Abstract
The One Health approach is used to assess health-associated risks resulting from human exposure to antibiotic-resistant bacteria (ARB) that pose a significant public health risk. In this approach, wastewater treatment plants (WWTPs) play an important role in reducing bacteria and antibiotic-resistant genes (ARGs) in the environment. The ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are of significant concern due to their ability to evade the effects of multiple antibiotics, including last-resort treatments such as carbapenems and glycopeptides. This study aimed to investigate the environmental surveillance of ESKAPE bacteria in wastewater and their adjacent receiving water bodies in Limpopo Province, South Africa. Methodology: Over a period of 6 months, all isolates were identified phenotypically, and genomic DNA was extracted using the QIAamp 96 DNA QIAcube® HT Kit. Species-specific PCR was performed, followed by Sanger sequencing. The relevant sequences were compared to NCBI GenBank references using BLAST for confirmation and to assess the potential human health-associated risks. Results: ESKAPE organisms identified phenotypically were confirmed using PCR in both WWTP samples. Bacteria such as Acinetobacter baumannii and Enterobacter spp. were not detected in upstream or downstream river samples, particularly during August and September. In December and January, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa were not detected in effluent samples at both sites. Phylogenetic analysis revealed a diverse range of clinically significant genera, including Pseudomonas, Klebsiella, Enterobacter, and Staphylococcus, with strains closely related to global clinical isolates. Many of the isolates were associated with resistance to carbapenems, fluoroquinolones, and aminoglycosides. In addition, some strains clustered with both methicillin-sensitive and methicillin-resistant lineages. Conclusions: The findings emphasise the urgent need for increased genomic surveillance in environmental settings affected by wastewater discharge and highlight the importance of integrated antimicrobial resistance monitoring that connects clinical and environmental health sectors.
1. Introduction
In recent years, ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) pathogens have become pandemic, necessitating continuous efforts to mitigate their risk, especially that of multidrug-resistant (MDR) strains [1]. According to Mustafa et al. [2], most ESKAPE pathogens are primarily Gram-negative, a characteristic that enhances their resistance due to their cell wall structure. This has led to the development of the priority list of these pathogens by the World Health Organisation (WHO) because they are a threat to human health and require an urgent development of new antibiotic drugs or a change in strategy in the use of existing ones [2]. Specifically, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae (resistant to carbapenems and third-generation cephalosporins), as well as vancomycin-resistant Enterococcus (VRE) and methicillin-resistant Staphylococcus aureus (MRSA), are considered high-priority ARB [2]. The increase in ARB and the spread of antibiotic-resistant genes (ARGs) is driven by excess usage of antibiotics, which form part of a cesspool in wastewater in clinical and other forms, such as stormwater [3,4,5,6,7,8,9]. The release of these antibiotics without prescribed regulations from health facilities, pharmaceuticals, and other settings as waste into wastewater treatment plants and other water surfaces places a huge pressure on existing bacteria [10,11]. Although there is robust regulation regarding the discharge of treating hospitals or other health facilities containing antibiotics into sewage water in countries like South Africa, non-adherence, such as illegal disposal, has played a role in worsening the situation. This has jeopardised the current wastewater pollution by disseminating ESKAPE pathogens into the aquatic environment [12]. Okafor and Nwodo [12] further reported that approximately two million people will die worldwide in the next two decades due to antibiotic-resistant pathogens if no mitigation action is in place. Vulnerable countries of the Global South, such as South Africa, where there is poor sanitation in rural areas, insufficient infrastructure, and a high immunocompromised population, are at risk of antibiotic resistance-related mortality. Santajit and Indrawattana [13] further reported that the growing number of ESKAPE pathogens is also driven by nosocomial infections, which place a huge burden on existing resources in both healthcare and public systems. These pathogens are also able to thrive in extreme environmental conditions due to their adaptability to different environmental conditions [14]. The authors further opined that these pathogens are responsible for high mortality and morbidity rates; thus, increased treatment costs, diagnostic uncertainties, and lack of trust in orthodox medicine. The justification of this study is pegged on the following: a risk of community-acquired infections for various water sources due to inadequate wastewater treatment. This drives pathogens and resistance genes into various water sources, thus calling for effective management of wastewater as part of achieving agendas 3 and 6 of the Sustainable Development Goals (SDGs) [15]. The unique reason for this study in this region is due to limited similar work, which underscores the need for environmental AMR surveillance, integrating environmental monitoring of ESKAPE pathogens within a One Health framework in this region, and also linking the performance capability of wastewater treatment plants in contributing to the environmental spread of high-priority, multidrug-resistant pathogens in this part of the region; finally, the data is generated from part of the antimicrobial resistance, which is crucial for risk assessment and policymaking [16,17]. Therefore, this study aims to conduct the environmental surveillance of ESKAPE bacteria in wastewater and their associated receiving water bodies from rural and peri-urban treated effluents and rivers within the Vhembe District.
