Abstract
The haloarchaea are unusual in possessing genes for multiple homologs to the ubiquitous single-stranded DNA binding protein (SSB or replication protein A, RPA) found in all three domains of life. Halobacterium salinarum contains five homologs: two are eukaryotic in organization, two are prokaryotic and are encoded on the minichromosomes, and one is uniquely euryarchaeal. Radiation-resistant mutants previously isolated show upregulation of one of the eukaryotic-type RPA genes. Here, we have created deletions in the five RPA operons. These deletion mutants were exposed to DNA-damaging conditions: ionizing radiation, UV radiation, and mitomycin C. Deletion of the euryarchaeal homolog, although not lethal as in Haloferax volcanii, causes severe sensitivity to all of these agents. Deletion of the other RPA/SSB homologs imparts a variable sensitivity to these DNA-damaging agents, suggesting that the different RPA homologs have specialized roles depending on the type of genomic insult encountered.
1. Introduction
The single-stranded DNA binding proteins (SSBs) and replication protein A (RPA) in archaea and eukaryotes) are ubiquitous elements of replication forks across all three domains of life. In addition to their role in coating and protecting lagging strand DNA, they are necessary for protection of single-stranded regions generated during other processes, including restarting stalled replication forks, homologous recombination, and DNA repair [1,2,3,4]. During these processes, SSB/RPA proteins recruit and interact with a variety of other cellular proteins, and their interactions change depending on the state of the DNA [1,4,5,6]. Through these interactions, SSB/RPAs orchestrate the process being carried out [1,2,4,5,6]. SSBs/RPAs are characterized by containing one or more ssDNA binding domains, oligonucleotide oligosaccharide-binding (OB) fold domains, and are thought to have originated from duplication of an ancestral SSB gene [7,8]. Typical bacterial SSBs are encoded by a single predominant essential gene and function as homotetramers [4,9,10]. Eukaryotic RPAs are heterotrimeric and encoded by genes RPA1, RPA2, and RPA3 [7,11]. The Archaea are more diverse in their repertoire. Like bacteria, the Crenarchaea typically contain a single gene with a single OB-fold; however, whether the functional protein is a monomer or multimer is unclear, as the OB-fold is more eukaryotic than prokaryotic [8,12]. The euryarchaea, on the other hand, often contain a unique RPA found only among the euryarchaea, and may possess multiple OB-fold-containing proteins, the most common of which contains two OB-folds and a zinc–finger domain [8,13]. The Haloarchaea, for example, typically contain genes for several types of proteins [8]. A bacterial or crenarchaeal protein with one OB-fold is encoded on the mini-chromosomes. At least one operon encoding proteins with homology to RPA1 and RPA2, as well as a third gene encoding a protein with no homology to RPA3, is found on the main chromosome [8,12]. Finally, the uniquely euryarchaeal RPA contains three to four OB-folds in a single open reading frame [8,12].
The extremely radiation- and UV-resistant model halophile Halobacterium salinarum provides a unique system for the study of the individual roles of multiple SSB/RPA proteins. In radiation-resistant mutants of Hbt. salinarum, one of the two eukaryotic-like operons present on the main chromosome was constitutively upregulated [14,15]. The three genes, rfa3, rfa8, and ral, were shown to be co-transcribed, and recent evidence has suggested that at least the Rfa3 and Rfa8 proteins interact in the cell [16]. Additional expression of this operon on a plasmid conferred increased radiation resistance, indicating that increased levels of these proteins are sufficient to increase resistance [16].
In this study, we have systematically deleted each of the genes with homology to SSB/RPA in Hbt. salinarum, except for the first gene (rfa2) in one of two operons encoding eukaryotic-type RPA proteins. The effect of these mutations on growth, as well as the phenotype of these mutants after exposure to DNA-damaging agents, has been tested. Interestingly, despite the high degree of homology between Haloferax volcanii and Hbt. salinarum regarding these genes, we were unable to delete rfa2, whereas the homolog was not essential in Hfx. volcanii [17]. The RPA/SSB proteins of the two halophiles and their shared features are shown in Table 1 [17]. However, deletion of the Hfx. volcanii homolog conferred DNA damage sensitivity. In contrast, we show that deletion of rfa1 in Hbt. salinarum confers an extreme DNA damage sensitivity, but the gene is not essential, whereas the homolog was shown to be essential in Hfx. volcanii [17]. These results suggest that despite the high degree of conservation among the haloarchaea regarding the group of single-stranded DNA binding proteins, there has been divergence of roles between these two related species regarding specific involvement in DNA repair pathways and replication.

