Abstract
Genomic clustering of functionally related genes is rare in yeasts and other eukaryotes with only few examples available. Here, we summarize our data on a nontelomeric MAL cluster of a non-conventional methylotrophic yeast Ogataea (Hansenula) polymorpha containing genes for α-glucosidase MAL1, α-glucoside permease MAL2 and two hypothetical transcriptional activators. Using genome mining, we detected MAL clusters of varied number, position and composition in many other maltose-assimilating non-conventional yeasts from different phylogenetic groups. The highest number of MAL clusters was detected in Lipomyces starkeyi while no MAL clusters were found in Schizosaccharomyces pombe and Blastobotrys adeninivorans. Phylograms of α-glucosidases and α-glucoside transporters of yeasts agreed with phylogenesis of the respective yeast species. Substrate specificity of unstudied α-glucosidases was predicted from protein sequence analysis. Specific activities of Scheffersomyces stipitis α-glucosidases MAL7, MAL8, and MAL9 heterologously expressed in Escherichia coli confirmed the correctness of the prediction—these proteins were verified promiscuous maltase-isomaltases. α-Glucosidases of earlier diverged yeasts L. starkeyi, B. adeninivorans and S. pombe showed sequence relatedness with α-glucosidases of filamentous fungi and bacilli.
1. Introduction
Maltose utilization in yeasts has been studied mostly in Saccharomyces as these yeasts are commonly used in brewing. The beer wort contains 50–60% of maltose, 15–20% maltotriose, and some other sugars, including isomaltose. These oligosaccharides are transported into the yeast cell, hydrolyzed to glucose by maltases and isomaltases, and fermented to ethanol and CO2 [1].
Ogataea (formerly Hansenula) polymorpha, Komagatella phaffii (formerly Pichia pastoris), and Candida boidinii are the most thoroughly studied methylotrophic yeasts [2,3,4]. We showed that one of these species—Ogataea polymorpha (Op)—can grow on maltose and sucrose using maltase (an α-glucosidase) for their hydrolysis [5,6]. In following studies, we focused on genetics, biochemistry, and regulation of utilization of α-glucosidic sugars in Op [7,8,9,10,11,12].
In Saccharomyces, α-glucosidic oligosaccharides are hydrolyzed by two specialized enzymes—maltases (EC 3.2.1.20) that degrade maltose and maltotriose (both α-1,4 linked), and isomaltases (EC 3.2.1.10) that degrade α-1,6 linked substrates such as isomaltose and palatinose [13,14,15]. These specific α-glucosidases (AGS) of Saccharomyces most probably evolved from a promiscuous ancestral protein ancMALS through gene duplications and further evolution of a duplicate [13]. Interestingly, we recently showed [12] that resurrected ancMALS and the Op maltase MAL1 are highly similar according to substrate usage and signature amino acids potentially involved in substrate binding. As Op belongs to an earlier diverged lineage of the ascomycetes [16], the presence of a living ‘twin’ of the hypothetical ancMALS protein supports the hypothesis raised in [13]. The Op MAL1 is a promiscuous enzyme with perfect catalytic properties towards a wide range of substrates: maltose, maltotriose, maltulose, sucrose, turanose, melezitose, isomaltose, isomalto-oligosaccharides, palatinose, and α-methylglucoside (α-MG). Therefore the MAL1 of O. polymorpha was regarded as maltase-isomaltase rather than a maltase [12].
In Saccharomyces cerevisiae, the genes required for utilization of α-glucosidic sugars are clustered in subtelomeric regions of the chromosomes forming so-called MAL clusters or loci. Genomic clustering of functionally related genes is rare in yeasts and other eukaryotes. Yet, aside from MAL loci, metabolic clusters for the utilization of galactose, allantoine, and nitrate are described in yeasts and filamentous fungi [17,18,19,20]. As emphasized in [21], metabolic gene clusters confer a survival advantage to the host when coinherited. As minimum, the MAL cluster of S. cerevisiae contains genes encoding a permease, an AG (maltase or isomaltase) and a transcriptional activator of the MAL genes. The number and composition of MAL clusters, as well as properties of encoded proteins, vary between the strains of S. cerevisiae [22,23,24]. In addition to Saccharomyces, the MAL loci have also been described for few other yeasts such as Scheffersomyces (Pichia) stipitis, an efficient fermenter of lignocellulosic sugars that has at least 35 loci of functionally linked genes [21,25]. However, the properties of the proteins encoded by S. stipitis MAL clusters have not been studied.
Clustering of MAL genes has also been shown for non-conventional yeasts Op [10] and Ogataea parapolymorpha (Opp) [26]. In the current work, we (i) searched for MAL clusters from the genomes of other non-conventional yeasts; (ii) performed the phylogenetic analysis of AGs and α-glucoside transporters (AGTs) encoded by the clusters; (iii) predicted substrate specificity of AGs using protein sequence analysis; and (iv) evaluated the correctness of the prediction by enzymatic analysis of three heterologously expressed AGs of S. stipitis.
2. Materials and Methods
2.1. The Yeasts and the Genomes
Yeast strains and genomes analyzed in the current study are listed in Table 1. Most of the genomes were accessed through the MycoCosm portal http://genome.jgi.doe.gov/programs/fungi/index.jsf [27]. Two Ogataea strains were studied: O. polymorpha (Op) and O. parapolymorpha (Opp) (Table 1). The first genome of Op (of RB11 strain, an odc1 derivative of CBS4732) was sequenced 15 years ago [28], but it has not yet been released to the public domain. The Op strains NCYC 495 (Table 1) and CBS 4732 mate, yield viable spores, and are almost identical in DNA sequence [29,30,31]. The genomes of Opp DL-1 [26], Meyerozyma guillermondii and Lodderomyces elongisporus [32] in MycoCosm originate from the National Center of Biolotechnology Information (NCBI). The genome of Debaryomyces hansenii is present as a copy from Génolevures Project in MycoCosm [33]. The genome sequence and gene predictions of Blastobotrys (Arxula) adeninivorans strain LS3 were obtained by Cécile Neuvéglise from MycoCosm. This genome of B. adeninivorans was originally sequenced by the Génolevures consortium [33]. The Schizosaccharomyces pombe genome in MycoCosm is a copy from www.pombase.org. MAL genes and clusters of S. cerevisiae S288C were used as a reference. S. stipitis CBS 6054 used in growth assay on sugars and cloning of the AG genes was kindly provided by Prof. A. Sibirny (Lviv, Ukraine).

Table 1.
Yeast strains and genomes analyzed in the current study.
2.2. Extraction of DNA and Protein Sequences and Analysis of Genomic Neighborhood of AG Genes to Detect MAL Clusters
Potential MAL genes were searched by using two different approaches. First, the Blast searches were run in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and MycoCosm websites against respective Op AG (MAL1) and AGT (MAL2) proteins to retrieve the genes encoding related proteins from other yeasts. Additionally, potential genes of interest were searched by their predicted function in the KOG (EuKaryotic Orthologous Groups) tab in MycoCosm webpage. Genes predicted to function in carbohydrate transport and metabolism were investigated further. The neighboring areas of the revealed genes were investigated using annotation data in the Synteny menu of MycoCosm. The MAL cluster was defined as a cluster comprising at least two potential MAL genes, with one of them encoding an AG. Due to that, not all potential MAL genes of studied species were covered in this study. For example, eight MAL clusters containing an AG gene were detected in Lipomyces starkeyi genome, but this yeast has several AG genes outside the clusters. pDRAW32 v1.1.107 (http://www.acaclone.com/) was used for analysis and visualization of the sequences of D. hansenii. Information on introns in the genes was extracted from MycoCosm and GenBank. Accession numbers and acronyms of the genes and proteins are given in Tables S1 and S2.
