Abstract
Effluents from wastewater treatment plants has been identified as a main point-source of antibiotics and antibiotic resistance genes in natural water environments. In this study, a typical municipal sewage treatment system in south China was taken as the research object to investigate the effects of each treatment unit on eight target antibiotics (sulfamethoxazole, sulfamethazine, tetracycline hydrochloride, oxytetracycline dihydrate, norfloxacin, ofloxacin, clarithromycin, roxithromycin), 17 antibiotic resistance genes (ARGs) and class 1 integron genes in the system using Accelerated Solvent Extraction-Solid phase extraction-Ultra high Performance Liquid Chromatography-Tandem mass spectrometry (ASE-SPE-UPLC-MS/MS) and real-time fluorescent quantitative PCR (qPCR) and the correlation between them. Seven antibiotics (mainly sulfonamides and tetracyclines, 4.19–141.97 ng·L−1) were detected in the influent, while only sulfamethoxazole, sulfamethazine, ofloxacin, and clarithromycin were detected in the effluent (3.11–16.61 ng·L−1). The tetracycline antibiotics in the wastewater treatment plant (WWTP) were transferred to the sludge phase by adsorption, in which tetracycline hydrochloride and oxytetracycline dihydrate were mostly removed in the aerobic and anaerobic stages, while sulfamethoxazole was mainly removed through biological transformation. Sul I was the most abundant resistance gene, but the WWTP had no obvious effect on its removal. Anaerobic treatment was found to play an important role in tetA, tetQ, and tetX removal. Moreover, correlation analysis revealed that the relative abundance of tetX was significantly correlated with clarithromycin (p = 0.039) and ofloxacin (p = 0.028), while that of tetQ was significantly correlated with sulfamethazine (p = 0.007) and sulfamethoxazole (p = 0.001), and that of tetC was significantly correlated with the class 1 integron gene (p = 0.014). Overall, the results presented herein provide a reference for improving the antibiotics and ARGs removal efficiency of WWTPs in south China.
1. Introduction
In 2013, the total amount of antibiotics used in China reached 162,000 tons, 48% (77,760 tons) of which were used in the medical field, while 52% (84,240 tons) were employed for veterinary [1]. In the United States, only 22,700 tons of antibiotics were used in 2009, of which 9600 tons were used for medicine, and the per capita use was less than 10% of that in China [2]. However, most antibiotics have a low metabolic rate after ingestion by humans and animals, and 50% to 90% of them are excreted in the form of maternal actives or conjugates through urine and feces [3]. When effluents treated by a sewage treatment plant enters a natural water body, the functional characteristics of these antibiotics can quickly stabilize, inducing bacteria to develop antibiotic resistance genes (ARGs) under the pressure of persistent selection [4]. This could promote the global spread of antibiotic resistance genes, and ultimately pose a major threat to the health and safety of aquatic and terrestrial organisms, as well as human beings [5]. Therefore, it is necessary to evaluate the distribution characteristics of antibiotics and resistance genes in wastewater treatment plants (WWTPs) and understand their propagation mechanisms.
The fate and removal efficiency of antibiotics, ARGs, and antibiotics resistant bacteria (ARB) by WWTPs has recently received increasing interest in China, and several studies have characterized their distribution in WWTPs in different regions [6,7,8]. Zhang et al. and Gao et al. investigated the degree of antibiotic contamination in different sewage treatment plants in the Jiulongjiang and Lanzhou regions of China, respectively [9,10]. In northern China, 30 ARGs that confer resistance to tetracyclines, sulfonamides, quinolones or macrolides were detected in two WWTPs, and 12 ARGs (8 tet, 2 sul, qnrB, and ermC) were discharged through the dewatered sludge and final effluent at higher rates than influent, illustrating overall proliferation of ARB [11]. Liu et al. reported the occurrence of chloramphenicol and tetracycline in urban sewage of Guiyang City (Southwest China), the concentration of CLA, OTC, and tetracycline hydrochloride (TC)fluctuated in the range of 0.16–65.20 ug·L−1, and found that untreated urban sewage could seriously affect river water and sediment [12]. Yuan et al. evaluated the occurrence and removal rules of ARB and ARGs (vanA, ampC, sul, Iand ereA genes) in a secondary municipal WWTP in Shanghai for a whole year, found that no statistical relevance between all ARB abundances and their corresponding genes, while most ARB and ARG proportions were positively related to the chemical oxygen demand (COD) and turbidity of the raw sewage (it may be an indirect link) [13]. However, few studies have studied both antibiotics and ARGs in Chinese sewage treatment systems, and the co-occurrence profiles and relevance among diverse ARGs and antibiotic harbored in different processing units within WWTPs have not been systematically explored.
At present, municipal WWTPs in China have the main purpose of removing suspended solids (SS), organic matter, and microbial contamination, but do not target the degradation of antibiotics and ARGs [14]. Nevertheless, as the main repository for this new type of trace contaminant entering the environment [15,16,17], WWTPs play a vital role in their risk management; therefore, it is necessary to conduct in-depth research regarding the pollution level and migration and transformation of antibiotics and ARGs in sewage treatment plants.
In this study, the concentration and abundance of eight typical antibiotics and 17 ARGs in each treatment unit of a WWTP in Shenzhen were investigated, and the reduction rules of each treatment process were analyzed in detail. Different media were then utilized to reveal the degradation mechanism of biodegradation and sludge adsorption. Finally, the correlations between ARGs and different antibiotics in this system were identified through correlation analysis to provide a technical reference for the improvement of the removal capacity of antibiotics and ARGs in sewage treatment systems in South China.