Table 1.
Halobacterium salinarum and Haloferax volcanii replication protein A (RPA)/single-stranded DNA binding (SSB) protein homologues, shared features, and percent identity.
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
Halobacterium salinarum ssp. NRC-1 and derivative mutant strains (Table 2) were grown in CM+ medium or minimal Grey and Fitt medium lacking uracil (GF-U) as appropriate [19,20]. All chemicals were purchased from ThermoFisher Scientific (Hampton, NH, USA) unless otherwise stated. Mevinolin and 5-fluoroorotic acid (5-FOA) were added to CM+ where appropriate for selection. Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA, USA) was used for plasmid construction and was grown in LB medium containing ampicillin at a concentration of 100 µg/mL where appropriate [21].

Table 2.
Hbt. salinarum strains used.
2.2. Transformation
Hbt. salinarum was transformed by an adaptation of the PEG method [20,22]: Late log-phase cells were harvested by centrifugation, and cell pellets were washed with spheroplasting solution (SPS; 2 M NaCl, 25 mM KCl, 50 mM Tris pH 8.8, 15% sucrose) then resuspended gently in fresh SPS. The S-layer was removed by addition of 0.5 M EDTA in SPS. Approximately 1 µg plasmid DNA in 2 M NaCl was added to the cells and incubated for 5 min. SPS containing 60% PEG-600 was added to the caps of each tube and vigorously mixed. Samples were incubated for 15 min, then recovery medium (CM+ containing 15% sucrose) was added, and samples were centrifuged. The supernatant was decanted, and samples were washed twice in recovery medium. Pellets were then resuspended in recovery medium and shaken at 37 °C for 1 to 3 days to allow for recovery of the cells. Cells were then centrifuged, resuspended in basal salts solution (BSS, 4.3 M NaCl, 81 mM MgSO4, 14 mM KCl, 5 mM Na3C6H5O7) or CM+, and plated on the appropriate growth medium. Plates were incubated at 42 °C for 7–21 days until colonies formed.
E. coli DH5α was made competent using calcium chloride treatment and transformed using established methods [23].
2.3. Plasmids and Strain Construction
Plasmid pMPK410 [24] was used to make uracil auxotrophic strains of Hbt. salinarum ssp. NRC-1 and derivative strain LH5 [14]. Plasmid DNA was transformed as described, and transformants were plated on CM+ containing mevinolin for selection of the integrated plasmid. Transformant colonies were streaked twice on GF-U, then grown to stationary phase in liquid CM+. Diluted cultures were spread on CM+ containing 5-FOA. Plates were incubated at 42 °C for 7–10 days for selection of de-integration of the plasmid and loss of the ura3 gene. Colonies were then purified once on CM+ with 5-FOA and once on CM+, and uracil auxotrophy was confirmed by plating on GF-U. Deletion of the ura3 gene was confirmed via PCR.
Plasmids for RPA gene deletion were constructed to generate in-frame deletions of each gene. Briefly, primers pairs were designed to amplify ~500 bp of upstream and ~500 bp of downstream sequence for homology for integration (Supplementary Table S1). One primer in each pair contained one engineered EcoRI site for cloning into the suicide vector. The primers bridging the deletion were designed with overlapping sequences. The upstream and downstream region amplicons were combined to create deletion fragments by overlap extension PCR. The resulting deletion fragments were confirmed by gel electrophoresis, then digested in PCR buffer with EcoRI (Fermentas, Waltham, MA, USA) followed by purification using the MiniElute PCR cleanup kit (Qiagen, Germantown, MD, USA). Digested deletion fragments were ligated to purified EcoRI-digested and dephosphorylated pMPK408 [24]; the ligation reaction was then transformed into DH5α. Recombinant plasmids were isolated from transformants using an alkaline lysis miniprep kit (Stratagene, San Diego, CA, USA), analyzed by restriction digestion using EcoRI, and verified through PCR.