2. Methods and Materials
2.1. Study Site
The study was carried out in the Vhembe District, located in Limpopo Province, South Africa, which is characterised by several water bodies supporting agricultural, industrial, and farming activities. The region faces challenges due to inadequate wastewater treatment facilities and pollution from non-point sources. The two types of WWTPs used for sampling in this study included the Thohoyandou WWTP 1 [a peri-urban WWTP] and the Malamulele WWTP 2 [a rural WWTP].
2.2. Sample Collection
The samples were collected over a six-month period between August 2024 to January 2025 once a month. The samples were obtained from both the influents and effluents of each of the two WWTPs, together with samples from the upstream and downstream points on the river where the effluent is pumped. Duplicates of approximately 500 mL of water samples were collected in a sterile plastic collection bottle and transported to the laboratory on ice for further analysis.
2.3. Isolation of ESKAPE Pathogens Using Culture Technique
To isolate ESKAPE pathogens from environmental samples, 20 mL of each sample was inoculated into 200 mL of Luria–Bertani (LB) broth (Oxoid, ThermoFisher Scientific, Johannesburg, South Africa) and incubated at 37 °C for 24 h [18]. Serial dilutions (10−1 to 10−6) of each sample were prepared in 0.85% (w/v) saline solution (Separation, SA), and 100 µL of each dilution was spread onto selective media plates for the following organisms: Enterococcus faecium, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Enterobacter spp. [19,20]. For the isolation of Staphylococcus aureus, 100 µL of each dilution was spread onto Mannitol Salt agar, and Baird-Parker agar was prepared [19]. For the isolation of Acinetobacter baumannii, an enrichment step was performed using Modified Preston broth (Oxoid, Lenexa, KS, USA), supplemented with polymyxin B, amphotericin B, trimethoprim, and rifampicin [21]. The presumptive colonies for each bacterium were identified based on small, red-maroon colonies for E. faecium; yellow zones on Mannitol Salt agar or black colonies with clear halos on Baird-Parker agar (S. aureus); purple-magenta, mucoid colonies (K. pneumoniae); cream, circular colonies (A. baumannii); fluorescent, yellow-green colonies (P. aeruginosa); and pink-red, mucoid colonies (Enterobacter spp.), as reported by Ekwanzala et al. [22] and Havenga et al. [23]. Pure cultures were further characterised using conventional PCR and sequencing.
2.4. Genomic DNA Extraction, Species-Specific PCR, and DNA Sequencing for ESKAPE Identification
Genomic DNA was extracted using the QIAamp 96 DNA QIAcube® HT Kit (QIAGEN, Johannesburg, South Africa) following the manufacturer’s instructions. The PCR mixtures for the identification of E. faecium, S. aureus, K. pneumoniae, P. aeruginosa, and Enterobacter spp. consisted of a final volume of 25 µL and contained 12.5 µL MyTaq™ HS Red Mix (Bioline, Celtic Molecular Diagnostics, Kenilworth, Cape Town, South Africa), 2.5 µL primer mix (forward and reverse), 8 µL PCR-grade water, and 2 µL template DNA. The PCR mixture for the identification of A. baumannii consisted of 20 μL of PCR, which included 2 μL of 10X buffer (Qiagen, Germany), 2 μL of forward and reverse primers, 2 μL of MgCl2, 4 μL of Q-solution, 0.4 μL of dNTPs, 0.1 μL of Taq polymerase, 7.5 μL of PCR-grade water, and 2 μL of genomic DNA.
For each PCR assay, sterile Milli-Q water, MyTaq, and primers without genomic DNA were used as a negative control (no band/s was expected in the gel), while genomic DNA extracted from E. faecium (Strain WHRC/SC/2015), S. aureus ATCC BAA-1026, K. pneumoniae ATCC 13883, A. baumannii ATCC 19606, P. aeruginosa ATCC 27853, and Enterobacter spp. (Strain LH/S/2017/N69) were used as positive controls. The primers employed in this study and the amplification cycles used are listed in Table 1. Amplification was performed on a T100TM Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA).
Following amplification, the PCR products were visualised using gel electrophoresis [80 Volts (V) for 80 min] on a 1% (w/v) agarose (SeaKem® LE Agarose, Lonza) gel [containing 0.5 µg/mL ethidium bromide (EtBr)], in 1X tris/acetate/ethylenediaminetetraacetic acid (TAE) buffer. The amplicon size for each ESKAPE isolate was determined using the Generuler™ 1 kb Plus DNA ladder (Thermo Fischer Scientific), whereafter the results were visualised under UV illumination using the MiniBIS Pro (DNR Bio-Imaging System, Neve Yamin, Israel) and the images were captured using the GelCapture Version 4.25 (DNR Bio-Imaging System).