2. Materials and Methods
2.1. Instruments and Reagents
An ultra-high performance liquid chromatography-mass spectrometry system (Acquity UPLC-Quattro Premier XE tandem, UPLC-MS/MS), Oasis HLB cartridge (6 mL/500 mg) and column (BEHC18, 1.7 μm, 2.1 × 50 mm) were provided by Waters (USA). Ultra-pure water system (arium® comfort II) was obtained from Sartorius (Germany). The real-time PCR system (StepOne Plus™) was obtained from Applied Biosystems (USA), and the Gel imaging system (GelDoc XR+) was provided by BioRad (USA).
Standard samples of eight antibiotics were obtained from Dr. Ehrenstorfer GmbH (Germany). Two sulfonamides (SAs), including sulfamethoxazole (SMX, 97%) and sulfamethazine (SMZ, 99%). Two tetracyclines (TCs), including tetracycline hydrochloride (TC, 97%) and oxytetracycline dihydrate (OTC, 97%). Two quinolones (QNs), including norfloxacin (NOR, 99%) and ofloxacin (OFX, 97%), and two macrolides (MLs), including clarithromycin (CLA, 99%) and roxithromycin (ROX, 97%). UPLC grade methanol and acetonitrile were obtained from Merck (Germany), and analytical reagent grade Na2EDTA and Na2HPO4 were obtained from Merck Beijing Chemical Works (Beijing). Power SYBR® Green PCR Master Mix was obtained from Tiangen Biotech (Beijing), and Premix Ex Taq™ Hot Start was obtained from Takara Biomedical Technology (Beijing) Co., Ltd (Beijing, China). The FastDNA Spin Kit for soil was provided by MP (USA). The primers used in this experiment were synthesized by Shanghai Shenggong Bioengineering Co., Ltd (Shanghai, China).
2.2. Description of Sampling Points and Sample Collection
The largest sewage treatment plant in Shenzhen, South China was taken as the research object. The plant utilizes a combined high-efficiency sedimentation tank + UV disinfection process to treat wastewater originating from Qiaocheng East Road in the Futian District, west to the Nanshan Qianhai Free Trade Zone and Mawan Port in the central and western regions. The service area is 103 square kilometers, and it serves about 2.1 million people with a processing scale of 73.6 × 104 m3·d−1. Sample collection was conducted from April to September of 2019, during which the samples were collected hourly within one sampling day a month from 8:00 am to 8:00 pm to generate a mixed sample. Sewage samples were collected from behind the inlet screen (Inf), the outlet of the swirling grit chamber (Sw-W), the outlet of the secondary settling tank (Ss-W), and the outlet of the UV disinfection device (UV-W). Sludge samples were taken from the anaerobic tank (An-S), aerobic tank (Ao-S), anoxic tank (Ox-S), secondary settling tank (Ex-S), and sludge dewatering device (Dw-S). Before entering the dehydration equipment, flocculant (PAC) needs to be added to the excess sludge, and then it will be stirred and flocculated to alum flowers. Finally, it’s pumped into the belt filter press to complete the dehydration process. Sampling locations are shown in Figure 1. For each sewage sampling point, 2000 mL samples were collected into polypropylene bottles that had been rinsed with water and methanol. For each sludge sampling point, about 50 g dewatered sludge, and untreated sludge samples were wrapped in prebaked aluminum foil and sealed in Ziplock bags. All collected samples were processed immediately after being shipped back to the laboratory within half an hour. The characteristics of the inlet and outlet can be found in Table 1.
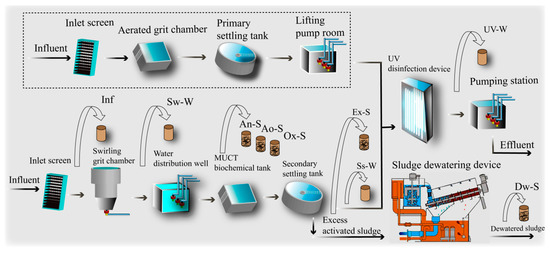
Figure 1.
Flowsheet of the wastewater treatment plant and sampling points. Inf, effluent of the grid screen as the influent of the main process. Sw-W, effluent of the swirling grit chamber. An-S, activated sludge in the anaerobic unit. Ao-S, activated sludge in the anoxic unit. Ox-S, activated sludge in the oxic unit. Ss-W, secondary settling tank effluent. Ex-S, excess sludge from the secondary settling tank. Dw-S, sludge treated by the dewatering device. UV-W, disinfection effluent treated by the UV device. The dotted line refers to the standby system, which was operated during heavy rain. The standby system was turned off during this study.

Table 1.
Physicochemical characteristics of raw wastewater and effluent.