Deletions of RPA genes were generated in a similar manner as uracil auxotrophs with the following changes. Transformants were plated on GF-U for selection of the integrated plasmid and grown at 42 °C for 7–10 days. Transformants were purified twice on GF-U, then grown to stationary phase in two separate cultures in either CM+ or GF-U. Presence of the integrated plasmid was confirmed by PCR amplification using DNA from cultures grown in GF-U to detect both the deletion allele and the wild-type allele using the appropriate upstream (UF) and downstream (DR) primers (Supplementary Table S1). The cultures grown in CM+ were plated on CM+ containing 5-FOA to select for loss of the integrated plasmid. Plates were incubated at 42 °C for 7–10 days. Colonies were then purified once on CM+ containing 5-FOA, then once on CM+. Purified colonies were grown in liquid CM+, and cells were harvested by centrifugation. Pellets were lysed in deionized water, and this lysate was used as DNA template for PCR amplification. Deletion of each RPA was confirmed by amplification with the appropriate UF and DR primers (Supplementary Table S1) followed by gel electrophoresis to screen each culture for either the deletion or wild-type allele.
2.4. DNA Damage Treatments
2.4.1. Ionizing Radiation
All irradiations were performed at the Idaho Accelerator Center (Idaho State University, Pocatello, ID, USA) using a pulsed S-band medical grade linear accelerator (LINAC) delivering 23 MeV electrons at 60 Hz with a pulse-width of 2 µs as described previously [25]. A 200 μL cell suspension in CM+ medium (approximately 1 × 107 cells) was irradiated at room temperature in polypropylene PCR tubes (Molecular Bioproducts, San Diego, CA, USA). After irradiation, cultures were 10-fold serially diluted to 10−5 in liquid CM+ and 10 μL of each dilution was spotted on a CM+ plate and allowed to dry. Plates were incubated at 42 °C for 7 days then analyzed for survival.
2.4.2. Ultraviolet Irradiation
Early log-phase cultures were serially diluted to 10−5 in liquid CM+, and 10 μL of each 10-fold dilution was spotted on a CM+ plate and allowed to dry. Plates were exposed to 254 nm UV-C at a power of 1.5 W/m2 for various times as indicated in each experiment. Treatment was performed in the dark to remove the effects of photoreactivation, and plates were incubated at 42 °C in the dark for 7 days, then analyzed for survival. For each strain, surviving fractions from triplicate experiments were averaged and plotted in Origin 2018 (OriginLab, Northampton, MA, USA) as described previously, with error bars representing the standard error of the mean [14].
2.4.3. Mitomycin C Treatment
Mitomycin C was added at a final concentration of 0.75 µg/mL to exponential phase broth cultures and incubated at 42 °C with shaking for 20 min. After exposure, cultures were pelleted and resuspended in fresh medium for recovery, then diluted and plated on solid CM+ as appropriate.
2.5. Reverse Transcription—Polymerase Chain Reaction and Transcript Analysis
cDNA was made using a microarray labeling and hybridization protocol developed previously [26]. Briefly, in a nuclease-free PCR tube, 10 µL of RNA and 1 µL of random hexamers were mixed thoroughly and the total volume was brought up to 16 µL. This reaction was incubated at 65 °C in a thermocycler for 10 min. Then, 9.8 µL of RT master mix (6 µL of 5X 1st strand buffer; 3 µL of 0.1 M dithiothreitol (DTT); 0.6 µL of deoxyribonucleotide triphosphate (dNTP) mix (25 mM dATP, dTTP, dGTP, and 10 mM dCTP); 0.2 µL of RNase out ribonuclease inhibitor, all chilled on ice for 2 min) was added to the previous reaction. Then, 2 µL of Superscript III Reverse Transcriptase (Invitrogen) was added, mixed by pipetting, and incubated for 10 min at room temperature. The reaction was then incubated at 42 °C in a thermal cycler for 110 min with the lid temperature set at 43 °C. The reaction was stopped by adding 1.5 µL 1 M NaOH and placed in a thermocycler at 65 °C for 10 min. The reaction was stopped with 1.5 µL 1 M HCl and quickly diluted with 150 µL neutralize tagment (NT) buffer from a NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany). The reactions were purified with the same clean up kit and eluted with nuclease free water.