2.3. Alignment of Gene and Protein Sequences for Identity Evaluation, Construction of Phylogenetic Trees, and Defining Signature Amino Acids
Gene sequences from domains 1 and 2 (D1/D2) of large subunit ribosomal RNA (rRNA) [16] were used to build a phylogenetic tree of yeast species. As we could not find the large subunit rRNA sequences in genomic scaffolds of Cyberlindnera fabianii YJS4271, respective D1/D2 sequence (KY107353.1) of C. fabianii type strain CBS 5640 was used instead.
MEGA 7.0 package [40] was used to calculate neighbor-joining phylogenetic trees [41]. The Dayhoff model [42] was used for the protein (AGs and AGTs) phylograms, and the maximum composite likelyhood model [43] for the rRNA genes, with 1000 bootstrap replicates in both cases. The sequences were aligned using ClustalW [44] to calculate the identity values between the proteins presented in Tables S3 and S4 and to define the signature amino acids [12,13] suggested crucial for sugar binding. The genes were translated using respective alternative genetic code in case of CTG clade yeasts.
2.4. Heterologous Expression of AGs Encoded in the Genome of Scheffersomyces stipitis and Substrate Specificity Assay of the Enzymes
Three AGs: MAL7, MAL8 and MAL9 encoded by S. stipitis MAL loci [21] were cloned from S. stipitis CBS 6054 genomic DNA isolated with a PowerSoil Kit (MoBio, Carlsbad, CA, USA). Plasmid pY6 [45], kindly provided by C. Michels (New York, NY, USA), was used as a source of S. cerevisiae maltase gene MAL62. The polymerase chain reaction (PCR)-amplified genes were first inserted into pJET vector (Thermo Fisher Scientific, Waltham, MA, USA) and further cloned into pURI3-Cter vector [46]. Recombinant Pfu polymerase (Thermo Fisher Scientific, Waltham, MA, USA) was used in amplification and cloning, for primers see Tables S5 and S6. Resulting plasmids were electroporated into Escherichia coli coli BL21 (DE3) for heterologous expression of the AG proteins. Respective E. coli transformants were grown in 200 mL of lysogeny broth (LB) ampicillin (0.15 mg/mL) medium at 37 °C on a shaker to an optical density (OD) at 600 nm of ~0.5. Then 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added, the temperature was shifted to 22 °C and bacteria were further grown for ~20 h. Cells were harvested by centrifugation (2400× g, 20 min), washed in maltase buffer (100 mM K-phosphate buffer with 0.1 mM ethylenediaminetetraacetic acid (EDTA), pH 6.5)), suspended in the same buffer and crude cell extracts were prepared as in [6]. The cell extracts were assayed for the hydrolysis of 1 mM PNPG (p-nitrophenyl-α-d-glucopyranoside) and 100 mM sucrose, maltose, α-MG and palatinose in maltase buffer at 30 °C. Initial velocity of the reaction was measured by recording p-nitrophenol release (in case of PNPG) or glucose release in case of other substrates [47]. The activity was normalized to protein concentration in the extracts measured using a BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) and expressed in micromoles of hydrolyzed substrate per min per mg of protein in the extract (µmol/mg∗min; U/mg). Hydrolysis of the above listed substrates by cell extract of E. coli BL21 (DE3) transformant carrying the empty pURI3-Cter vector was assayed to register possible background activity.
2.5. Assay of Scheffersomyces stipitis Growth Ability on Sugars and Evaluation of Hydrolysis of α-Glucosidic Sugars by S. stipitis Cell Extracts
S. stipitis was grown in 0.67% BD Difco Yeast Nitrogen Base (YNB) medium (Thermo Fisher Scientific, Waltham, MA, USA) without amino acids supplemented with His (50 mg/L), Trp (50 mg/L), and 0.2% of a sugar (glucose, maltose, maltotriose, maltulose, sucrose, turanose, palatinose, xylose, or α-MG) on Greiner 96-well flat-bottom transparent polystyrol microplates (Greiner Bio-One, Frickenhausen, Germany) under agitation for 20 h at 30 °C. Cells grown overnight on 0.2% glucose were used for inoculation. Optical density of the culture at 600 nm was measured at the beginning and at the end of the experiment using an Infinite M200 PRO microplate reader (Tecan Group Ltd., Männedorf, Switzerland) equipped with Tecan i-control 1.7 software.
For enzymatic assay, S. stipitis was grown till mid-exponential growth phase in His- and Trp-supplemented YNB medium containing 1% of either glucose, maltose, sucrose, palatinose, or xylose. Sugars were added to growth medium from filter-sterilized (pore size 0.22 µm) solutions. The cells were collected by centrifugation at 4 °C, washed twice in maltase buffer and disrupted by using glass-beads. Supernatants of disrupted cells [6] were used as crude cell extract to measure specific activity of hydrolysis of PNPG, maltose, sucrose, palatinose and α-MG as described above.
3. Results
3.1. MAL Genes are Clustered in Ogataea polymorpha (Op) and O. parapolymorpha (Opp)
Sequencing of MAL genes in inserts of genomic library clones of Op CBS 4732 [8,10] revealed a four-gene MAL cluster. The genes for AG (MAL1) and AGT (MAL2) were shown adjacent sharing a bi-directional promoter [10] (Figure 1) as in the case of S. cerevisiae [48]. The MAL clusters of Op (in chromosome 1) and Opp (in chromosome 7) occurred similar (see Table S7 regarding the synteny). The sequence identity of the AGTs encoded in MAL clusters of Op and Opp was 87% and respective value for AGs was 98%. The MAL clusters of Op and Opp were not subtelomeric—the distance of Op MAL cluster from the chromosome end was about 410,000 bp, and the respective distance for Opp was 380,000 bp.
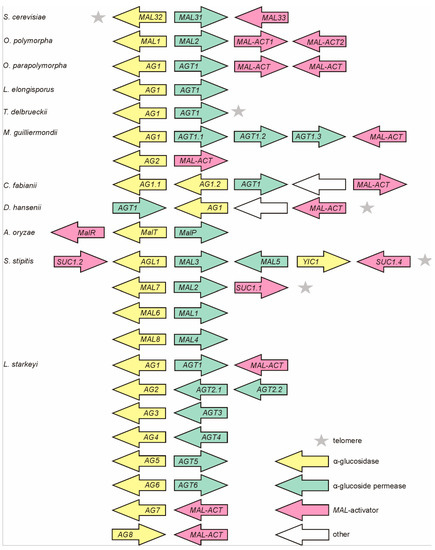
Figure 1.
MAL loci and their composition in Ogataea polymorpha (Op) NCYC 495, Ogataea parapolymorpha (Opp) DL-1 and other non-conventional yeasts. Genes (potentially) encoding α-glucosidases (AG), α-glucoside transporters (AGT), and MAL-activators (MAL-ACT) were retrieved from the genomic databases (see Table 1). The MAL clusters of Scheffersomyces stipitis [21], Aspergillus oryzae [50], and Saccharomyces cerevisiae (MAL32—maltase; MAL31—α-glucoside permease; MAL33—MAL-activator) are presented for comparison. Accession numbers and annotation data of the AG and AGT genes are given in Tables S1 and S2.