2.3. Extraction and Determination of Target Antibiotics
Solid phase extraction-Ultra high Performance Liquid Chromatography-Tandem mass spectrometry (SPE-UPLC-MS/MS) methods. Sewage samples were filtered through GF/F glass fiber filters (0.45 μm, Whatman, UK) to remove the suspended impurities, after which antibiotic compounds were extracted by solid phase extraction (SPE) as previously described. After the sludge sample was sent back to the laboratory, it was first centrifuged and concentrated, then frozen at −20 °C for 12 h, then put it into the vacuum freeze dryer for 24 h to constant weight, and finally the dried sludge was ground and sieved with an agate mortar (80 mesh). The sludge samples were purified by ultrasonic extraction (USE) and then extracted by SPE. Samples were analyzed for eight target antibiotics were analyzed by UPLC-MS/MS. A detailed description of the analytical procedures and optimal parameters of extracts for quantification can be found in Zhang et al. [18]. Liquid phase conditions: the mobile phase consisted of eluent A (0.2% formic acid in acetonitrile) and eluent B (0.1% formic acid in ultrapure water), flow rate is 0.2 mL/min, injection volume is 5 mL, column temperature is 30 °C. The gradient elution procedure is as follows: 0–8 min: 10% A, 90% B; 8–12 min: 12%–20% A, 88%–80% B; 12–14 min: 20%–45% A, 80%–55% B; 14–15 min: 45%–100% A, 55%–0% B. Mass spectrometry conditions. A Quattro Premier XE triple quadrupole mass spectrometer equipped with an electrospray ionization (ESI) source and it’s operated at the ion source temperature of 120 °C and positive ionization (PI) mode. The capillary and nozzle voltages were 2.5 kV and 0.2 V, respectively. Quantification of each target compound was performed in multiple reaction monitoring mode (MRM). The mass spectrometry parameters of the target antibiotic are shown in Table 2.

Table 2.
Experimental parameters of electrospray ionization tandem mass spectrometry for the determination of antibiotics and surrogates.
Quality assurance. To quantify the recovery, detection limit, and relative standard deviation of the detection method, a recovery experiment was conducted. For each set of samples, at least one program blank and one independent inspection standard were run to check for background contamination and evaluate the instrument performance. Six sewage samples and six sludge samples were analyzed, three of which were spiked with the antibiotic mixed standard solution to determine the relative standard deviation (RSD). The method detection limits (MDL) were calculated by S/N = 3. The recovery rates of target antibiotics in sewage and sludge samples ranged from 74% to 96% and 82% to 97%, respectively, with an RSD < 3.2%. The MDL ranged from 0.2 ng·L−1 to 1.5 ng L−1 and 0.1 ng·L−1 to 1.2 ng·L−1, respectively. Quantitative analysis of the linear equation concentration range of 0.1–1000 ng·L−1, the coefficient of determination (R2) is greater than 0.999. These results indicated that the sensitivity and accuracy of the method could satisfy the subsequent analysis of antibiotic residues. The recovery and MDL are shown in Table 3.

Table 3.
Recovery (%), coefficient of determination (R2), method detection limits (MDL), and method quantitation limits (MQL) of the target antibiotics.
2.4. DNA Extraction and PCR
DNA extraction. After the sample was passed through a 0.45 μm filter, the solid trapped on the filter was washed off with PBS, and 1.0 mL of the mixture was transferred to a 2.5 mL centrifuge tube and centrifuged at 14,000 × g for 5 minutes, and then stripped off the sludge at the bottom of the tube by PBS. This process was repeated three times, after which a FastDNA Spin Kit for soil was used to extract DNA from the resulting pellet, according to the manufacturer’s instructions. According to previous reports of ARGs in this region [13,19,20], 17 representative ARGs were selected for PCR qualitative analysis, including nine TCs (tetA, tetB, tetC, tetD, tetM, tetO, tetQ, tetW, tetX), four SAs (sul I, sul II, sul III, sul A), two MLs (ereA, ermB), qnrS, blaPSE-1, class 1 integron gene (intI 1), and the bacterial 16S rRNA. The primer information is shown in Table 4. PCR was conducted in 20 μL reaction mixtures containing 10.0 μL Premix Ex Taq™ Hot Start Version, 1.0 μL forward primers, 1.0 μL reverse primers, 7.5 μL ddH2O, and 0.5 μL DNA extract. The PCR thermal cycling conditions were as follows: pre-denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s and annealing for 30 seconds and then final extension at 72 °C for 60 s. The PCR products were stored at −20 °C until they were stained with Acid Gen dye and checked by 1.5% (weight/volume) agarose gel electrophoresis. Finally, the specificity of the PCR products was further evaluated by the GelDoc XR+ system.

Table 4.
ARGs, intI 1, and 16S rRNA genes primer information for PCR and real-time PCR.
2.5. Quantification of ARGs by Real-Time qPCR
Based on the results of PCR, intI 1, the bacterial 16S rRNA gene and eight ARGs (tetA, tetC, tetQ, tetW, tetX, sul I, sul II, sul III) were selected for quantitative detection using SYBR Green II real-time PCR. The primer information is shown in Table 4. The qPCR was conducted in 20 μL reaction mixtures composed of 10.0 μL Power SYBR® Green PCR Master Mix (Tiangen Biotech, Beijing), 0.4 μL forward primers, 0.4 μL reverse primers, 8.2 μL ddH2O, and 1.0 μL DNA extract. The qPCR thermal cycling conditions were as follows: pre-denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing for 30 s (annealing temperature is shown in Table 4), and an extension at 72 °C for 30 s. Each sample was analyzed in triplicate to reduce the error.
2.6. Data Analysis
Statistical analysis and calculation of the data were performed using the Excel 2007 software, the correlation between ARGs and antibiotic concentrations at different sampling points was analyzed using IBM SPSS Statistics 17.0, and a statistically significant level of significance was found at 5% (p < 0.05). The drawing was done using Origin Pro 2018 (originlab, MA, USA). The sampling point plotting was done using the Adobe Illustrator CC 2017 software.
