S.; Dittmer, N.; Pappan, K.; Cui, F.; Castaneto, M.; Poulain, J.; Dossat, C.; et al. Predicted Effector Molecules in the Salivary Secretome of the Pea Aphid (Acyrthosiphon pisum): A Dual Transcriptomic/Proteomic Approach. J. Proteome Res. 2011, 10, 1505–1518. [Google Scholar] [CrossRef]
- Cilia, M.; Fish, T.; Yang, X.; Mclaughlin, M.; Thannhauser, T.W.; Gray, S. A Comparison of Protein Extraction Methods Suitable for Gel-Based Proteomic Studies of Aphid Proteins. J. Biomol. Tech. 2009, 20, 201–215. [Google Scholar]
- Cilia, M.; Tamborindeguy, C.; Fish, T.; Howe, K.; Thannhauser, T.W.; Gray, S. Genetics Coupled to Quantitative Intact Proteomics Links Heritable Aphid and Endosymbiont Protein Expression to Circulative Polerovirus Transmission. J. Virol. 2011, 85, 2148–2166. [Google Scholar] [CrossRef]
- Yu, W.; Liu, J.; Colangelo, C.; Gulcicek, E.; Zhao, H. A New Protocol of Analyzing Isotope Coded Affinity Tag Data from High Resolution LC-MS Spectrometry. Comput. Biol. Chem. 2007, 31, 215–221. [Google Scholar] [CrossRef]
- Kellermann, J.; Lottspeich, F. Isotope-coded protein label. Methods Mol. Biol. 2012, 893, 143–153. [Google Scholar] [CrossRef]
- Rauniyar, N.; Yates, J.R. Isobaric Labeling-Based Relative Quantification in Shotgun Proteomics. J. Proteome Res. 2014, 13, 5293–5309. [Google Scholar] [CrossRef]
- Zhao, J.; Chi, Y.; Zhang, X.-J.; Lei, T.; Wang, X.-W.; Liu, S.-S. Comparative proteomic analysis provides new insight into differential transmission of two begomoviruses by a whitefly. Virol. J. 2019, 16, 32. [Google Scholar] [CrossRef]
- Deshoux, M.; Monsion, B.; Uzest, M. Insect cuticular proteins and their role in transmission of phytoviruses. Curr. Opin. Virol. 2018, 33, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein-Protein Interaction Detection: Methods and Analysis. Int. J. Proteomics 2014, 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Seddas, P.; Boissinot, S.; Strub, J.-M.; Van Dorsselaer, A.; Van Regenmortel, M.H.V.; Pattus, F. Rack-1, GAPDH3, and actin: Proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 2004, 325, 399–412. [Google Scholar] [CrossRef]
- Badillo-Vargas, I.E.; Chen, Y.; Martin, K.M.; Rotenberg, D.; Whitfield, A.E. Discovery of novel thrips vector proteins that bind to the viral attachment protein of the plant bunyavirus, tomato spotted wilt virus. J. Virol. 2019, 93, e00699-19. [Google Scholar] [CrossRef] [PubMed]
- Kanakala, S.; Ghanim, M. Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of Tomato yellow leaf curl virus. Front. Plant Sci. 2016, 7, 1702. [Google Scholar] [CrossRef]
- Kumari, S.; Roy, S.; Singh, P.; Singla-Pareek, S.L.; Pareek, A. Cyclophilins: Proteins in search of function. Plant Signal. Behav. 2013, 8, e22734. [Google Scholar] [CrossRef]
- von Hahn, T.; Ciesek, S. Cyclophilin polymorphism and virus infection. Curr. Opin. Virol. 2015, 14, 47–49. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Q.; Cao, W.; Yang, F.; Ma, L.; Liu, W.; Zhang, K.; Liu, X.; Zhu, Z.; Zheng, H. Foot-and-mouth disease virus nonstructural protein 2B interacts with cyclophilin A, modulating virus replication. FASEB J. 2018, 32, 6706–6723. [Google Scholar] [CrossRef]
- Montero-Astúa, M.; Ullman, D.E.; Whitfield, A.E. Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virology 2016, 493, 39–51. [Google Scholar] [CrossRef]