Interestingly, we detected hypothetical AGT and AG genes outside of the above-mentioned MAL cluster (Table S8) in both Ogataea species. However, we considered that these genes were not required for the growth of these yeasts on α-glucosidic sugars. If either the MAL1 or MAL2 gene of the MAL locus of Op was disrupted, the cells lost the ability to assimilate maltose, sucrose, turanose, maltotriose, maltulose, melezitose, isomaltose, palatinose, and isomalto-oligosaccharides [12]. Regarding Opp, Agaphonov et al. showed that disruption of the Opp MAL1 (a homologue of Op MAL1) resulted in colonies not growing on maltose [49]. Two MAL clusters have been also detected in Aspergillus oryzae, with only one of them proven functional [50].
3.2. MAL Clusters Are Also Present in the Genomes of Other Non-Conventional Yeasts
In addition to Op and Opp, MAL clusters have been identified in some other non-conventional yeasts such as S. stipitis [21] and Kluyveromyces lactis [51,52] as well as in Aspergillus fungi [50]. In current study, MAL clusters of varied content, position, and number were detected in the genomes of L. elongisporus, Torulaspora delbrueckii, M. guillermondii, C. fabianii, D. hansenii, and Lipomyces starkeyi (Figure 1) but not in B. adeninivorans and S. pombe. L. starkeyi was most special with eight MAL clusters discovered. Notably, most of the MAL genes of L. starkeyi had introns (see Tables S1 and S2). In S. stipitis, introns were present only in putative MAL-activator genes SUC1.1, SUC1.2, and SUC1.4, no introns were reported for MAL genes of other studied yeasts. Subtelomeric positioning was verified for MAL loci of D. hansenii and T. delbrueckii (Figure 1), and for two MAL clusters of S. stipitis [21].
3.3. MAL-Activator Genes Are Often Genomically Clustered with AG and AGT Genes
MAL-activator is a positive transcriptional regulator of MAL genes containing an N-terminal Zn(2)-Cys(6) DNA-binding domain [53,54,55]. Many laboratory strains of S. cerevisiae (such as S288C and W303-1A) fail to grow on maltose because of a defective MAL-activator allele [23,56,57]. Until now, functionality of the MAL-activators of S. cerevisiae (MALx3 proteins) and Candida albicans (SUC1 protein) has been proven [53,54,55,58]. Figure 1 (pink arrows) depicts potential MAL-activator genes in MAL clusters of yeasts and Aspergillus. Notably, while most MAL clusters have a divergently positioned pair of AG and AGT genes, the position and transcription direction of MAL-activator genes varies between the clusters. The MAL loci of both Op and Opp contain two potential MAL-activator genes with functionality and roles being yet not known. Our search did not detect potential MAL-activator genes in MAL clusters of L. elongisporus and T. delbrueckii.
3.4. Phylograms of AGTs and AGs Encoded by the MAL Clusters Largely Agree with Phylograms of Yeast Species
Our dataset of yeast genomes encompasses Ascomycota species of varied evolutionary age. The Ascomycota phylum is comprised of three monophyletic subphyla [59]: the Saccharomycotina, the Pezizomycotina (comprising filamentous fungi such as Aspergillus and Neurospora), and the Taphrinomycotina (syn. Archaeascomycota, comprising for example S. pombe). Schizosaccharomyces is most distant from other Ascomycota—evolutionary distance between Schizosaccharomyces and Saccharomyces is about 1 billion years, which is ~25% of the age of the Earth. The Saccharomycotina subphylum contains four major clades: (i) the Saccharomycetaceae; (ii) the CTG yeasts; (iii) the methylotrophs; and (iv) the basal group to the Saccharomycotina. The basal group that includes Lipomyces and Blastobotrys that were studied in current work is very heterogeneous according to their genomic signatures and proteomes [60].
Phylogenesis of AGTs and AGs was assayed as described in the Materials and Methods section. Proteins encoded in MAL clusters (Figure 1) were all included except for YIC1 (see Figure 1), a 823 aa protein annotated as a GH31 family α-glucosidase/xylosidase. In the case of B. adeninivorans and S. pombe in which we did not detect the MAL clusters, either proteins which have been experimentally studied (Sut1 and Mal1 of S. pombe) or closest homologues of respective Op proteins were added to the dataset. As the Sut1 transporter of S. pombe has a very low identity to other AGTs (Table S4), it was excluded from the phylogram. The Sut1 [61] is a fungal homologue of plant sucrose transporters that also has a considerable similarity to sugar transporters of bacteria.
In general, the phylogram of AGs coincided well with that of AGTs and D1/D2 sequences of ribosomal large subunit RNA of respective yeasts (Figure 2), suggesting that the MAL genes have evolved concomitantly with the yeast species. AGTs and AGs of Op and Opp clustered most closely with respective proteins of C. fabianii. AGTs and AGs of CTG yeasts (S. stipitis, L. elongisporus and M. guillermondii) [60,62] formed distinct clusters close to respective proteins of methylotrophs. AGTs and AGs of S. pombe, B. adeninivorans and L. starkeyi were revealed as early diverged representatives of these proteins. The phylogram of yeasts (Figure 2B) illustrates early branching of these three species. According to the literature, the family Lipomycetaceae is the earliest-branching lineage of Saccharomycotina, followed by a clade containing also B. adeninivorans [59].
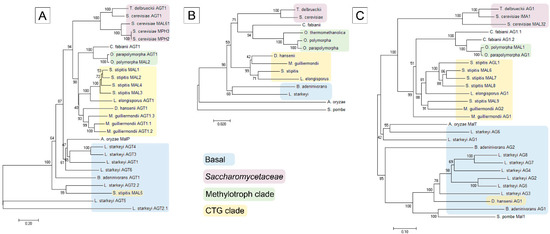
Figure 2.
Phylogram of α-glucoside transporters (AGT) (A); α-glucosidases (AG) (C); and respective host species (B). See the Materials and Methods section for details. The scale bar in panels (A,C) indicates the number of substitutions per amino acid site, the scale bar in panel (B) indicates the number of base substitutions per site. The subgroups of Saccharomycotina according to [60] are designated by different background coloring.
In two cases, we detected clustering of AGs and AGTs that did not match the evolution of yeast species—the MAL5 permease of S. stipitis and AG1 of D. hansenii clustered within respective proteins of basal clade to Saccharomycotina.