3. Results and Discussion
3.1. Antibiotic Contamination Level
3.1.1. Distribution and Concentration Variation of Antibiotics in the Aqueous Phase
The test results of eight target antibiotics in the wastewater phase and sludge phase of the sewage treatment plant are shown in Figure 2. The concentrations of the antibiotics in the wastewater are shown in Figure 2a. NOR was not detected in the influent water, while the other seven antibiotics were detected at concentrations of 4.19–141.97 ng·L−1, although the overall level was low. The concentration of OFX in the influent was the highest, which may be related to the large consumption of sulfonamides in this region [31]. Additionally, the OFX levels were close to those reported in a prior investigation of sulfa antibiotics in seven wwtps in the Ebro River Basin (27.2–592 ng·L−1) [32]. The concentration of TC in the influent was 4.92 ng·L−1, which was lower than that observed result by Watkinson in the Brisbane (Australia) wastewater treatment plant (0.08–1.50 ng·L−1), indicating that the TC pollution level in the area was lower than that of Brisbane [33]. The concentration of SMX in the influent of sewage treatment plants in Hong Kong [34] and Guizhou [35] was at the μg level, which was much higher than the levels observed in this study. According to McCardell, the CLA content in the influent of three sewage treatment plants in Switzerland was 57–330 ng·L−1 [36], which also was much higher than the results of this study. Among quinolones, the total concentration of the inflow water was much lower than that obtained from monitoring data (2573 ± 241 ng·L−1) of Jia et al. in a Beijing sewage treatment plant [37]. Based on these findings, the degree of antibiotic contamination in the WWTP investigated in this study was relatively low, and the influent concentration of various antibiotics in different regions was significantly different, and it indicates that the level of antibiotics contamination in the region is at low risk.

Figure 2.
Median concentration of eight investigated antibiotics in the sludge phase (b) and wastewater phase (a). ∑TCs total concentration of tetracyclines (TC and OTC). ∑MLs total concentration of macrolides (ROX and CLA). ∑QNs total concentration of quinolones (NOR and OFX). ∑SAs total concentration of sulfonamides (SMZ and SMX).
After the raw water passed through the aerated grit chamber, the concentration of various antibiotics decreased slightly, which may have been a result of adsorption onto the surface of large particles, after which it was removed with the sedimentation of particles or degraded by aeration and photolysis [38,39]. Under the irradiation of sunlight, antibiotics undergo photoisomerization reaction, which can promote the decomposition of antibiotics [40]. The SMX concentration of the secondary settling tank effluent and the UV disinfection tank effluent were below the MDL, and the concentration of the remaining sludge was only 6.75 μg·kg−1, indicating that biodegradation plays a leading role in the removal of SMX. SAs are mainly anionic or neutral in sewage with a pH of about 7.0; therefore, they can generate electrostatic repulsion with activated sludge via the negatively charged surfaces, thereby hindering the occurrence of adsorption. In addition, the lgKow (octanol-water partition coefficient, Kow) of SAs was less than 2.5 and its Kd (distribution coefficient, Kd) was less than 150 L·kg−1 [41]; therefore, the activated sludge composed of negatively charged colloidal particles has a weak adsorption capacity for SAs [42], and biodegradation and conversion are the main removal routes for such antibiotics in wastewater plants. We also found that the concentration of OFX and SMZ increased slightly after effluent from the secondary settling tank passed through the UV disinfection tank. Under the action of UV photons, the structure of the combination of substances changed to form a new free body, which may have caused substances in the matrix to interact and change so that more targets would be released into the liquid phase, eventually resulting in an increase of its concentration in the effluent. [43] At the same time, five antibiotics (CLA, OFX, SMX, and SMZ) were detected in the final effluent at levels between 3.11 ng·L−1 and 16.61 ng·L−1, while the other three target antibiotics were undetectable.
3.1.2. Distribution and Concentration Variation of Antibiotics in the Sludge Phase
The concentrations of antibiotics in sludge are shown in Figure 2b. Antibiotics are brought into the processing unit by effluent from the aerated sand settling tank and sludge from the secondary settling tank. The adsorption of activated sludge and the transformation and degradation of microorganisms play a key role in the removal of most antibiotics [7,44]. When compared with the previous treatment unit, the concentrations of TC, OTC, NOR, and ROX of aerobic and anaerobic sludge samples increased significantly, indicating that the TCs and QNs in the influent of the secondary settling tank were not actually removed, but shifted into the sludge. The ionic interaction and surface complexation mechanism of TCs significantly enhance their adsorption in sewage treatment systems, and QNs can adsorb sludge flocs by the electrostatic interaction of ferric and aluminum salt coagulants in sewage, thereby producing a high sludge adsorptive capacity [45,46]. Specifically, the concentration of TC in the aerobic unit sludge was higher than that of OTC, while the OTC concentration was higher than that of TC in the anaerobic unit sludge, which may have been because TC was in the fast adsorption stage under aerobic conditions [47], at which time its adsorption amount on the activated sludge had not reached the adsorption equilibrium state, so it was mainly adsorbed in the aerobic unit. Under anaerobic conditions, OTC can be complexed with metal ions and exchanged with cations. In addition, the dilution of reflux sludge and anaerobic biodegradation have an auxiliary promoting effect, so it is mainly adsorbed in anaerobic units. Overall, the average removal of antibiotics from the biochemical treatment stage and the dehydration process was higher than that of the grid, grit chamber, and secondary settling tank.