- Liu, W.; Gray, S.; Huo, Y.; Li, L.; Wei, T.; Wang, X. Proteomic Analysis of Interaction between a Plant Virus and Its Vector Insect Reveals New Functions of Hemipteran Cuticular Protein. Mol. Cell. Proteomics 2015, 14, 2229–2242. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Zhou, Y. Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res. 2018, 247, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Mar, T.; Liu, W.; Wang, X. Proteomic analysis of interaction between P7-1 of Southern rice black-streaked dwarf virus and the insect vector reveals diverse insect proteins involved in successful transmission. J. Proteomics 2014, 102, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gao, X.-W. The Cuticle Protein Gene MPCP4 of Myzus persicae (Homoptera: Aphididae) Plays a Critical Role in Cucumber Mosaic Virus Acquisition. J. Econ. Entomol. 2017, 110, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel, J.F.; Verbeek, M.; van der Wilk, F. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J. Gen. Virol. 1994, 75, 2559–2565. [Google Scholar] [CrossRef]
- Bandla, M.D.; Campbell, L.R.; Ullman, D.E.; Sherwood, J.L. Interaction of Tomato Spotted Wilt Tospovirus (TSWV) Glycoproteins with a Thrips Midgut Protein, a Potential Cellular Receptor for TSWV. Phytopathology 1998, 88, 98–104. [Google Scholar] [CrossRef]
- Kikkert, M.; Meurs, C.; van de Wetering, F.; Dorfmüller, S.; Peters, D.; Kormelink, R.; Goldbach, R. Binding of Tomato Spotted Wilt Virus to a 94-kDa Thrips Protein. Phytopathology 1998, 88, 63–69. [Google Scholar] [CrossRef]
- Linz, L.B.; Liu, S.; Chougule, N.P.; Bonning, B.C. In Vitro Evidence Supports Membrane Alanyl Aminopeptidase N as a Receptor for a Plant Virus in the Pea Aphid Vector. J. Virol. 2015, 89, 11203–11212. [Google Scholar] [CrossRef]
- Li, C.; Cox-Foster, D.; Gray, S.M.; Gildow, F. Vector Specificity of Barley Yellow Dwarf Virus (BYDV) Transmission: Identification of Potential Cellular Receptors Binding BYDV-MAV in the Aphid, Sitobion avenae. Virology 2001, 286, 125–133. [Google Scholar] [CrossRef]
- Li, S.; Xiong, R.; Wang, X.; Zhou, Y. Five Proteins of Laodelphax striatellus Are Potentially Involved in the Interactions between Rice Stripe Virus and Vector. PLoS ONE 2011, 6, e26585. [Google Scholar] [CrossRef]
- Chavez, J.D.; Cilia, M.; Weisbrod, C.R.; Ju, H.-J.; Eng, J.K.; Gray, S.M.; Bruce, J.E. Cross-linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus-plant interactions. J. Proteome Res. 2012, 11, 2968–2981. [Google Scholar] [CrossRef]
- Webster, C.G.; Thillier, M.; Pirolles, E.; Cayrol, B.; Blanc, S.; Uzest, M. Proteomic composition of the acrostyle: Novel approaches to identify cuticular proteins involved in virus-insect interactions. Insect Sci. 2017, 24, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Ohnesorge, S.; Bejarano, E.R. Begomovirus coat protein interacts with a small heat-shock protein of its transmission vector (Bemisia tabaci). Insect Mol. Biol. 2009, 18, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, J.; Fu, S.; Li, C.; Zhu, Z.-R.; Zhou, X. Rice stripe tenuivirus nonstructural protein 3 hijacks the 26S proteasome of the small brown planthopper via direct interaction with regulatory particle non-ATPase subunit 3. J. Virol. 2015, 89, 4296–4310. [Google Scholar] [CrossRef] [PubMed]
- Mulot, M.; Monsion, B.; Boissinot, S.; Rastegar, M.; Meyer, S.; Bochet, N.; Brault, V. Transmission of Turnip yellows virus by Myzus persicae Is Reduced by Feeding Aphids on Double-Stranded RNA Targeting the Ephrin Receptor Protein. Front. Microbiol. 2018, 9, 457. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.-H. C1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef]