Representative amplicons for each of the ESKAPE isolates were purified and concentrated using the Wizard® SV Gel and PCR Clean-up System (Promega Corp), as per the manufacturer’s instructions, and sent for sequencing using the BigDye Terminator Version 3.1 Sequencing Kit (Applied Biosystems®, Fairland, Johannesburg, South Africa). The FinchTV version 1.4.0 software was used to examine the chromatogram of each sequence, whereafter sequence identification for each isolate was carried out using the National Centre for Biotechnology Information (NCBI), Basic Local Alignment Search Tool (BLAST) [24]. Sequences of the representative isolates showing a similarity of ≥97% to E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp. on the NCBI database were recorded as positive similarity hits.

Table 1.
Primers and conventional PCR cycling parameters used detection and identification of ESKAPE pathogens from wastewater and river samples.
Table 1.
Primers and conventional PCR cycling parameters used detection and identification of ESKAPE pathogens from wastewater and river samples.
| Organism | Primer Name | Primer Sequence (5′-3′) | PCR Cycling Parameters | Gene (Size bp) | Reference |
|---|---|---|---|---|---|
| E. faecium | pstS1-F | TTGAGCCAAGTCGAAGCTGGAG | 95 °C for 15 min; 35 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s; final extension at 72 °C for 5 min | pstS (583) | Homan et al. [25] |
| pstS2-R | CGTGATCACGTTCTACTTCC | ||||
| S. aureus | vicK1 | CTAATACTGAAAGTGAGAAACGTA | 94 °C for 5 min; 35 cycles at 94 °C for 40 s, 50 °C for 40 s, and 72 °C for 60 s; final extension at 72 °C for 5 min | vicK (289) | Liu et al. [26] |
| vicK2 | TCCTGCACAATCGTACTAAA | ||||
| K. pneumoniae | Kpn-F | GTGCGATGCGGTCTTTG | 95 °C for 15 min; 45 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 90 s; final extension at 72 °C for 5 min | phoE (398) | Kaushik and Balasubramanian [27] |
| Kpn-R | GGGCGAACTGAACTGATG | ||||
| A. baumannii | A.b_hyp F | CGTCGGTCGGATCCGTGTA | 95 °C for 15 min; 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; final extension at 72 °C for 5 min | A.b_hyp (545) | Havenga et al. [23] |
| A.b hyp R | AAGTAAAGTGGCAGGCGCTT | ||||
| P. aeruginosa | kpd1 | GCCCACGACCAGTTCGAC | 95 °C for 15 min; 30 cycles at 94 °C for 15 s, 54 °C for 15 s, and 72 °C for 15 s; final extension at 72 °C for 5 min | rhIB (226) | Bodour et al. [28] |
| kpd2 | CATCCCCCTCCCTATGAC | ||||
| Enterobacter spp. | ENB-F | AGTGGAACGGTCTGGAAAGG | 95 °C for 15 min; 45 cycles at 95 °C for 10 s, 56 °C for 20 s, and 72 °C for 20 s; final extension at 72 °C for 5 min | 23S rRNA (154) | Patel et al. [29] |
| ENB-R | TCGGTCAGTCAGGAGTATTTAGC |
3. Results and Discussions
3.1. Molecular Detection of ESKAPE Organisms from WWTPs 1 and 2
Table 2 shows the PCR results for the detection of ESKAPE organisms that were initially detected using culture methods from individual samples obtained from the two WWTPs (influents and effluents) and the rivers (upstream and downstream) where the WWTPs discharge their final effluents. Most influent samples tested positive for ESKAPE organisms from both WWTP influent samples using PCR throughout the study. Most influent samples tested positive for ESKAPE organisms throughout the study, likely due to sewage spills, improper hospital waste disposal, anthropogenic activities, and agricultural waste. According to studies by Nyenje et al. [30] and Hrenovic et al. [31], the common sources of these organisms into various wastewater influents are sewage spills, improper deposition of hospital waste, anthropogenic activities, and improper disposal of agricultural waste, which is similar to the findings of this study. Marutescu et al. [32] further reported that these organisms can rapidly develop antimicrobial resistance through selective processes such as selective pressure by the excess use of agricultural activities, animal rearing, and medical therapy by humans, which was also observed along the adjacent rivers and WWTPs. According to Pärnänen et al. [33] and Reichert et al. [34], wastewater also receives numerous human antibiotic-resistant genes (ARGs), metals, antibiotics, and other related chemicals that are formed within the sewer lines through commensal microbiota. This increases the accumulation and dissemination of antibiotic-resistant organisms into the environment.

Table 2.
The identification of the ESKAPE organisms.
In August, most organisms identified as presumptive positives using culture-based techniques were also detected via PCR in two WWTP samples (Table 2).