3.2. Pollution Level of Antibiotic Resistance Genes
3.2.1. Occurrence of Antibiotic Resistance Genes in Influent and Effluent
The target antibiotics in the influent and effluent were detected by PCR (Table 5). According to the qualitative test results of PCR, 11 target ARGs were detected positive in the influent, including seven tetracyclines, three sulfonamides, and a class of integrons, while macrolides and quinolone resistance genes were negative. Eight ARGs were detected positive in effluent, including four tetracyclines, three sulfonamides, and one type of integrant. The tetA, tetB, tetC, tetX, sul I, sul II, sul III, intI 1 were detected in both the inlet and outlet, while tetD, tetQ, tetW were detected only in the inlet, indicating that process can reduce tetQ and tetW coding genes of tetracycline ribosomal protective proteins, and the removal of the tetracycline antibiotic modification gene tetX and sulfonamide ARGs was limited.

Table 5.
Detection of the target antibiotic resistance genes in the influent and effluent.
3.2.2. Relative Abundance and Distribution Characteristics of ARGs
To compare the relative abundances in different samples, the ARGs detected by qPCR were normalized to 16S rRNA (Figure 3). The results revealed that the sul I was the dominant sulfonamide resistance gene, and its relative abundance was 10−5.44–10−4.38, while the relative abundance of sul II and sul III was very low (negligible). Compared with the influent, the relative abundance of sul in the effluent of secondary clarifier and dewatered sludge decreased only slightly, indicating that the sewage treatment system had little effect on reduction of the sul gene, which was consistent with the results of a study by Wen et al. that showed the relative abundance of the sul gene remained stable in each treatment unit of a WWTP [48]. Moreover, the relative abundance of tetC, tetQ and tetX in influent was on the same order of magnitude (10−5.68, 10−5.59, and 10−5.62, respectively), which were about half order of magnitude higher than tetA. The relative abundance of tetW was extremely low in this WWTP, contrary to the results reported by Chen et al., who found high levels in rural domestic sewage treatment systems [49]. The tetA, tetQ, and tetX in anaerobic activated sludge decreased significantly, suggesting that the anaerobic treatment process played an important role in their removal. Conversely, the relative abundance of tetC in anaerobic and anoxic units increased significantly, returning to the original level in the oxic unit, which may be attributed to microbial adsorption, wastewater receiving capacity, or variation in total bacterial abundance [50]. In addition, tetA generally increased with the treatment process, reaching a maximum level in the excess sludge, then decreased to the influent level after dehydration. These findings indicate that polyacrylamide and dehydration processes help remove tetA, which is consistent with the results reported by Munir et al. [25]. It should be noted that the relative abundance of a class of integrons and tetC increased sharply in the effluent of the secondary clarifier (10−4.65 and 10−5.06, respectively) to levels obviously higher than in other treatment units. The high variability and negative removal of this antibiotic removal effect were because of the conversion of metabolites and the release of antibiotics from suspended solids in the effluent. [51]

Figure 3.
Enrichment of nine investigated genes (sul I, sul II, sul III, tetA, tetC, tetQ, tetX, tetW, intI 1) relative to 16S rRNA through each stage of WWTP. ∑tet total relative abundance of tetracyclines gens (tetA, tetC, tetQ, tetX and tetW). ∑sul total relative abundance of sulfonamide gens (sul I, sul II, and sul III).
3.3. Correlation Analysis between Antibiotics and ARGs
The rapid proliferation of ARGs in aquatic environments is affected by many factors, with the existence of antibiotics being one of the primary ones. Therefore, it is necessary to analyze the relationship between the relative abundance of ARGs and the concentration of antibiotics. Correlation analysis of ARGs and the corresponding antibiotics in the collected WWTP samples are shown in Figure 4. As shown in Figure 4c,d, the relative abundance of tetQ was significantly positively correlated with SMZ concentration (r = 0.892, p = 0.007) and SMX concentration (r = 0.946, p = 0.001), respectively. According to the finding, there is no significant correlation between the abundance of tet-ARGs and the concentration of tetracyclines but is strongly correlated with the concentration of sulfonamides, which may be the result of the co-selection of ARGs by antibiotics in the environment [52]. As shown in Figure 4a,b, tetX was positively correlated with CLA (r = 0.779, p = 0.039) and OFX (r = 0.809, p = 0.028), respectively, while tetX was positively correlated with ROX (r = 0.891, p = 0.007; Figure 4e) and there was no correlation with TCs. These findings illustrate that antibiotic concentration is not the only factor affecting ARGs abundance. In fact, ARGs may also be affected by light [53], temperature [54], heavy metals concentrations [55], and other pollutants and environmental factors. In addition, no significant correlation was observed between other genes (including sul) and antibiotic content, which is consistent with a previous survey of antibiotics and ARGs in a sewage treatment plant and polluted river in Beijing [56]. Furthermore, a significant positive correlation was found between tetC and the relative abundance of a class of integrons (r = 0.855, p = 0.014; Figure 4f), supporting the view that a class of integrons,, including mobile gene elements (MGEs) can mediate horizontal metastasis (HGT) of ARGs [57], which might provide some information to prove that intI 1 plays a key role in the acquisition and prevalence of ARGs among various microorganisms via the horizontal gene transfer pathway. Taken together, these findings indicate that the correlation between ARGs and antibiotics is not common in all WWTP processes and that it may be the result of co-selection of antibiotics and environmental factors for ARGs in wastewater. [58] Therefore, to better describe and understand the relationship between ARGs and antibiotics, additional studies investigating the selective pressures caused by persistence and evolution should be conducted. Detailed correlation analysis data can be found in Table S1.