- Cilia, M.; Howe, K.; Fish, T.; Smith, D.; Mahoney, J.; Tamborindeguy, C.; Burd, J.; Thannhauser, T.W.; Gray, S. Biomarker discovery from the top down: Protein biomarkers for efficient virus transmission by insects (Homoptera: Aphididae) discovered by coupling genetics and 2-D DIGE. Proteomics 2011, 11, 2440–2458. [Google Scholar] [CrossRef]
- Tamborindeguy, C.; Bereman, M.S.; DeBlasio, S.; Igwe, D.; Smith, D.M.; White, F.; MacCoss, M.J.; Gray, S.M.; Cilia, M. Genomic and proteomic analysis of Schizaphis graminum reveals cyclophilin proteins are involved in the transmission of cereal yellow dwarf virus. PLoS ONE 2013, 8, e71620. [Google Scholar] [CrossRef]
- Kruse, A.; Fattah-Hosseini, S.; Saha, S.; Johnson, R.; Warwick, E.; Sturgeon, K.; Mueller, L.; MacCoss, M.J.; Shatters, R.G.; Heck, M.C. Combining ’omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS ONE 2017, 12, e0179531. [Google Scholar] [CrossRef]
- Low, T.Y.; Mohtar, M.A.; Ang, M.Y.; Jamal, R. Connecting Proteomics to Next-Generation Sequencing: Proteogenomics and Its Current Applications in Biology. Proteomics 2018, 19, 1800235. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 59–73. ISBN 978-1-4939-7013-1. [Google Scholar]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Statistics for Biology and Health; Springer: New York, NY, USA, 2005; pp. 397–420. ISBN 978-0-387-29362-2. [Google Scholar]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. [Google Scholar] [CrossRef]
- Choi, M.; Chang, C.-Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Pei, G.; Chen, L.; Zhang, W. WGCNA Application to Proteomic and Metabolomic Data Analysis. In Met Enzymol; Elsevier: Amsterdam, The Netherlands, 2017; Volume 585, pp. 135–158. ISBN 978-0-12-809742-7. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ding, X.; Xiao, J.; Jiménez-Gόngora, T.; Liu, R.; Lozano-Durán, R. Inference of a Geminivirus−Host Protein−Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis. Viruses 2017, 9, 275. [Google Scholar] [CrossRef] [PubMed]
- Schuchman, R.; Kilianski, A.; Piper, A.; Vancini, R.; Ribeiro, J.M.C.; Sprague, T.R.; Nasar, F.; Boyd, G.; Hernandez, R.; Glaros, T. Comparative Characterization of the Sindbis Virus Proteome from Mammalian and Invertebrate Hosts Identifies nsP2 as a Component of the Virion and Sorting Nexin 5 as a Significant Host Factor for Alphavirus Replication. J. Virol. 2018, 92, e00694-18. [Google Scholar] [CrossRef] [PubMed]
- Chand, K.; Biswas, S.K.; Mondal, B. Purification of infective bluetongue virus particles by immuno-affinity chromatography using anti-core antibody. VirusDisease 2016, 27, 98–101. [Google Scholar] [CrossRef]
- Han, S.; Li, J.; Ting, A.Y. Proximity labeling: Spatially resolved proteomic mapping for neurobiology. Curr. Opin. Neurobiol. 2018, 50, 17–23. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L. Recent advances in proximity-based labeling methods for interactome mapping. F1000Research 2019, 8, 135. [Google Scholar] [CrossRef]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006, 1, 1278–1286. [Google Scholar] [CrossRef]
| Insect Vector | Virus | Antigen | ELISA Type | Reference |
|---|---|---|---|---|
| Rhopalosiphum padi and Rhopalosiphum maidis | Barley yellow dwarf virus (BYDV) | Antiserum made by injecting PAV and MAV (two isolates transmitted by R. padi) to rabbits | Double antibody sandwich (DAS) ELISA | Lister and Rochow 1979 [23] |
| Aphis gossypii | Plum pox potyvirus (PPV)-D and M serotypes | PPV-D- and PPV-M-specific monoclonal antibodies | Double antibody sandwich indirect (DASI) ELISA | Olmos et al. 1997 [24] |