3.5. Analysis of Yeast AGs for Signature Amino Acids: Prediction of Substrate Specificity
In most cases, the AGs encoded by MAL clusters (Figure 1) have not been studied for functionality and properties. However, substrate specificity of an AG can, at least to some extent, be predicted on the basis of amino acid residues residing close to the active site pocket [12,13,63,64]. Crystal structure is available for S. cerevisiae (Sc) isomaltase IMA1 and its catalytically inactive mutant. Respective structures in complex with maltose [64] or isomaltose and maltose [65], have revealed the amino acids Y158, V216, G217, S218, L219, M278, Q279, D307, and E411 bordering the substrate-binding pocket. These nine amino acids are variable between the maltases, isomaltases, and maltase-isomaltases, and have been used as a signature sequence for AGs [12,13,63,64]. A shared signature sequence for Sc IMA1 and IMA2 was YVGSLMQDE (see Table 2). Mutation of Sc IMA1 has shown that Val216 (shown in bold in the signature sequence) is of key importance for the ability for the enzyme to use isomaltose and isomaltose-like sugars [63,65]. All Sc isomaltases had a Val at respective positions (Table 2) and they hydrolyze isomaltose and isomaltose-like sugars (for example palatinose and α-MG), but not maltose [66]. If Val216 was mutated to a Thr (Sc maltases have a Thr at this position), the ability to hydrolyze maltose emerged in IMA1 and isomaltose hydrolysis concomitantly decreased [63]. Mutation of Gly217 and Ser219 in IMA1 to maltase-specific amino acids Ala and Gly, respectively, had additional effects on substrate specificity, increasing the maltose/isomaltose hydrolyzing ratio [63]. Substitution of Gln279 in IMA1 with Ala also shifted substrate specificity—hydrolysis of isomaltose was strongly reduced and a low maltose-hydrolyzing activity emerged [63,65]. Maltase-isomaltases of Op (MAL1) and L. elongisporus (we designated it Le AG1) use both maltose- and isomaltose-like substrates [12,13]. Similarly to Sc maltase, these proteins had Thr, Ala, and Gly at positions 216–217–218 (Sc IMA1 numbering) respectively (Table 2). Thus, these amino acids are assumed to be required for the hydrolysis of maltose and maltose-like substrates by both maltases and maltase-isomaltases. Mutation of Thr200 (equivalent of Val216 of IMA1) in Op maltase-isomaltase to a Val strongly reduces hydrolysis of maltose-like substrates, making the enzyme more similar to isomaltases [12,13].

Table 2.
Signature amino acids of α-glucosidases (AGs) and prediction of substrate specificity. Background coloring is as follows: maltases (yellow), isomaltases (blue), maltase-isomaltases (green), enzymes not studied experimentally before this study (white). The key position of Sc IMA1 (Val216) is shown in bold. Characteristic motifs T216V217, A216I217, and V216I217 of AGs are shown in red, green, and blue letters, respectively. Bs, Bacillus stearothermophilus; Bt, Bacillus thermoglucosidasius; Ao, Aspergillus oryzae; Fo, Fusarium oxysporum; An, Aspergillus niger. See Table S1 for acronyms of AGs.
For prediction of function (substrate specificity) of AGs of non-conventional yeasts, the protein sequences were aligned and signature amino acids were extracted from the alignment. AGs of Sc, Aspergillus, Fusarium and Bacillus AGs were used for comparison (Table 2).
The AG of C. fabianii (Cf AG1.2), the closest neighbor of Op MAL1 in the phylogenesis tree (Figure 2C), has a signature HTAGLVGDN differing from that of Op MAL1 only in the first position, suggesting that AG1.2 of C. fabianii is a maltase-isomaltase. Cf AG1.1 was predicted by us as an isomaltase—its signature sequence has six matches with that of isomaltase IMA5 of Sc. We also suggested that MAL6, MAL7, MAL8, MAL9, and AGL1 of S. stipitis are all maltase-isomaltases. Their signature amino acids had the highest number of matches with respective positions of experimentally studied maltase-isomaltases of O. polymorpha and L. elongisporus [12,13]. According to our prediction, the AG2 of M. guillermondii is a maltase-isomaltase and AG1 is an isomaltase. Signature sequences of AGs of S. pombe, L. starkeyi, and B. adeninivorans significantly differed from those described above. These signature sequences were grouped according to three characteristic motifs. A T216V217 motif (IMA1 numbering) shown in red letters, was detected in Ls AG1, Ls AG6, and Ba AG2. The MalT protein of A. oryzae encoded in the MAL cluster (Figure 1) had a T216V217 motif in the signature sequence. According to [50] the MalT protein had PNPG-hydrolyzing activity, was induced at maltose growth, and was defined by the authors as a maltase. The Ba AG2 is also a maltase (our unpublished data). Therefore we suggested that AGs with a T216V217 motif in the signature sequence are maltases. An A216I217 motif (Table 2, in green letters) was detected in signature sequence of two experimentally characterized maltases—Mal1 of S. pombe [67] and α-1,4-glucosidase (maltase; BAA12704.1) of B. stearothermophilus. If Ala200 (equivalent to Val216 of IMA1) of B. stearothermophilus maltase was mutated to Val, the ability to hydrolyze maltose was lost [68]. Due to the presence of a A216I217 motif in the signature sequence, the Ls AG2, AG4, and AG5 were predicted as maltases. A V216I217 motif (shown in blue letters) is present in three experimentally studied AGs presented in Table 2: oligo-1,6-glucosidase (isomaltase) of B. thermoglucosidasius [68] and two isomaltases of filamentous fungi—AgdC of Aspergillus niger and Foagl1 of F. oxysporum [69]. If Val200 (corresponds to Val216 of IMA1) in B. thermoglucosidasius isomaltase was mutated to Ala, the ability to hydrolyze maltose emerged in the enzyme [68]. We predict that AG3, AG7, and AG8 of L. starkeyi, AG1 of D. hansenii and AG1 of B. adeninivorans are all isomaltases.
3.6. Substrate Specificity Evaluation of Scheffersomyces stipitis AGs: Verifying the Prediction
3.6.1. S. stipitis Assimilates Both Maltose-Like and Isomaltose-Like Sugars
Several AGs from non-conventional yeasts, including five AGs of S. stipitis, were predicted by us as maltase-isomaltases (Table 2) meaning that they should enable the yeast to assimilate a wide range of α-glucosidic sugars. According to the CBS data, S. stipitis CBS 6054 assimilates glucose, galactose, xylose, sucrose, maltose, trehalose, melezitose, α-MG and few other sugars. However, the ability of S. stipitis to grow on many α-glucosidic sugars such as turanose, maltulose, maltotriose, isomaltose, and palatinose has not been previously studied. We assayed the growth of S. stipitis on these sugars using glucose, maltose, sucrose, xylose, and α-MG as reference substrates. All these sugars supported aerobic growth of S. stipitis showing that it assimilates not only sugars that are cleaved by maltases (maltose, maltotriose, turanose, maltulose), but also those cleaved by isomaltases (isomaltose, palatinose, α-MG) [12,13]. Therefore, this yeast must have enzymatic capacity to hydrolyze both maltose- and isomaltose-like sugars.
3.6.2. Cell Extracts of S. stipitis Hydrolyzed both Maltose-Like and Isomaltose-Like Sugars
S. stipitis was grown on glucose, sucrose, maltose, palatinose and xylose as a single carbon source, all supplemented at 1%. Cell extracts were assayed for the hydrolysis of 1 mM PNPG and 100 mM maltose, sucrose, palatinose and α-MG (Figure 3).
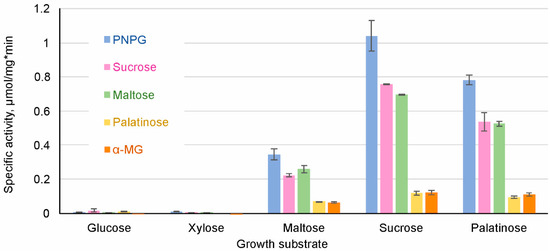
Figure 3.
Specific activity of hydrolysis of α-glucosidic substrates in cell extracts of S. stipitis grown on different sugars. α-MG, α- methylglucoside; PNPG, p-nitrophenyl-α-d-glucopyranoside.