Figure 4.
Correlation analysis of the ARG/16S rRNA gene and antibiotic residue concentration in collected samples. △, sewage sample Inf; ☐, sewage sample Sw-W; ▲, sludge sample An-S; ◆, sludge sample Ox-S; ■, sewage sample Ss-W; ●, sludge sample Ex-S; ○, sludge sample Dw-S. (a) tetX and CLA; (b) tetQ and SMX; (c) tetQ and SMZ; (d) tetQ and SMX; (e) tet and ROX; (f) tetC and intI 1.
4. Conclusions
Four classes of eight commonly used antibiotics in the sewage and sludge of a WWTP in south China were analyzed by SPE-UPLC-MS/MS, and four classes of 17 commonly used ARGs were detected using PCR and qPCR. SMX, SMZ, and OFX were the dominant antibiotics (4.19–141.97 ng·L−1) in the influent, while the concentrations of TC, OTC, CLA, and ROX in the sludge (6.05–22.52 μg·kg−1) were higher than those in the sewage (3.11–8.94 ng·L−1). Based on the correlation analysis, the relationship between different ARGs and antibiotics was investigated. The relative abundance of tetracycline resistance genes (tetX and tetQ) was significantly correlated with the concentration of sulfonamide antibiotics (SMZ and SMX) (0.779 < r < 0.946, 0.001 < p < 0.039). There was also a correlation between tetC and a class of integrons (r = 0.855, p = 0.014). The results presented herein provide a reference for improving our understanding of the fate and pollution level of antibiotics and ARGs of WWTPs in South China. This study has revealed the correlation between the tet gene and sulfonamide antibiotic concentrations, tet, and int I 1, and elaborated their treatment effects at different stages of WWTP. However, no significant correlation between ARGs and corresponding antibiotics was observed. Thus, further studies are needed to investigate the effects of these antibiotics on the proliferation and transmission behavior of ARGs in sewage treatment plants. Those findings will substantially expand our current knowledge of antibiotics and ARGs in WWTPs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4441/11/12/2495/s1, Table S1: Correlation analysis of ARG/16S rRNA gene and the corresponding antibiotic residue concentration in collected samples.
Author Contributions
Conceptualization—H.L. and X.Z.; methodology—X.Z.; software—H.L.; validation—H.L. and X.Z.; formal analysis—H.L.; investigation—H.H. and J.Z.; resources—H.H. and J.Z.; data curation—H.L.; writing-original draft preparation—H.L.; writing-review and editing—X.Z.; visualization—H.L.; supervision—H.H. and J.Z.; project administration—H.H. and J.Z.; funding acquisition—X.Z. and J.Z.
Funding
This research was funded by the China Post-doctoral Science Foundation, grant number 2019M653086; the Scientific Research Foundation of Shenzhen Water (Group) Co. Ltd., grant number 20180004, and the Key Technology Assessment and Standardization of Urban Water Supply System Operation Management, grant number 2017ZX07501002.
Acknowledgments
The authors acknowledge the use of the StepOne Plus qPCR instrument at the School of Life Sciences, Sun Yat-Sen University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luo, Y.; Mao, D.; Rysz, M.; Zhou, Q.; Zhang, H.; Xu, L.; Alvarez, P.J.J. Trends in antibiotic resistance genes occurrence in the Haihe River. Environ. Sci. Technol. 2010, 44, 7220–7225. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the aquatic environment-a review-part II. Chemosphere 2009, 75, 435–441. [Google Scholar] [CrossRef]
- De Souza Santos, L.V.; Teixeira, D.C.; Jacob, R.S.; Amaral, M.C.; Lange, L.C. Evaluation of the aerobic and anaerobic biodegradability of the antibiotic norfloxacin. Water Sci. Technol. 2014, 70, 265–271. [Google Scholar] [CrossRef]
- Corno, G.; Yang, Y.; Eckert, E.M.; Fontaneto, D.; Fiorentino, A.; Galafassi, S.; Zhang, T.; Di Cesare, A. Effluents of wastewater treatment plants promote the rapid stabilization of the antibiotic resistome in receiving freshwater bodies. Water Res. 2019, 158, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.T.; Shuai, D.M. Research highlights: Antibiotic resistance genes: From wastewater into the environment. Environ. Sci. Water Res. Technol. 2015, 1, 264–267. [Google Scholar] [CrossRef]
- Peng, X.Z.; Zhang, K.; Tang, C.M.; Huang, Q.X.; Yu, Y.Y.; Cui, J.L. Distribution pattern, behavior, and fate of antibacterials in urban aquatic environments in South China. J. Environ. Monit. 2011, 13, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Ying, G.G.; Liu, S.; Zhao, J.L.; Yang, B.; Chen, Z.F.; Lai, H.J. Occurrence and fate of eleven classes of antibiotics in two typical wastewater treatment plants in South China. Sci. Total Environ. 2013, 452–453, 365–376. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.M.; Hansen, L.T.; Jamieson, R.C.; Neudorf, K.D.; Yost, C.K.; Tong, A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci. Total Environ. 2018, 643, 292–300. [Google Scholar] [CrossRef]