Specific transcripts were amplified by RT-PCR using transcript primers (Supplementary Table S2). DNA, RNA, and cDNA were used as template. For the reactions with the primers amplifying the rfa2/rfa7 junction, the following parameters were used: 1 cycle of 98 °C for 30 s; 29 cycles of 98 °C for 10 s, 66 °C for 10 s, and 72 °C for 15 s; and 1 cycle of 72 °C for 5 min. For the reactions with the primers amplifying the rfa7/yhcR junction: 1 cycle of 98 °C for 30 s; 29 cycles of 98 °C for 10 s, 63 °C for 10 s, and 72 °C for 15 s; and 1 cycle of 72 °C for 5 min.
2.6. Quantitative PCR
RNA was isolated from 5 mL of culture with an Invitrogen PureLink™ RNA Mini Kit. To ensure that no residual DNA was present, the Invitrogen™ Ambion™ TURBO DNA-free kit was also used before conversion to cDNA. Concentrations of each cDNA sample were determined using a ThermoFisher Scientific NanoDrop 2000, and 2 µg of template was used in the Applied Biosystems (Foster City, CA, USA) High Capacity RNA-to-cDNA kit.
All qPCR reactions were performed in with three biological and three technical replicates on a PikoReal Real Time PCR System (ThermoFisher Scientific) with TaqMan® Gene Expression primers and probes (Supplementary Table S3) under the following conditions: 95 °C for 10 min; (95 °C for 1 min, 60 °C for 30 s) for 35 cycles; and 72 °C for 7 min [26]. Analysis was completed using PikoReal Software 2.2 (ThermoFisher Scientific).
Standard curves were obtained using serially diluted gene targets of known concentrations that were amplified via PCR (Supplementary Table S1). Triplicate samples of each dilution were used as template in qPCR and the recorded Cq was graphed versus DNA concentration. Standards for the gene target were included with all experimental sample plates to ensure consistent readings.
2.7. Statistical Analysis
For colony size analysis and DNA damage experiments, statistical significance was determined by analysis of variance (ANOVA) with post-hoc Tukey Honestly Significant Difference (HSD) test. Details are noted in the figure captions for each experiment.
For quantitative PCR analysis, statistical significance was determined using R version 3.4.2. [27]; a two-factor ANOVA followed by the post-hoc Tukey HSD test was performed for each gene, and a one-way ANOVA followed by the post-hoc Tukey Honest Significance Difference test was performed for each gene’s treatment and to determine significance between absolute levels of each gene during normal growth.
3. Results
To determine the roles of each of the SSB/RPA homologs present in the genome of Hbt. salinarum, an in-frame deletion allele of each gene was created in a Halobacterium suicide vector containing a selectable ura3 gene, and recombinant integrants were selected in an otherwise wild-type Δura3 background (LH101). From these integrants, 5-FOAr excisants were selected, where the integrated plasmid had been lost through a second recombination event (the “pop-in, pop-out” method; [24]). Except for Δrfa2, two excisants—one containing the deletion allele and one containing the restored wild-type allele—were purified for each gene. Each was verified through PCR using the forward primer from the upstream region and the reverse primer from the downstream region. The individual genes deleted were: of the two eukaryotic-type RPAs, each gene within the rfa3 operon (rfa3, rfa8, and ral) and the second and third genes in the putative rfa2 operon (rfa7 and yhcR); the uniquely euryarchaeal rfa1; and each of the duplicate SSB-type genes on the mini-chromosomes, rfa5 and rfa6. (Figure 1). In addition, qPCR was performed on LH101 (no deletion), LH128 (ΔyhcR), LH134 (Δrfa3), LH136 (Δrfa8), LH140 (Δrfa1), and LH142 (Δrfa6) to verify lack of expression, and all levels were below detection (data not shown).
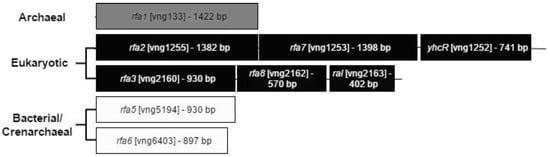
Figure 1.