The extracts of cells grown on either glucose or xylose had only negligible ability to hydrolyze PNPG, sucrose, maltose, palatinose and α-MG (Figure 3). High AG activity was observed in the cells grown on maltose, palatinose, or sucrose. Sucrose was the most potent inducer of AG activity, followed by palatinose (an isomaltose-like sugar). In some yeasts such as S. cerevisiae, sucrose can be hydrolyzed also by invertase (β-fructofuranosidase). AGs and invertases can be differentiated by the inability of AGs to hydrolyze raffinose [70]. As we did not detect raffinose-hydrolyzing activity in cell extracts of S. stipitis, sucrose hydrolysis measured in S. stipitis extracts was due to AG activity.
Repression of AGs by glucose has been described in several yeasts such as S. cerevisiae, O. polymorpha, and C. albicans [5,71,72]. Our data showed that xylose also repressed AGs expression. In accordance with our data, Jeffries et al. [25] detected the expression of only one AG gene (MAL8) in xylose-grown S. stipitis, and the level of expression was very low. Expression of MAL8 was not detected in glucose-grown S. stipitis [25].
3.6.3. MAL7, MAL8, and MAL9 of S. stipitis Proven to be Maltase-Isomaltases
As mentioned above, S. stipitis AGs were predicted by us in silico as maltase-isomaltases (Table 2). To verify this prediction, we chose three of them (MAL7, MAL8, and MAL9) for heterologous expression in E. coli to study their substrate specificity. Extracts of recombinant E. coli were used as crude preparations to assay enzymatic hydrolysis of PNPG, maltose, sucrose, palatinose, and α-MG as described in Materials and Methods. The extract of E. coli transformant carrying the maltase gene MAL62 of S. cerevisiae was used as a reference for maltase activity. In agreement with our earlier data [6], E. coli revealed no background AG activity. Results of substrate specificity assay are presented in Figure 4. Data regarding Op MAL1 (a maltase-isomaltase) and Sc IMA1 (an isomaltase) were taken from the literature [12,66].

Figure 4.
Substrate specificity of S. stipitis MAL7, MAL8, and MAL9 (lower panels) with O. polymorpha MAL1 (maltase-isomaltase), S. cerevisiae MAL62 (maltase), and IMA1 (isomaltase) (upper panels) used as references. Specific activity (µmol/mg·min) of each enzyme preparation with 100 mM sucrose was taken for 100%. Data on Sc IMA1 were calculated from Table 2 of [66], data on Op MAL1 were taken from Table 1 of [12]. The asterisk shows nondetectable or only negligible activity with corresponding substrates. Standard deviation values of the mean are given on the bars. See [66] for standard deviation data of Sc IMA1.
Substrate specificity assay of S. stipitis MAL7, MAL8, and MAL9 (Figure 4) confirmed our prediction—all these enzymes hydrolyzed not only maltose-like substrates (maltose, sucrose) but also isomaltose-like substrates (palatinose, α-MG). S. cerevisiae MAL62 used as a reference, behaved as a typical maltase using only maltose-like substrates.
4. Discussion
4.1. The Natural Habitat of Non-Conventional Yeasts Possessing MAL Genes Contains α-Glucosidic Sugars
Yeasts typically prefer sugars to other carbon sources and therefore their natural habitats are sugar-rich. Plant sap and berries are rich in sucrose, whereas degradation of plant starch yields a variety of α-glucosidic oligosaccharides such as maltose, maltotriose, and isomaltose [12]. Ogataea species that use α-glucosidic sugars as well as methanol, have been isolated from spoiled plant material, plant leaves, soil, and insect gut. Living plant leaves emit methanol [73], raising from turnover of cell-wall pectin, and methanol is also released in soil due to the degradation of plant pectin and lignin [12]. C. fabianii has been isolated from alcoholic beverages, sugar cane (very rich in sucrose) and clinical material. The isolate YJS4271 analyzed in this work, was isolated from olives in Spain [33]. M. guillermondii is a flavinogenic yeast that grows on n-alkanes and pentoses, but also on α-glucosidic sugars [35,74]. Intriguingly, riboflavin metabolism and α-glucoside utilization are somehow linked in M. guillermondii. So, riboflavin permease was synthesized only in media containing α-glucosides, including sucrose, maltose, α-MG, and melezitose, whereas cells grown on other substrates (glucose, fructose) did not take up riboflavin [74]. S. stipitis can be isolated from the gut of passalid beetles that inhabit and degrade white-rotten hardwood [21]. According to our best knowledge, the current study is the first one on the metabolism of α-glucosidic sugars in S. stipitis. D. hansenii is a halo- and cryotolerant marine yeast found in cheese, dairy and brine [75]. A lipid-accumulating yeast L. starkeyi CBS 1807, from which we detected eight genomic MAL clusters (Figure 1), was isolated from soil, which is considered a primary habitat of this yeast [76]. Bacterial and fungal soil residents assumingly support L. starkeyi with α-glucosidic oligosaccharides arising from starch-rich plant residues. B. adeninivorans strains have been isolated from soil and wood hydrolysates. This yeast has great biotechnological potential because of metabolism of rare substrates such as n-butanol and plant phenolics. Metabolism of α-glucosidic oligosaccharides has not been assayed in B. adeninivorans. S. pombe has been isolated from fermented beverages, fruits, kombucha, and molasses used to produce rum and tequila [77]. Our current study shows that all above-mentioned yeasts have genes for transport and hydrolysis of α-glucosidic sugars. Mostly, these genes were found as constituents of genomic MAL clusters.
4.2. A Bi-Directional Promoter between the AGT and AG Genes Contributes to Balance the Transport and Further Metabolism of Disaccharides
In most MAL clusters of yeasts (Figure 1) as well as of numerous Aspergillus species [50], the genes for AGT and AG occur divergently positioned, sharing a bi-directional promoter. This genomic setup has been preserved in evolution as it enables tightly coordinated expression of both genes. Co-induction of these genes from a shared promoter by maltose and co-repression by glucose has been reported for S. cerevisiae, O. polymorpha and A. oryzae [10,50,72]. Induction of the promoter by maltose was shown to be stronger in AG direction in Op, whereas the opposite was true for Sc [72,78]. The AGT activity has been shown first required to provide intracellular maltose for the induction of MAL genes [79,80]. As S. cerevisiae strains usually have several MAL loci, they most likely have sufficiently high basal AGT activity to ensure the induction. Our data on Op indicated that monosaccharides (glucose and fructose) produced at intracellular hydrolysis of disaccharides repress the MAL1 promoter and their phosphorylation is obligatory for the repression. At the same time, temporary accumulation of unphosphorylated hydrolysis products was shown to activate the promoter [11]. The growth of Op on disaccharides is assumingly complicated [11], because hydrolysis products of disaccharides that promote initial derepression of MAL genes will later cause repression when their phosphorylated species accumulate. Therefore, expression of AGT and AG proteins has to be finely adjusted with further glycolytic flux to provide an appropriate expression level of MAL genes. In good accordance with this, it was shown that S. cerevisiae cells (i) lyse if the transport and intracellular hydrolysis of maltose are not balanced [81]; and (ii) if grown at maltose limitation and then exposed to excess maltose, get rid of some intracellular glucose by efflux via glucose transporters [82].