- Zhang, H.; Du, M.; Jiang, H.; Zhang, D.; Lin, L.; Ye, H.; Zhang, X. Occurrence, seasonal variation and removal efficiency of antibiotics and their metabolites in wastewater treatment plants, Jiulongjiang River Basin, South China. Environ. Sci. Process. Impacts 2015, 17, 225–234. [Google Scholar] [CrossRef]
- Gao, J.H.; Wang, Z.W.; Zhang, H.Y. Occurrence and the fate of typical antibiotics in sewage treatment plants in Lanzhou. Acta Sci. Circumst. 2016, 36, 3765–3773. [Google Scholar]
- Mao, D.Q.; Yu, S.; Ryszc, M.; Luo, Y.; Yang, F.X.; Li, F.X.; Hou, J.; Mu, Q.H.; Alvarez, P.J.J. Concurrence of aqueous and gas phase contamination of groundwater in the Wattenberg oil and gas field of northern Colorado. Water Res. 2015, 88, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, G.P.; Liu, C.Q.; Li, L.; Xiang, M. The occurrence of chloramphenicol and tetracyclines in municipal sewage and the Nanming River, Guiyang City, China. J. Environ. Monit. 2009, 11, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.B.; Guo, M.T.; Yang, J. Monitoring and assessing the impact of wastewater treatment on release of both antibiotic-resistant bacteria and their typical genes in a Chinese municipal wastewater treatment plant. Environ. Sci. Process. Impacts 2014, 16, 1930–1937. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence and removal of antibiotics in a municipal wastewater reclamation plant in Beijing, China. Chemosphere 2013, 92, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Czekalski, N.; Gascón Díez, E.; Bürgmann, H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Zhang, J.; Zong, D.; Chang, A.; Meng, F.; Wang, L.; Deng, W.; Guan, Y. Determination of common antibiotics in aquatic environment by solid-phase extraction and ultra pressure liquid chromatographytandem mass spectrometry (UPLC-MS/MS). Environ. Chem. 2015, 34, 1446–1452. [Google Scholar]
- Pan, M.; Chu, L.M. Occurrence of antibiotics and antibiotic resistance genes in soils from wastewater irrigation areas in the Pearl River Delta region, southern China. Sci. Total Environ. 2017, 624, 145–152. [Google Scholar] [CrossRef]
- Zou, S.; Zhu, C.; He, Z.; Luan, T.; Xu, W.; Zhang, G. Preliminary Studies on the Pollution Levels of Antibiotic Resistance Genes in the Water of Beijiang River, South China. Asian J. Ecotoxicol. 2009, 5, 655–660. [Google Scholar]
- Christine, M.; John, M.F.; Sadjia, B.; François, S.a.; Roger, C.L.; Roland, B.; Luke, M.; Serge, L.; Josée, H. Antimicrobial Resistance Genes in Enterotoxigenic Escherichia coli O149:K91 Isolates Obtained over a 23-Year Period from Pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liang, X.; Nie, X.; Huang, X.; Zou, S.; Li, X. The role of class I integrons in the dissemination of sulfonamide resistance genes in the Pearl River and Pearl River Estuary, South China. J. Hazard. Mater. 2015, 282, 61–67. [Google Scholar] [CrossRef]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China. J. Hazard. Mater. 2012, 235–236, 178–185. [Google Scholar]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef]
- Just, N.A.; Létourneau, V.; Kirychuk, S.P.; Singh, B.; Duchaine, C. Potentially pathogenic bacteria and antimicrobial resistance in bioaerosols from cage-housed and floor-housed poultry operations. Ann. Occup. Hyg. 2012, 56, 440–449. [Google Scholar]
- Luo, Y.; Xu, L.; Rysz, M.; Wang, Y.; Zhang, H.; Alvarez, P.J. Occurrence and transport of tetracycline, sulfonamide, quinolone, and macrolide antibiotics in the Haihe River Basin, China. Environ. Sci. Technol. 2011, 45, 1827–1833. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Zhang, H.; Luo, Z.; Yan, C. Occurrence, distribution, and seasonal variation of estrogenic compounds and antibiotic residues in Jiulongjiang River, South China. Environ. Sci. Pollut. Res. Int. 2012, 19, 1392–1404. [Google Scholar] [CrossRef] [PubMed]
- García-Galán, M.J.; Díaz-Cruz, M.S.; Barceló, D. Occurrence of sulfonamide residues along the Ebro River basin: Removal in wastewater treatment plants and environmental impact assessment. Environ. Int. 2011, 37, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, A.J.; Murby, E.J.; Costanzo, S.D. Removal of antibiotics in conventional and advanced wastewater treatment: Implications for environmental discharge and wastewater recycling. Water Res. 2007, 41, 4164–4176. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.W.; Minh, T.B.; Murphy, M.B.; Lam, J.C.; So, M.K.; Martin, M.; Lam, P.K.; Richardson, B.J. Distribution, fate and risk assessment of antibiotics in sewage treatment plants in Hong Kong, South China. Environ. Int. 2012, 42, 1–9. [Google Scholar] [CrossRef]
- Hu, J.; Juan, Z.; Shaoqi, Z.; Pan, W.; Tsang, Y.F. Occurrence and fate of antibiotics in a wastewater treatment plant and their biological effects on receiving waters in Guizhou. Process Saf. Environ. Prot. 2018, 113, 483–490. [Google Scholar] [CrossRef]