In September, a similar trend was observed, except that organisms such as Acinetobacter baumanii and Enterobacter spp. were not detected from upstream and downstream samples in both WWTPs, possibly due to effective wastewater treatment or dilution effects in the river. According to Huddleston [35], these ESKAPE microorganisms can thrive through horizontal gene transfer with other microorganisms, making them a perfect candidate for the spread of virulent and resistance determinants. This is possible when wastewater is not treated adequately before being discharged into various water sources or when untreated water is used for irrigation purposes. In a study on runoff water in China after a period of heavy rainfall, several multiple multi-resistance microbes were detected including Pseudomonas aeruginosa, Klebsiella spp., and Acinetobacter spp. [36]. The authors further reported similar findings seen within the water systems, mainly rivers, puddles around hospital environments, effluents and influents of wastewater treatment plants in South Africa, and similar cases after severe storms [36]. Based on these findings, especially in the case of storms, there are high chances that these pathogens are washed directly into the river water system, in addition to being transported through sewer lines into wastewater treatment plants and ultimately into the wider aquatic ecosystem [36].
In October, most ESKAPE pathogens were detected in both WWTP samples, except at the downstream site of the river near WWTP 1, where none of the organisms were identified (Table 2). This might be due to the effectiveness of tertiary treatment by dosing enough chlorine into the final effluent before discharge; thus, more residual chlorine was effective for a longer period in the downstream. Bacteria like Staphylococcus aureus and Pseudomonas aeruginosa were not detected in any of the sampling points in WWTP 1. In WWTP 2, most organisms were detected, except for Staphylococcus aureus, at both the influent and effluent points of the same plant in November (Table 2). These findings are similar to a study conducted in Tunisia by Hassen et al. [37], which isolated K. pneumoniae and other ESKAPE organisms from river and wastewater samples collected between 2017 and 2018, respectively. Hosu et al. [38] further reported that P. aeruginosa was detected in both abattoir wastewater and surface water in the Eastern Cape of South Africa when they investigated the implications of antibiotic resistance genes on public health. The findings also indicate that the wastewater and polluted rivers ecosystem favoured the survival of the ESKAPE organisms, particularly those with MDR profiles and carrying ESBL-associated genes such as blaCTX-M, blaSHV, and blaTEM [39].
In December, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, and Pseudomonas aeruginosa were not detected in any of the effluent samples, except for Enterobacter spp., which was detected in WWTP 1, suggesting effective treatment processes. This plant also reported that none of the ESKAPE organisms were detected in the downstream samples. In WWTP 2, only Acinetobacter baumanii was not detected in any of the samples. Similarly, Staphylococcus aureus was not detected in either upstream or downstream samples in the same month (Table 2). Most of the bacteria were undetectable in the final treated effluents because of adequate tertiary chlorine dosage in WWTP 1. The presence of Enterobacter spp. in WWTP 1 aligns with studies reporting high levels of this bacterium in hospital wastewater and farm soils due to poor sewage systems or non-hospital contamination sources by Ebomah and Okoh [17], when they evaluated the presence of genes related to Enterobacter spp. in various environmental sources in South Africa, including hospital wastewater effluents, wastewater treatment plants final effluents, surface waters (rivers, dams, and canals), irrigation water, farm soil, and vegetable sources. Interestingly, they reported that hospital wastewater contains the highest number of Enterobacter spp., followed by farm soils, and this was because poor sewage systems or some other source of contamination independent of hospital sources [17]. In a study conducted by Galarde-López et al. [40], where they compared the prevalence of ESKAPE organisms in both untreated influent and treated final effluents, they reported that K. pneumoniae and Enterobacter spp. were predominant in the raw wastewater, while Staphylococcus aureus, Acinetobacter baumanii, and Pseudomonas aeruginosa were found in the treated effluents. Ma et al. [41] reported that domination by these bacteria (K. pneumoniae and Enterobacter spp.) in raw wastewater is driven by the presence of faeces from human settlements. These ESKAPE organisms in the final treated effluents can bypass and survive the treating processes through the formation of biofilm that assists them in displaying intrinsic tolerance [42]. In the month of January, Acinetobacter baumannii was not detected throughout the two plants, and none of the ESKAPE pathogens were detectable in the effluents of the plants (Table 2). In the downstream of WWTP 1, all the organisms were undetectable, while Staphylococcus aureus was not detected in either the downstream or upstream samples of the two plants (Table 2). This indicates that these two WWTPs were effective in treating their final effluents.
3.2. Phylogenetic Analysis
To explore the evolutionary relationships and potential transmission pathways of the bacterial strains identified in this study, a comprehensive phylogenetic analysis was conducted, incorporating both locally recovered isolates and globally referenced sequences obtained from the NCBI GenBank database. The analysis revealed that several of the environmental strains were closely related to clinical strains previously reported worldwide, suggesting possible links between environmental and clinical reservoirs of antimicrobial-resistant pathogens. The nucleotide sequences of the ESKAPE strains obtained from this investigation were submitted to NCBI GenBank under the following accession numbers: PV240204, PV240205, PV240206, PV240207, PV240208, PV240209, PV240210, PV240211, PV240212, PV240213, PV240214, PV240215, PV240216, PV240217, PV240218, PV240219, PV240220, PV240221, PV240222, and PV240223.