4.3. How have the MAL Clusters Emerged and Evolved?
Among early diverged yeasts S. pombe, B. adeninivorans and L. starkeyi, MAL clusters were found only in L. starkeyi. MAL clusters have also been described in Aspergillus species [50] (belonging to Pezizomycotina, a subfamily of Ascomycota). We hypothesize that initially, a two-gene cluster comprised of divergently positioned AG and AGT genes was formed, and only after that was a MAL-activator gene added. As emphasized in [21], a divergent orientation (as in the case of AG and AGT genes) is typical for the genes that assumingly have evolved in longest association with one another. It is possible that the MAL clusters formed already before the separation of Pezizomycotina and Ascomycotina. Blastanalysis of AGs of L. starkeyi MAL clusters indicated that they were most similar to respective (putative) enzymes of Pezizomycotina species—Aspergillus, Fusarium, Penicillium, and others. Interestingly, the MAL cluster genes of L. starkeyi that belong to the most basal lineage of Saccharomycotina, were highly intronated. The genome of L. starkey is extremely rich in introns, with approximately three introns per protein-encoding gene [60]. Introns were also detected in AG genes of filamentous fungi A. niger and F. oxysporum (Table S1). According to [83,84], introns—a specific genetic feature of eukaryotes—existed at a high density in ancestral fungi and already in the last common ancestor of all extant eukaryotes. In many lineages of modern fungi (for example Saccharomyces and methylotrophs), massive loss of introns has been reported [60].
The bacterial origin of at least some MAL cluster genes can also be considered. According to Gabriško [85], although with low bootstrap values, fungal AGs of the glycoside hydrolase (GH) family 13 (GH13) always root deeply in the prokaryotic group, hinting that they may have bacterial ancestry. He also considers a possibility of (ancient) horizontal transfer of AG genes from bacteria. As bacteria do not have introns in protein-encoding genes, gain of introns (ancestral fungi are intron-rich) is expected. So, Da Lage et al. [84] showed that if an α-amylase gene from an actinobacterium was horizontally transferred to already intron-rich ancestor of Agaricomycotina fungi, introns were inserted soon after the transfer to adjust with requirements of the splicing machinery of the host.
Our current study (Table 2) clearly shows that AGs of early diverged yeasts Lipomyces, Blastobotrys, and Schizosaccharomyces have more similarity to AGs of filamentous fungi (Pezizomycotina) and bacteria (Bacillus) than to AGs of more recently diverged yeast lineages. Intriguingly, the MAL1 gene of O. polymorpha also has some features of a bacterial gene—its promoter region is perfectly recognized in bacteria as possesses two pairs of sigma 70-like sequences [8]. Due to that, the MAL1 promoter was successfully applied for overexpression of foreign proteins in E. coli [86].
4.4. Evolution of AGs: Repeated Changes in Substrate Specificity
We suppose that substrate specificity of AGs has been repeatedly altered during the evolution. According to the literature data (see Introduction) and our current study, specialized AGs—maltases and isomaltases—are found not only in ‘modern’ yeasts (S. cerevisiae), but also in early diverged Saccharomycotina species L. starkeyi and B. adeninivorans. In O. polymorpha, L. elongisporus [12,13], and S. stipitis (Figure 4) having intermediate position in Ascomycota phylogenesis (Figure 2B), promiscuous AGs have been described. Gabriško [85] suggested that (i) the common ancestor of the Ascomycota phylum had two specialized AG genes; (ii) in the subphylum Saccharomycotina, one of these genes (coding for isomaltase) was lost and the other one (coding for maltase) was retained and further duplicated in the lineage; (iii) in Pezizomycotina (filamentous fungi) evolution, both maltase and isomaltase genes were retained and one of them (coding for isomaltase) was duplicated. Considering that, filamentous fungi are expected to have more isomaltases than maltases. According to Gabriško [85], in distinct lineages of the Saccharomycotina, isomaltases evolved repeatedly from maltases, meaning that maltase-isomaltases found in O. polymorpha, L. elongisporus, and S. stipitis may represent ‘half-way’ variants of AGs evolution. As shown in [13] subsequent divergent evolution of promiscuous AGs can give rise to specific AGs—maltases and isomaltases. Most likely, the spectrum and amount of α-glucosidic oligosaccharides in yeasts’ habitat act as main driving force in substrate specificity evolution of AGs.
5. Concluding Remarks
Genome mining of non-conventional yeasts revealed MAL clusters that encode proteins most of which have not been characterized. AGs encoded by MAL clusters and elsewhere in the genome are perfect objects to study protein evolution, but they may also have biotechnological value. We detected eight MAL clusters in a lipid-accumulating yeast L. starkeyi, all encoding a potential AG protein. Since now, only one paper on an AG of L. starkeyi [87] has been published. It would be highly interesting to purify and biochemically characterize AGs of early diverged yeasts (L. starkeyi and B. adeninivorans) for their comparison with AGs of filamentous fungi and bacteria. S. stipitis has mostly been studied from the aspect of metabolism of lignocellulosic sugars and production of biofuel [25]. We showed that aside from xylose, glucose, mannose, galactose, cellobiose, and xylose oligomers derived from lignocellulosic material, S. stipitis assimilates numerous α-glucosidic sugars. This knowledge can be used for biotechnological purposes when selecting substrates for cultivation of S. stipitis.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/7/354/s1, Table S1: α-glucosidases (AGs) used in current study, Table S2: α-glucoside transporters (AGTs) used in current study Table S3: Identity matrix of α-glucosidases (AGs) of yeasts and bacilli, Table S4: Identity matrix of yeast α-glucoside transporters (AGTs), Table S5. Primers for amplification of Scheffersomyces stipitis α-glucosidase genes of the MAL loci, Table S6. Primers for cloning the Scheffersomyces stipitis α-glucosidase genes to pURI3-Cter vector, Table S7. Synteny between the Ogataea polymorpha NCYC 495 and Ogataea parapolymorpha DL-1 genomes according to MycoCosm data. Forward synteny between the chromosomes is shown on white background, reverse syntheny on grey background, Table S8. Hypothetical permease and α-glucosidase genes outside the MAL locus in Ogataea polymorpha and Ogataea parapolymorpha with their genomic neighbors indicated. Transcription direction is indicated by arrow.
Author Contributions
Conceptualization, T.A. and K.V.; Methodology, K.V., K.P. and T.A; Investigation, K.V., K.P. and T.V.; Resources, T.A.; Writing-Original Draft Preparation, K.V., K.P., T.A.; Writing-Review & Editing, K.V., K.P.,T.A and T.V.; Visualization, K.V. and K.P.; Supervision, T.A.; Funding Acquisition, T.A. All authors read and approved the manuscript.
Funding
This work was supported by the Estonian Research Council (grant number PUT1050) to T.A.