- McArdell, C.S.; Molnar, E.; Suter, M.J.; Giger, W. Occurrence and fate of macrolide antibiotics in wastewater treatment plants and in the Glatt Valley watershed, Switzerland. Environ. Sci. Technol. 2003, 37, 5479–5486. [Google Scholar] [CrossRef]
- Jia, A.; Wan, Y.; Xiao, Y.; Hu, J. Occurrence and fate of quinolone and fluoroquinolone antibiotics in a municipal sewage treatment plant. Water Res. 2012, 46, 387–394. [Google Scholar] [CrossRef]
- Lorenzo, F.; Navaratnam, S.; Edge, R.; Allen, N.S. Primary photophysical properties of moxifloxacin—A fluoroquinolone antibiotic. Photochem. Photobiol. 2008, 84, 1118–1125. [Google Scholar] [CrossRef]
- Samira, B.; Amin, T.Y.; Trong-On, D. Photocatalytic pathway toward degradation of environmental pharmaceutical pollutants: Structure, kinetics and mechanism approach. Catal. Sci. Technol. 2017, 7, 4548–4569. [Google Scholar]
- Werner, J.J.; Chintapalli, M.; Lundeen, R.A.; Wammer, K.H.; Arnold, W.A.; McNeill, K. Environmental photochemistry of tylosin: Efficient, reversible photoisomerization to a less-active isomer, followed by photolysis. J. Agric. Food Chem. 2007, 55, 7062–7068. [Google Scholar] [CrossRef]
- Stevens-Garmon, J.; Drewes, J.E.; Khan, S.J.; McDonald, J.A.; Dickenson, E.R. Sorption of emerging trace organic compounds onto wastewater sludge solids. Water Res. 2011, 45, 3417–3426. [Google Scholar] [CrossRef]
- Golet, E.M.; Alder, A.C.; Giger, W. Environmental exposure and risk assessment of fluoroquinolone antibacterial agents in wastewater and river water of the Glatt Valley Watershed, Switzerland. Environ. Sci. Technol. 2002, 36, 3645–3651. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, X.; Wen, X. Progress of the studies on occurrence and fate of new emerging micro-pollutants-PPCPs in municipal wastewaters. Chin. J. Environ. Eng. 2007, 1, 1–9. [Google Scholar]
- De Jesus Gaffney, V.; Cardoso, V.V.; Cardoso, E.; Teixeira, A.P.; Martins, J.; Benoliel, M.J.; Almeida, C.M.M. Occurrence and behaviour of pharmaceutical compounds in a Portuguese wastewater treatment plant: Removal efficiency through conventional treatment processes. Environ. Sci. Pollut. Res. Int. 2017, 24, 14717–14734. [Google Scholar] [CrossRef]
- Xia, Z.; Xiao, W.; Zhong, C.; Hao, X.; Qing, Z. Microbial community structure and pharmaceuticals and personal care products removal in a membrane bioreactor seeded with aerobic granular sludge. Appl. Microbiol. Biotechnol. 2015, 99, 425–433. [Google Scholar] [CrossRef]
- Peng, X.; Tan, J.; Tang, C.; Yu, Y.; Wang, Z. Multiresidue determination of fluoroquinolone, sulfonamide, trimethoprim, and chloramphenicol antibiotics in urban waters in China. Environ. Toxicol. Chem. 2008, 27, 73–79. [Google Scholar] [CrossRef]
- Chao, S.; Xue, S.; Peng, X.; Yun, W.a.n.g.; Shu, W. Investigation of fate and behavior of tetracycline in nitrifying sludge system. RSC Adv. 2015, 5, 87333–87340. [Google Scholar]
- Wen, Q.; Yang, L.; Duan, R.; Chen, Z. Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environ. Pollut. 2016, 212, 34–40. [Google Scholar] [CrossRef]
- 49Chen, H.; Zhang, M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 2013, 55, 9–14. [Google Scholar]
- Goyal, N.; Jain, S.C.; Banerjee, U.C. Comparative studies on the microbial adsorption of heavy metals. Adv. Environ. Res. 2003, 7, 311–319. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473–474, 619–641. [Google Scholar] [CrossRef] [PubMed]
- Binh, C.T.; Heuer, H.; Gomes, N.C.; Kotzerke, A.; Fulle, M.; Wilke, B.M.; Schloter, M.; Smalla, K. Short-term effects of amoxicillin on bacterial communities in manured soil. FEMS Microbiol. Ecol. 2007, 62, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Engemann, C.A.; Keen, P.L.; Knapp, C.W.; Hall, K.J.; Graham, D.W. Fate of tetracycline resistance genes in aquatic systems: Migration from the water column to peripheral biofilms. Environ. Sci. Technol. 2008, 42, 5131–5136. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Cha, J.; Carlson, K.H.; Pruden, A. Response of antibiotic resistance genes (ARG) to biological treatment in dairy lagoon water. Environ. Sci. Technol. 2007, 41, 5108–5113. [Google Scholar] [CrossRef] [PubMed]
- Mondragón, V.A.; Llamas-Pérez, D.F.; González-Guzmán, G.E.; Márquez-González, A.R.; Padilla-Noriega, R.; Durán-Avelar Mde, J.; Franco, B. Identification of Enterococcus faecalis bacteria resistant to heavy metals and antibiotics in surface waters of the Mololoa River in Tepic, Nayarit, Mexico. Environ. Monit. Assess. 2011, 183, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef]
- Che, Y.; Xia, Y.; Liu, L.; Li, A.D.; Yang, Y.; Zhang, T. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 2019, 7, 44. [Google Scholar] [CrossRef]
- McKinney, C.W.; Loftin, K.A.; Meyer, M.T.; Davis, J.G.; Pruden, A. tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 2010, 44, 6102–6109. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).