Using maximum likelihood methods and a curated alignment of conserved bacterial gene sequences, we constructed a circular phylogenetic tree (Figure 1), which revealed a pattern of relatedness between the study strains and the reference strains isolated from clinical and environmental sources worldwide. The resulting topology displays a wide taxonomic extent, encompassing clinically significant genera including Pseudomonas, Klebsiella, Enterobacter, and Staphylococcus, among others. Notably, the current study strains were widely distributed across the tree, indicating a high degree of microbial diversity in the sampled environments, which included surface waters and wastewater sources. Despite this diversity, many strains were tightly clustered with known multidrug-resistant (MDR) reference strains, indicating shared evolutionary ancestry and possible recent divergence from globally circulating lineages.
The phylogenetic tree revealed a strongly supported and clearly defined Pseudomonas aeruginosa clade, comprising several strains identified in this study. These strains (Figure 1) clustered closely with previously reported clinical isolates, indicating high genetic similarity and suggesting possible links between environmental and clinical sources. These strains (Figure 1) were closely related to well-known clinical strains previously reported around the world (Accessions: AY956935.1, GU393321.1, L37763.1), many of which are well known to harbour resistance to carbapenems, fluoroquinolones, and aminoglycosides [43]. The tight clustering suggested that the Pseudomonas aeruginosa strains in the current study samples likely represented clonally related lineages that have disseminated globally. Their presence in environmental matrices may reflect the environmental persistence of this species, well-documented for its metabolic versatility, biofilm-forming capacity, and resistance to desiccation and disinfectants. These findings raise concern about environmental compartments functioning as reservoirs and amplifiers of Pseudomonas aeruginosa multidrug-resistant (MDR) strains [8].
Klebsiella pneumoniae strains from the current study formed several distinct but well-supported phylogenetic clusters. Multiple study strains (Figure 1) aligned closely with reference sequences representing globally important strains previously reported around the world, including those associated with the Cefotaximase-Munich (CTX-M) type extended-spectrum β-lactamases (ESBLs) and Klebsiella pneumoniae carbapenemase (KPC) type carbapenemases (NZ-JAHUYW010001071.1, OL656096.1). The observations were particularly alarming, as they suggest the environmental persistence or introduction of Klebsiella pneumoniae strains that are frequently implicated in hospital outbreaks and are often refractory to treatment [41]. The phylogenetic proximity indicated a low level of genetic divergence from globally dominant multidrug-resistant (MDR) strains and supports the hypothesis of anthropogenic contamination, likely through hospital effluent, improperly treated wastewater, or contaminated runoff. Enterobacter strains from the current study clustered with Enterobacter cloacae, E. hormaechei, and E. asburiae reference strains, suggesting a shared evolutionary lineage. These organisms are increasingly recognised for their role in nosocomial infections, particularly in neonatal units and among immunocompromised individuals [43]. The strains (Figure 1) showed high sequence similarity to global reference strains previously reported (MW480239.1, LC498102.1), suggesting potential genetic conservation of resistance genes across strains from different geographical origins. Given the ubiquitous nature of Enterobacter in soil and water, their detection in the present study pointed to the interface between natural ecosystems and anthropogenically impacted environments.
The study strains (Figure 1) clustered within a defined clade included both methicillin-sensitive and methicillin-resistant strains, as previously reported strains (LC855698.1, PP390184.1, PP939095.1). The close phylogenetic grouping between the environmental Staphylococcus strains and Clinical methicillin-resistant Staphylococcus aureus (MRSA) strains raises concerns about the environmental spread of human-associated pathogens, potentially via wastewater discharge, direct human contact, or bio-aerosolization [1]. These findings highlighted the necessity of environmental surveillance, especially in peri-urban and rural areas with insufficient sanitation infrastructure. The Acinetobacter clade in the tree revealed several of the study strains (Figure 1) clustering with Acinetobacter baumannii reference previously reported strains (JEVWO1000226, NZ JBALGP010000114.1), widely recognised for their association with ventilator-associated pneumonia and bloodstream infections [44].
These strains are frequently found in units of the number of base substitutions per site. This analysis involved 97 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1982 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 [45]. Multidrug-resistant (MDR) strains and their evolutionary proximity to the study strains were indicative of either clonal expansion or genetic convergence through horizontal gene transfer. Given that Acinetobacter baumannii is known for its remarkable environmental resilience, its presence in water bodies contaminated with wastewater suggested silent dissemination routes with implications for both human and animal health. The phylogenetic relationships revealed in this analysis highlighted the interconnectedness of environmental and clinical microbial populations. The genetic closeness between many of our strains and globally reported strains suggested that environmental reservoirs may serve as channels for the persistence and spread of MDR pathogens [2]. This supports the One Health concept, which recognises that human, animal, and environmental health are interlinked [46]. The presence of high-priority pathogens, including P. aeruginosa, K. pneumoniae, and A. baumannii in the study environmental samples, all exhibiting close evolutionary ties to globally referenced strains, highlighted the global dimension of antimicrobial resistance (AMR) [32]. The current tree-based evidence provides molecular confirmation of the potential for long-distance dissemination of resistant lineages, either through trade, human mobility, or environmental contamination pathways [1]. The phylogenetic reconstruction provides valuable insights into the evolutionary relationships of bacterial strains isolated in this study. It revealed notable local genetic diversity alongside clear global relatedness, with several isolates showing strong phylogenetic affiliations to known multidrug-resistant (MDR) strains previously reported in healthcare settings worldwide [47]. These findings underline the urgent need for enhanced genomic surveillance in environmental reservoirs, particularly those influenced by wastewater discharge, and highlight the importance of integrative antimicrobial resistance (AMR) monitoring frameworks that connect clinical, environmental, and public health domains.