Acknowledgments
We thank Lele Liiv for major contribution to isolation and initial characterization of the O. polymorpha α-glucosidase.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Stewart, G.G. Saccharomyces species in the production of beer. Beverages 2016, 2, 34. [Google Scholar] [CrossRef]
- Yurimoto, H. Molecular basis of methanol-inducible gene expression and its application in the methylotrophic yeast Candida boidinii. Biosci. Biotechnol. Biochem. 2009, 73, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Weninger, A.; Hatzl, A.-M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 2016, 235, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kickenweiz, T.; Glieder, A.; Wu, J.C. Construction of a cellulose-metabolizing Komagataella phaffii (Pichia pastoris) by co-expressing glucanases and β-glucosidase. Appl. Microbiol. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Alamäe, T.; Liiv, L. Glucose repression of maltase and methanol-oxidizing enzymes in the methylotrophic yeast Hansenula polymorpha: Isolation and study of regulatory mutants. Folia Microbiol. 1998, 43, 443–452. [Google Scholar] [CrossRef]
- Liiv, L.; Pärn, P.; Alamäe, T. Cloning of maltase gene from a methylotrophic yeast, Hansenula polymorpha. Gene 2001, 265, 77–85. [Google Scholar] [CrossRef]
- Kramarenko, T.; Karp, H.; Järviste, A.; Alamäe, T. Sugar repression in the methylotrophic yeast Hansenula polymorpha studied by using hexokinase-negative, glucokinase-negative and double kinase-negative mutants. Folia Microbiol. 2000, 45, 521–529. [Google Scholar] [CrossRef]
- Alamäe, T.; Pärn, P.; Viigand, K.; Karp, H. Regulation of the Hansenula polymorpha maltase gene promoter in H. polymorpha and Saccharomyces cerevisiae. FEMS Yeast Res. 2003, 4, 165–173. [Google Scholar] [CrossRef]
- Karp, H.; Järviste, A.; Kriegel, T.M.; Alamäe, T. Cloning and biochemical characterization of hexokinase from the methylotrophic yeast Hansenula polymorpha. Curr. Genet. 2003, 44, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Tammus, K.; Alamäe, T. Clustering of MAL genes in Hansenula polymorpha: Cloning of the maltose permease gene and expression from the divergent intergenic region between the maltose permease and maltase genes. FEMS Yeast Res. 2005, 5, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Suppi, S.; Michelson, T.; Viigand, K.; Alamäe, T. Repression vs. activation of MOX, FMD, MPP1 and MAL1 promoters by sugars in Hansenula polymorpha: The outcome depends on cell’s ability to phosphorylate sugar. FEMS Yeast Res. 2013, 13, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Visnapuu, T.; Mardo, K.; Aasamets, A.; Alamäe, T. Maltase protein of Ogataea (Hansenula) polymorpha is a counterpart to the resurrected ancestor protein ancMALS of yeast maltases and isomaltases. Yeast 2016, 33, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Voordeckers, K.; Brown, C.A.; Vanneste, K.; van der Zande, E.; Voet, A.; Maere, S.; Verstrepen, K.J. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012, 10, e1001446. [Google Scholar] [CrossRef] [PubMed]
- Naumoff, D.G.; Naumov, G.I. Discovery of a novel family of α-glucosidase IMA genes in yeast Saccharomyces cerevisiae. Dokl. Biochem. Biophys. 2010, 432, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Teste, M.-A.; Francois, J.M.; Parrou, J.-L. Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. J. Biol. Chem. 2010, 285, 26815–26824. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Mateo, R.Q.; Kolecka, A.; Theelen, B.; Robert, V.; Boekhout, T. Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Res. 2015, 15, fov050. [Google Scholar] [CrossRef] [PubMed]
- Ávila, J.; González, C.; Brito, N.; Machín, M.F.; Pérez, D.; Siverio, J.M. A second Zn(II)2Cys6 transcriptional factor encoded by the YNA2 gene is indispensable for the transcriptional activation of the genes involved in nitrate assimilation in the yeast Hansenula polymorpha. Yeast 2002, 19, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Wolfe, K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005, 37, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Slot, J.C.; Rokas, A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. USA 2010, 107, 10136–10141. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Gaillardin, C.; Czernicka, M.; Durrens, P.; Martin, T.; Böer, E.; Gabaldón, T.; Cruz, J.A.; Talla, E.; Marck, C.; et al. The complete genome of Blastobotrys (Arxula) adeninivorans LS3—A yeast of biotechnological interest. Biotechnol. Biofuels 2014, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.W.; Van Vleet, J.R.H. Pichia stipitis genomics, transcriptomics, and gene clusters. Fems Yeast Res. 2009, 9, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; Naumova, E.S.; Michels, C.A. Genetic variation of the repeated Mal loci in natural populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics 1994, 136, 803–812. [Google Scholar] [PubMed]
- Brown, C.A.; Murray, A.W.; Verstrepen, K.J. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. CB 2010, 20, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, F.; Vidgren, V.; Ruohonen, L.; Gibson, B. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016, 16, fow053. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.W.; Grigoriev, I.V.; Grimwood, J.; Laplaza, J.M.; Aerts, A.; Salamov, A.; Schmutz, J.; Lindquist, E.; Dehal, P.; Shapiro, H.; et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007, 25, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V.; Eldarov, M.A.; Kadnikov, V.V.; Beletsky, A.V.; Schneider, J.; Mardanova, E.S.; Smekalova, E.M.; Zvereva, M.I.; Dontsova, O.A.; Mardanov, A.V.; et al. Genome sequence and analysis of methylotrophic yeast Hansenula polymorpha DL1. BMC Genom. 2013, 14, 837. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Ramezani-Rad, M.; Hollenberg, C.P.; Lauber, J.; Wedler, H.; Griess, E.; Wagner, C.; Albermann, K.; Hani, J.; Piontek, M.; Dahlems, U.; et al. The Hansenula polymorpha (strain CBS4732) genome sequencing and analysis. FEMS Yeast Res. 2003, 4, 207–215. [Google Scholar] [CrossRef]
- Lahtchev, K.L.; Semenova, V.D.; Tolstorukov, I.I.; van der Klei, I.; Veenhuis, M. Isolation and properties of genetically defined strains of the methylotrophic yeast Hansenula polymorpha CBS4732. Arch. Microbiol. 2002, 177, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Göker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, A.; Lahtchev, K. The methylotrophic yeast Ogataea (Hansenula) polymorpha as a model organism for studying reproductive isolation. Genet. Plant Physiol. 2016, 6, 14–26. [Google Scholar]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.J.; Martin, T.; Nikolski, M.; Cayla, C.; Souciet, J.-L.; Durrens, P. Génolevures: Protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 2009, 37, D550–D554. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Savini, V.; Lanoue, A.; Simkin, A.J.; Crèche, J.; Giglioli-Guivarc’h, N.; Clastre, M.; Courdavault, V.; Sibirny, A.A. Candida guilliermondii: Biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr. Genet. 2013, 59, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Wood, V.; Gwilliam, R.; Rajandream, M.-A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Freel, K.C.; Sarilar, V.; Neuvéglise, C.; Devillers, H.; Friedrich, A.; Schacherer, J. Genome sequence of the yeast Cyberlindnera fabianii (Hansenula fabianii). Genome Announc. 2014, 2, e00638-14. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.L.; Armisén, D.; Proux-Wéra, E.; ÓhÉigeartaigh, S.S.; Byrne, K.P.; Wolfe, K.H. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA 2011, 108, 20024–20029. [Google Scholar] [CrossRef] [PubMed]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 Genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0. Molecular Biology and Evolution. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Charron, M.J.; Read, E.; Haut, S.R.; Michels, C.A. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 1989, 122, 307–316. [Google Scholar] [PubMed]