| Myzus persicae | Beet yellow virus (BVY) | BVY-specific monoclonal antibody | Triple antibody sandwich (TAS) ELISA | Stevens et al. 1997 [25] |
| Myzus persicae | Alfa mosaic virus (AMV) | Glutaraldehyde-fixed AMV antisera raised in rabbits | Indirect double antibody sandwich (IDAS) ELISA | Ahoonmanesh et al. 1990 [26] |
| Bemisia tabaci | Lettuce infectious yellows virus (LIYV) | Coat protein (CPm) | Double and triple antibody sandwich (DAS and TAS) ELISA | Chen et al. 2011 [9] |
| Peregrinus maidis | Maize stripe virus (MStpV) | Purified non-capsid protein (Falk 1983) | Double antibody sandwich (DAS) ELISA | Falk et al. 1987 [27] |
| Peregrinus maidis | Maize stripe tenuivirus (MStV) | MStV-US * [28] | Indirect ELISA | Ammar et al. 1995 [29] |
| Cicadulina chinai | Maize yellow stripe virus (MYSV) | MYSV * [30] | Direct antigen coating (DAC) ELISA | Ammar et al. 2007 [31] |
| Cicadulia mbila | Maize streak virus (MSV) | MSV | Indirect double antibody sandwich (IDAS) ELISA | Reynaud and Peterschmitt 1992 [32] |
| Frankliniella occidentalis | Tomato spotted wilt virus (TSWV) | Nucleocapsid and glycoprotein (GN) | Triple antibody sandwich (TAS) ELISA | Margaria et al. 2014 [33] |
| Frankliniella occidentalis | Tomato zonate spot virus (TZSV) | TZSV * | Double antibody sandwich (DAS) ELISA | Yin et al. 2014 [34] |
| Bemisia tabaci | Tomato yellow leaf curl Thailand virus (TYLCTHV) | Recombinant coat protein (CP) | Triple antibody sandwich (TAS) ELISA | Seepiban et al. 2017 [35] |
| Approaches to Study Protein-Protein Interactions | Pros/Cons | Examples of Insect Vector–Plant Virus System Using the Approach |
|---|---|---|
| In Vitro Techniques | ||
| One- or Two-Dimensional Virus Overlay/Far Western Blot Assays | Pros:
| Myzus persicae–Potato leaf virus [70] Frankliniella occidentalis–Tomato spotted wilt virus [60,71,72] Myzus persicae–Beet western yellow virus [59] Acyrthosiphon pisum–Pea enation mosaic virus I [73] Sitobion avenae–Barley yellow dwarf virus-MAV [74,75] Laodelphax striatellus–Rice stripe virus [69] |
| Coimmunoprecipitation | Pros:
| Sogatella furcifera–Southern rice black-streaked dwarf virus [68] |
| Cross-Linking/Protein Interaction Reporter Technology | Pros:
| Myzus persicae–Potato leafroll virus [76] |
| Protein or Peptide Arrays | Pros:
| Aphis pisum–Cucumber mosaic virus [77] |
| In Vivo Techniques | ||
| Yeast Two-Hybrid | Pros:
| Bemisia tabaci–Tomato yellow leaf curl sardinia virus [78] Spodoptera furcifera–Southern rice black-streaked dwarf virus [67] Myzus persicae–Cucumber mosaic virus [69] Laodelphax striatellus–Rice stripe virus [66,67,79] Myzus persicae–Turnip yellow virus [80] Frankliniella occidentalis–Tomato spotted wilt virus [60] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mittapelly, P.; Rajarapu, S.P. Applications of Proteomic Tools to Study Insect Vector–Plant Virus Interactions. Life 2020, 10, 143. https://doi.org/10.3390/life10080143
Mittapelly P, Rajarapu SP. Applications of Proteomic Tools to Study Insect Vector–Plant Virus Interactions. Life. 2020; 10(8):143. https://doi.org/10.3390/life10080143
Chicago/Turabian StyleMittapelly, Priyanka, and Swapna Priya Rajarapu. 2020. "Applications of Proteomic Tools to Study Insect Vector–Plant Virus Interactions" Life 10, no. 8: 143. https://doi.org/10.3390/life10080143
APA StyleMittapelly, P., & Rajarapu, S. P. (2020). Applications of Proteomic Tools to Study Insect Vector–Plant Virus Interactions. Life, 10(8), 143. https://doi.org/10.3390/life10080143