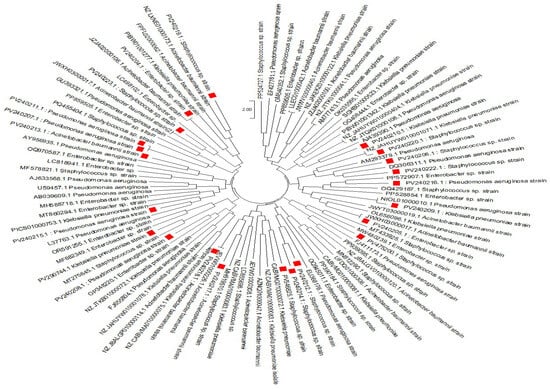
Figure 1.
The evolutionary history was inferred using the Neighbour-Joining method [48]. The optimal tree is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree [49]. The evolutionary distances were computed using the Maximum Composite Likelihood method [50] and are expressed in units of base substitutions per site. This analysis involved 97 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). The final dataset comprised a total of 1982 positions. Only bootstrap values > 70% were shown. Evolutionary analyses were conducted in MEGA11 [46]. The red squares represent the current strains that we obtained from our study vs the reference strain found in Genbank.
4. Conclusions
The results from this study showed that several environmental strains closely related with globally significant clinical reference sequences, including those harbouring CTX-M-type extended-spectrum β-lactamases (ESBLs) and KPC-type carbapenemases, based on phylogenetic analysis. Notably, environmental Staphylococcus strains clustered with clinical methicillin-resistant Staphylococcus aureus (MRSA), raising concern about the environmental spread of clinically relevant pathogens, potentially through wastewater discharge, direct human interaction, or aerosolization. These findings reinforce the need for expanded genomic surveillance and integrated AMR monitoring strategies, particularly in peri-urban and rural settings where sanitation infrastructure is limited. A One Health approach that bridges environmental, clinical, and public health domains is essential to address the growing threat of AMR. The study further recommended an extensive review and continuation of policy/infrastructure development, public health action against risks of using untreated river water for domestic purposes, and capacity building/research to track the evolution and spread of ESKAPE pathogens and resistance genes in aquatic environments in rural settings as part of achieving SDGs.
Author Contributions
Conceptualization, N.P., M.C.R. and L.O.K.; methodology, N.P., M.C.R., R.B., L.O.K. and A.N.T.; validation, N.P., M.C.R., R.B., L.O.K. and A.N.T.; formal analysis, N.P., M.C.R. and L.O.K.; investigation, N.P., M.C.R., R.B., L.O.K. and A.N.T.; resources, N.P.; writing—original draft preparation, N.P., M.C.R., R.B., L.O.K. and A.N.T.; writing—review and editing, N.P., M.C.R., R.B., L.O.K. and A.N.T.; visualization, N.P., M.C.R. and L.O.K.; supervision, N.P., M.C.R. and L.O.K.; project administration, N.P., M.C.R. and L.O.K.; funding acquisition, N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by: The Water Research Commission [Project C20222023-00991].