- Curiel, J.A.; de las Rivas, B.; Mancheño, J.M.; Muñoz, R. The pURI family of expression vectors: A versatile set of ligation independent cloning plasmids for producing recombinant His-fusion proteins. Protein Expr. Purif. 2011, 76, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Ernits, K.; Viigand, K.; Visnapuu, T.; Põšnograjeva, K.; Alamäe, T. Thermostability measurement of an α-glucosidase using a classical activity-based assay and a novel Thermofluor method. Bio-Protocol 2017, 7. [Google Scholar] [CrossRef]
- Michels, C.A.; Needleman, R.B. The dispersed, repeated family of MAL loci in Saccharomyces spp. J. Bacteriol. 1984, 157, 949–952. [Google Scholar] [PubMed]
- Agaphonov, M.; Romanova, N.; Choi, E.-S.; Ter-Avanesyan, M. A novel kanamycin/G418 resistance marker for direct selection of transformants in Escherichia coli and different yeast species. Yeast 2010, 27, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Takizawa, M.; Suyama, H.; Shintani, T.; Gomi, K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fairhead, C.; Dujon, B. Structure of Kluyveromyces lactis subtelomeres: Duplications and gene content. FEMS Yeast Res. 2006, 6, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Leifso, K.R.; Williams, D.; Hintz, W.E. Heterologous expression of cyan and yellow fluorescent proteins from the Kluyveromyces lactis KlMAL21–KlMAL22 bi-directional promoter. Biotechnol. Lett. 2007, 29, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Change, Y.S.; Dubin, R.A.; Perkins, E.; Forrest, D.; Michels, C.A.; Needleman, R.B. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr. Genet. 1988, 14, 201–209. [Google Scholar] [CrossRef]
- Kelly, R.; Kwon-Chung, K.J. A zinc finger protein from Candida albicans is involved in sucrose utilization. J. Bacteriol. 1992, 174, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Pougach, K.; Voet, A.; Kondrashov, F.A.; Voordeckers, K.; Christiaens, J.F.; Baying, B.; Benes, V.; Sakai, R.; Aerts, J.; Zhu, B.; et al. Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nat. Commun. 2014, 5, 4868. [Google Scholar] [CrossRef] [PubMed]
- Day, R.E.; Higgins, V.J.; Rogers, P.J.; Dawes, I.W. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast 2002, 19, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Day, R.E.; Rogers, P.J.; Dawes, I.W.; Higgins, V.J. Molecular analysis of maltotriose transport and utilization by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 5326–5335. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Michels, C.A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr. Genet. 1988, 14, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Zhou, X.; Kominek, J.; Kurtzman, C.P.; Hittinger, C.T.; Rokas, A. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 Genes Genomes Genet. 2016, 6, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.A.; Louis, E.J. Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 2017, 206, 717–750. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Ward, J.M. Functional characterization of the α-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol. Microbiol. 2001, 39, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Honda, H.; Taniguchi-Morimura, J.; Iwasaki, S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature 1989, 341, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Nakayama, A.; Yamamoto, Y.; Tabata, S. Val216 decides the substrate specificity of α-glucosidase in Saccharomyces cerevisiae: Substrate specificity of α-glucosidase. Eur. J. Biochem. 2004, 271, 3414–3420. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose: Crystal structure of isomaltase. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae. J. Biosci. Bioeng. 2011, 112, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Petitjean, M.; Teste, M.-A.; Kooli, W.; Tranier, S.; François, J.M.; Parrou, J.-L. Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio 2014, 4, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Ni, X.; Yao, S. Cloning and overexpression of a maltase gene from Schizosaccharomyces pombe in Escherichia coli and characterization of the recombinant maltase. Mycol. Res. 2008, 112, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y.; Tanaka, H.; Takemura, R.; Yokogawa, T.; Shimonaka, A.; Matsui, H.; Kashiwabara, S.-I.; Watanabe, K.; Suzuki, Y. Molecular determinants of substrate recognition in thermostable alpha-glucosidases belonging to glycoside hydrolase family 13. J. Biochem. 2007, 142, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Eisawa, H.; Ogawa, S.; Yamazaki, N.; Maekawa, K.; Yamaguchi, T.; Sato, S.; Shiota, K.; Yoshida, T. Characterization of three fungal isomaltases belonging to glycoside hydrolase family 13 that do not show transglycosylation activity. J. Appl. Glycosci. 2017, 64, 9–13. [Google Scholar] [CrossRef]
- Sainz-Polo, M.A.; Ramírez-Escudero, M.; Lafraya, A.; González, B.; Marín-Navarro, J.; Polaina, J.; Sanz-Aparicio, J. Three-dimensional structure of Saccharomyces Invertase: Role of a non-catalytic domain in oligomerization and substrate specificity. J. Biol. Chem. 2013, 288, 9755–9766. [Google Scholar] [CrossRef] [PubMed]
- Geber, A.; Williamson, P.R.; Rex, J.H.; Sweeney, E.C.; Bennett, J.E. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J. Bacteriol. 1992, 174, 6992–6996. [Google Scholar] [CrossRef] [PubMed]
- Bell, P.J.L.; Bissinger, P.H.; Evans, R.J.; Dawes, I.W. A two-reporter gene system for the analysis of bi-directional transcription from the divergent MAL6T-MAL6S promoter in Saccharomyces cerevisiae. Curr. Genet. 1995, 28, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Keppler, F.; Hamilton, J.T.G.; Braß, M.; Röckmann, T. Methane emissions from terrestrial plants under aerobic conditions. Nature 2006, 439, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. MMBR 2011, 75, 321–360. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Randhawa, A.; Ganesan, K.; Raghava, G.P.S.; Mondal, A.K. Draft genome sequence of salt-tolerant yeast Debaryomyces hansenii var. hansenii MTCC 234. Eukaryot. Cell 2012, 11, 961–962. [Google Scholar] [CrossRef] [PubMed]
- Angerbauer, C.; Siebenhofer, M.; Mittelbach, M.; Guebitz, G.M. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour. Technol. 2008, 99, 3051–3056. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An ancient yeast for young geneticists: A primer on the Schizosaccharomyces pombe model system. Genetics 2015, 201, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Tanouye, L.; Michels, C.A. The UASMAL is a bidirectional promotor element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr. Genet. 1992, 22, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bali, M.; Medintz, I.; Michels, C.A. Intracellular maltose is sufficient to induce MAL gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Alamäe, T. Further study of the Hansenula polymorpha MAL locus: Characterization of the α-glucoside permease encoded by the HpMAL2 gene. FEMS Yeast Res. 2007, 7, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Poolman, B. Proton-solute coupling mechanism of the maltose transporter from Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 14375. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.L.A.; De Winde, J.H.; Pronk, J.T. Hxt-carrier-mediated glucose efflux upon exposure of Saccharomyces cerevisiae to excess maltose. Appl. Environ. Microbiol. 2002, 68, 4259–4265. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Dietrich, F.S.; Roy, S.W. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007, 8, R223. [Google Scholar] [CrossRef] [PubMed]
- Da Lage, J.-L.; Binder, M.; Hua-Van, A.; Janeček, Š.; Casane, D. Gene make-up: Rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evol. Biol. 2013, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Gabriško, M. Evolutionary history of eukaryotic α-glucosidases from the α-amylase family. J. Mol. Evol. 2013, 76, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Visnapuu, T.; Mäe, A.; Alamäe, T. Hansenula polymorpha maltase gene promoter with sigma 70-like elements is feasible for Escherichia coli-based biotechnological applications: Expression of three genomic levansucrase genes of Pseudomonas syringae pv. tomato. Process Biochem. 2008, 43, 414–422. [Google Scholar] [CrossRef]
- Kelly, C.T.; Moriarty, M.E.; Fogarty, W.M. Thermostable extracellular α-amylase and α-glucosidase of Lipomyces starkeyi. Appl. Microbiol. Biotechnol. 1985, 22, 352–358. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).