Operon structure and relatedness of the RPA homologues in Hbt. salinarum. The Archaeal homolog Rfa1 contains three OB-folds, and is unique to the euryarchaeal lineage [8] The two eukaryotic homologs are each found in three-gene operons, with the first and second genes showing homology to eukaryotic RPA1 and RPA2, respectively. The three genes of the rfa3 operon (rfa3, rfa8, and ral) have been shown to be co-transcribed [14]. Similar evidence for the rfa2 operon is presented in this report.
3.1. Effect of Replication Protein A Deletions on Growth
rfa1, rfa3 and rfa8 Deletions Affect Growth
The strain carrying a deletion of rfa3 (LH134), but not rfa8 (LH136) or ral (LH138) within the rfa3 operon, as well as the strain carrying Δrfa1 (LH140), showed reduced colony size in comparison to their isogenic counterparts. This discrepancy in growth can be seen in Figure 2, where the diameter of colonies of strains carrying each of the three deletion alleles are compared to wild-type. Strains carrying Δral (LH138), Δrfa7 (LH166), ΔyhcR (LH128), Δrfa5 (LH110), or Δrfa6 (LH142) also formed normal-sized colonies, and are not shown. The growth defect was more pronounced in LH140 (Δrfa1) than in LH134 (Δrfa3).
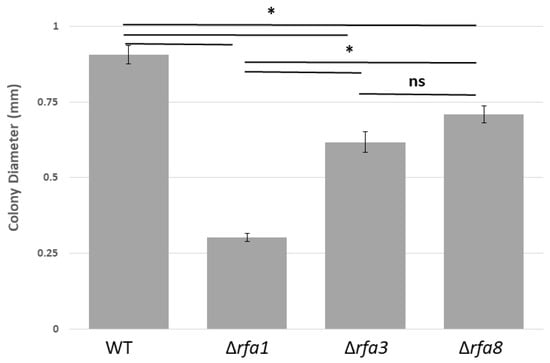
Figure 2.
Colony size comparison between LH101 (wild-type: WT) and LH140 (Δrfa1), LH134 (Δrfa3), and LH136 (Δrfa8). Plates were incubated for 7 days. Colony diameter was measured using ImageJ (Version 1.8.0_112, National Institutes of Health, Bethesda, MD, USA) [28]. * p < 0.05, analysis of variance (ANOVA) with Tukey’s Honest Significant Difference (HSD), n = 22–40 colonies total over 2 biological replicates. Error bars represent the standard error of the mean (SEM). ns = not significant.
3.2. Deletion of rfa2, rfa7, and yhcR
3.2.1. Co-Transcription of the rfa2, rfa7, and yhcR Genes
By comparison to the rfa3 operon, and from sequence analysis, rfa2, rfa7, and yhcR appear to also potentially form an operon. Prior to attempting to create deletion strains, we performed transcript analysis using primers that would allow for amplification of the two intergenic regions, rfa2-rfa7 (383 bp) and rfa7-yhcR (541 bp), from cDNA made from total RNA. The predicted size amplicon was present for both regions when either cDNA or genomic DNA, but not total RNA, was used as template, demonstrating the existence of a transcript containing the intergenic region, and thus confirming co-transcription of the three genes under normal growing conditions (Figure 3).
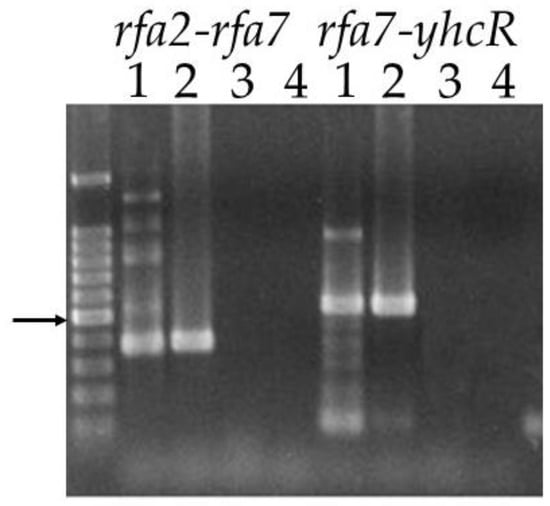
Figure 3.