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
Funding for this study was received from the Water Research Commission [Project C20222023-00991]. We express our gratitude for approving the study to the Vhembe District Municipality and the Department of Water and Sanitation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, S.S.; Batool, R.; Kamran, M.; Javed, H.; Jamil, N. Evaluating the role of wastewaters as reservoirs of antibiotic-resistant ESKAPEE bacteria using phenotypic and molecular methods. Infect. Drug Resist. 2022, 15, 5715–5728. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.M.; Lin, M.F.; Liao, P.C.; Yeh, H.W.; Chang, B.V.; Tang, T.K.; Cheng, C.; Sung, C.H.; Liou, M.L. Comparison of antimicrobial resistance patterns between clinical and sewage isolates in a regional hospital in Taiwan. Lett. Appl. Microbiol. 2009, 48, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Khan, A.U.; Wahid, A.; Butt, Z.A.; Farhan, M.; Ahmad, F. Role of hospital effluents in the contribution of antibiotics and antibiotic-resistant bacteria to the aquatic environment. Pak. J. Nutr. 2012, 11, 1177. [Google Scholar] [CrossRef]
- Rabbani, M.A.G.; Howlader, M.Z.H.; Kabir, Y. Detection of multidrug-resistant (MDR) bacteria in untreated wastewater disposals of hospitals in Dhaka City, Bangladesh. J. Glob. Antimicrob. Res. 2017, 10, 120–125. [Google Scholar] [CrossRef]
- Giebułtowicz, J.; Tyski, S.; Wolinowska, R.; Grzybowska, W.; Zaręba, T.; Drobniewska, A.; Wroczyński, P.; Nałęcz-Jawecki, G. Occurrence of antimicrobial agents, drug-resistant bacteria, and genes in the sewage-impacted Vistula River (Poland). Environ. Sci. Pol. Res. 2018, 25, 5788–5807. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef]
- Pandey, R.; Mishra, S.K.; Shrestha, A. Characterisation of ESKAPE pathogens with special reference to multidrug resistance and biofilm production in a Nepalese hospital. Infect. Drug Resist. 2021, 14, 2201–2212. [Google Scholar] [CrossRef]
- Nishiyama, M.; Praise, S.; Tsurumaki, K.; Baba, H.; Kanamori, H.; Watanabe, T. Prevalence of antibiotic-resistant bacteria ESKAPE among healthy people estimated by monitoring of municipal wastewater. Antibiotics 2021, 10, 495. [Google Scholar] [CrossRef]
- Kruse, H. Indirect transfer of antibiotic resistance genes to man. Acta Vet. Sci. Suppl. 1999, 92, 59–65. [Google Scholar]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Okafor, J.U.; Nwodo, U.U. Molecular characterisation of antibiotic resistance determinants in Klebsiella pneumoniae isolates recovered from hospital effluents in the Eastern Cape province, South Africa. Antibiotics 2023, 12, 1139. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Mapipa, Q.; Digban, T.O.; Nnolim, N.E.; Nontongana, N.; Okoh, A.I.; Nwodo, U.U. Molecular characterisation and antibiotic susceptibility profile of Acinetobacter baumannii recovered from hospital wastewater effluents. Cur. Microbiol. 2022, 79, 123. [Google Scholar]
- Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial resistance development pathways in surface waters and public health implications. Antibiotics 2022, 11, 821. [Google Scholar] [CrossRef]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater surveillance of antibiotic-resistant bacterial pathogens: A systematic review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef]
- Ebomah, K.E.; Okoh, A.I. An African perspective on the prevalence, fate and effects of carbapenem resistance genes in hospital effluents and wastewater treatment plant (WWTP) final effluents: A critical review. Heliyon 2020, 6, e03899. [Google Scholar] [CrossRef]
- Sandman, K.; Ecker, C. Pseudomonas isolation and identification: An introduction to the challenges of polyphasic taxonomy. J. Microbiol. Biol. Educ. 2014, 15, 287–291. [Google Scholar] [CrossRef]
- Clements, T.; Reyneke, B.; Strauss, A.; Khan, W. Persistence of viable bacteria in solar pasteurised harvested rainwater. Water Air Soil Pol. 2019, 230, 130. [Google Scholar] [CrossRef]
- O’Connor, K.; Morrissette, M.; Strandwitz, P.; Ghiglieri, M.; Caboni, M.; Liu, H.; Khoo, C.; D’Onofrio, A.; Lewis, K. Cranberry extracts promote the growth of Bacteroidaceae and decrease the abundance of Enterobacteriaceae in a human gut simulator model. PLoS ONE 2019, 14, e0224836. [Google Scholar] [CrossRef] [PubMed]
- Kassamali, Z.; Prince, R.A.; Danziger, L.H.; Rotschafer, J.C.; Rhomberg, P.R.; Jones, R.N. Microbiological assessment of polymyxin B components tested alone and in combination. Antimicrob. Agents Chemother. 2015, 59, 7823–7825. [Google Scholar] [CrossRef] [PubMed]
- Ekwanzala, M.D.; Dewar, J.B.; Kamika, I.; Momba, M.N.B. Tracking the environmental dissemination of carbapenem-resistant Klebsiella pneumoniae using whole genome sequencing. Sci. Total Environ. 2019, 691, 80–92. [Google Scholar] [CrossRef]
- Havenga, B.; Ndlovu, T.; Clements, T.; Reyneke, B.; Waso, M.; Khan, W. Exploring the antimicrobial resistance profiles of the WHO critical priority list bacterial strains. BMC Microbiol. 2019, 19, 303. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Homan, W.L.; Tribe, D.; Poznanski, S.; Li, M.; Hogg, G.; Spalburg, E.; Van Embden, J.D.; Willems, R.J. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002, 40, 1963–1971. [Google Scholar] [CrossRef]
- Liu, C.I.; Liu, G.Y.; Song, Y.; Yin, F.; Hensler, M.E.; Jeng, W.Y.; Nizet, V.; Wang, A.H.J.; Oldfield, E. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 2008, 319, 1391–1394. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, R.; Balasubramanian, R. Assessment of bacterial pathogens in fresh rainwater and airborne particulate matter using Real-Time PCR. Atmos. Environ. 2012, 46, 131–139. [Google Scholar] [CrossRef]
- Bodour, A.A.; Drees, K.P.; Maier, R.M. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl. Environ. Microbiol. 2003, 69, 3280–3287. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.B.; Shanker, R.; Gupta, V.K.; Upadhyay, R.S. Q-PCR-based culture-independent enumeration and detection of Enterobacter: An emerging environmental human pathogen in riverine systems and potable water. Front. Microbiol. 2016, 7, 172. [Google Scholar] [CrossRef]
- Nyenje, M.E.; Tanih, N.F.; Green, E.; Ndip, R.N. Current status of antibiograms of Listeria ivanovii and Enterobacter cloacae isolated from ready-to-eat foods in Alice, South Africa. Int. J. Environ. Res. Public Health 2012, 9, 3101–3114. [Google Scholar] [CrossRef]
- Hrenovic, J.; Ivankovic, T.; Ivekovic, D.; Repec, S.; Stipanicev, D.; Ganjto, M. The fate of carbapenem-resistant bacteria in a wastewater treatment plant. Water Res. 2017, 126, 232–239. [Google Scholar] [CrossRef]
- Marutescu, L.G.; Popa, M.; Gheorghe-Barbu, I.; Barbu, I.C.; Rodríguez-Molina, D.; Berglund, F.; Blaak, H.; Flach, C.F.; Kemper, M.A.; Spießberger, B.; et al. Wastewater treatment plants, an “escape gate” for ESCAPE pathogens. Front. Microbiol. 2023, 14, 1193907. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, 9124. [Google Scholar] [CrossRef]
- Reichert, G.; Hilgert, S.; Fuchs, S.; Azevedo, J.C.R. Emerging contaminants and antibiotic resistance in the different environmental matrices of Latin America. Environ. Pollut. 2019, 255, 113140. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef]
- Almakki, A.; Jumas-Bilak, E.; Marchandin, H.; Licznar-Fajardo, P. Antibiotic resistance in urban runoff. Sci. Total Environ. 2019, 667, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic characterisation of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from wastewater and river water in Tunisia: Predominance of CTX-M-15 and high genetic diversity. Environ. Sci. Pol. Res. 2020, 27, 44368–44377. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.; Okuthe, G.E.; Apalata, T. Molecular Detection of Antibiotic-Resistant Genes in Pseudomonas aeruginosa from Nonclinical Environment: Public Health Implications in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 2021, 2021, 8861074. [Google Scholar] [CrossRef]
- Aguilar-Salazar, A.; Martínez-Vázquez, A.V.; Aguilera-Arreola, G.; de Jesus de Luna-Santillana, E.; Cruz-Hernández, M.A.; Escobedo-Bonilla, C.M.; Lara-Ramírez, E.; Sánchez-Sánchez, M.; Guerrero, A.; Rivera, G.; et al. Prevalence of ESKAPE bacteria in surface water and wastewater sources: Multidrug resistance and molecular characterisation, An updated review. Water 2023, 15, 3200. [Google Scholar] [CrossRef]
- Galarde-López, M.; Velazquez-Meza, M.E.; Godoy-Lozano, E.E.; Carrillo-Quiroz, B.A.; Cornejo-Juárez, P.; Sassoé-González, A.; Ponce-de-León, A.; Saturno-Hernández, P.; Alpuche-Aranda, C.M. Presence and Persistence of ESKAPEE Bacteria before and after Hospital Wastewater Treatment. Microorganism 2024, 12, 1231. [Google Scholar] [CrossRef]
- Ma, X.; Dong, X.; Cai, J.; Fu, C.; Yang, J.; Liu, Y.; Zhang, Y.; Wan, T.; Lin, S.; Lou, Y.; et al. Metagenomic analysis reveals changes in bacterial communities and antibiotic resistance genes in an eye speciality hospital and a general hospital before and after wastewater treatment. Front. Microbiol. 2022, 13, 848167. [Google Scholar]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Vennema, H.; Deforche, K.V.D.; Avoort, H.V.D.; Peñaranda, S.; Oberste, M.S.; Vinjé, J.; Koopmans, M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180332. [Google Scholar] [CrossRef]
- Hernández-García, M.; Castillo-Polo, J.A.; Cordero, D.G.; Pérez-Viso, B.; García-Castillo, M.; Saez de la Fuente, J.; Morosini, M.I.; Cantón, R.; Ruiz-Garbajosa, P. Impact of ceftazidime-avibactam treatment in the emergence of novel KPC variants in the ST307-Klebsiella pneumoniae high-risk clone and consequences for their routine detection. J. Clin. Microbiol. 2022, 60, e02245-21. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbour-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).