Co-transcription of rfa2, rfa7 and yhcR. Primers bracketing the intergenic regions rfa2-rfa7 and rfa7-yhcR were used in reactions with different templates. Lane 1: cDNA; Lane 2: genomic DNA; Lane 3: total RNA; Lane 4: no template. Size marker is 1 kb PLUS DNA ladder (Gold Biotechnology, St. Louis, MO, USA). Arrow indicates 500 bp.
3.2.2. rfa2 May Be Essential
Although in Hfx. volcanii, strains carrying a deletion of the rfa2 homolog were obtained [17], we were unable to isolate strains in a wild-type background that carried this deletion in Hbt. salinarum. Over 1000 FOAr excisants were analyzed, including those with extremely compromised growth, and all contained the wild-type allele. Strains carrying deletions in either rfa7 or yhcR were readily isolated. Significantly, the Δrfa2 deletion allele was recovered in LH102, which is the Δura3 derivative of a previously described radiation-resistant mutant, LH5 [14]. LH5 has been shown to have increased expression of the rfa3 operon; this increased expression appeared to compensate for the defect imposed by the deletion of rfa2. This construct was verified as the others, using the forward primer of the upstream region and the reverse primer from the downstream region and verifying that the deletion allele (1053 bp) instead of the wild-type allele (2581 bp) was amplified (Supplementary Figure S1). However, qPCR analysis of LH154 showed reduction, but not absence, of rfa2 transcript in this strain (Table 3). Given the fluid nature of the Hbt. salinarum genome and its many active insertion elements [29], it is possible that a region amplifiable only by the internal, but not external, primers was rearranged to another transcriptional unit within the chromosome.

Table 3.
Expression of rfa2 in suspected rfa2 deletion strain relative to the wild-type. Expression was normalized to eef2 expression.
3.3. Effect of Deletions on Survival to Ionizing Radiation, Ultraviolet C, and Mitomycin C
3.3.1. Δrfa3, Δrfa8, and Δrfa1 Confer Ionizing Radiation Sensitivity
Because upregulation of the rfa3 operon has been shown to confer increased ionizing radiation resistance, but not increased resistance to UV [30], strains containing deletions were tested for survival after exposure to 23 MeV electrons from a LINAC, and UV-C, treatment.
Deletion strains were tested in exponential phase against the treatments as described. Based on the involvement of rfa3 upregulation in ionizing radiation resistance, it was expected that deletion of the genes would affect survival. As seen in Figure 4, strains harboring deletions in either rfa3 or rfa8, but not ral, had reduced survival to e-beam exposure compared to wild-type (representative wild-type shown), with comparable 10-fold reductions (D10) in survival at approximately 1000 Gy versus approximately 7000 Gy in the wild-type. The strain carrying Δrfa1 had a more severe survival defect, with a D10 of approximately 500 Gy. All other deletion strains tested (Δrfa5, Δrfa6, ΔyhcR) had survival comparable to the wild-type (data not shown). LH166 (Δrfa7) was not available for testing.
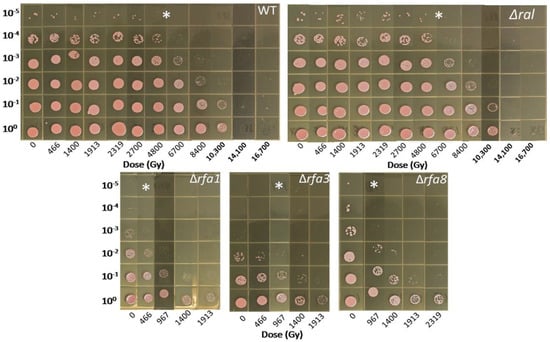
Figure 4.
Survival of Hbt. salinarum strains to Ionizing radiation exposure. LH101 (WT), LH138 (Δral), LH140 (Δrfa1), LH134 (Δrfa3), and LH136 (Δrfa8) were exposed to increasing doses of electron beam radiation. * indicates dose at which each strain reaches the approximate D10. LH110 (Δrfa5), LH142 (Δrfa6), and LH128 (ΔyhcR) showed wild-type survival and are not shown.
3.3.2. Ultraviolet C Survival is Similar to Ionizing Radiation
Each strain, as well as LH166 (Δrfa7), were also tested against UV-C; survival showed a similar pattern to IR. The rfa3 and rfa8 deletion strains had moderately reduced survival, with 10% survival at approximately 25 J/m2 (Figure 5). The rfa1 deletion strain (LH140) had extremely reduced survival, with 10% survival at approximately 2 J/m2. All other deletion strains (Δral, Δrfa7, ΔyhcR, Δrfa5, and Δrfa6) had wild-type levels of survival (Supplementary Figure S2).
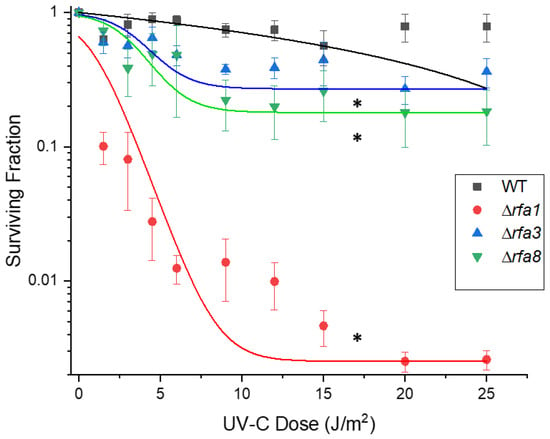
Figure 5.
Survival of Hbt. salinarum deletion strains to UV-C damage. LH101 (WT), LH140 (Δrfa1), LH134 (Δrfa3), and LH136 (Δrfa8). * p < 0.05 compared to LH101; ANOVA with Tukey’s HSD, n = 3. Error bars represent +/− standard error of the mean. Mutant strain survival curves were fitted to the Boltzmann function in Origin 2018 (OriginLab, Northampton, MA, USA).
3.3.3. Δrfa5, Δrfa6, Δrfa7, and ΔyhcR Strains Are Also Sensitive to Mitomycin C
Given the overlapping repair pathways for IR and UV damage, it was not unexpected that certain deletions would show similar phenotypes when challenged with those. To test an additional type of damage, DNA crosslinking, all deletion strains were treated with mitomycin C and survival at various times post-treatment was measured. Similarly to the IR and UV treatments, the Δrfa1, Δrfa3, and Δrfa8 strains were the most affected by this treatment and did not survive past 30 min exposure, while Δral showed no significantly increased sensitivity (Figure 6a). Interestingly, although LH110 (Δrfa5) and LH42 (Δrfa6) showed no difference in growth rate, colony size, IR-resistance, or UV resistance compared to the wild-type, mitomycin C treatment induced a ~15-fold decrease in survival in both deletion strains compared to the wild-type (Figure 6b). Even more drastic was the ~100-fold decrease in survival at 120 min of LH128 (ΔyhcR), but not LH166 (Δrfa7).
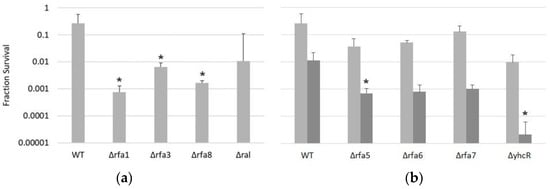
Figure 6.
Survival of Hbt. salinarum deletion strains to mitomycin C treatment. (a) Survival after 30 min exposure for LH101 (WT), LH140 (Δrfa1), LH134 (Δrfa3), LH136 (Δrfa8), and LH138 (Δral). (b) Survival after 30 (light grey) and 120 min (dark grey) exposure for LH101 (WT), LH110 (Δrfa5), LH142 (Δrfa6), LH166 (Δrfa7), and LH128 (ΔyhcR). * p < 0.05 compared to LH101, ANOVA with Tukey’s HSD, n = 3. Error bars represent +/− standard error of the mean.
3.4. Expression Levels of Replication Protein A Genes
3.4.1. Replication Protein A Genes are Expressed Under Normal Growing Conditions
The DNA damage survival results suggest that the different RPA genes in Hbt. salinarum are involved in several aspects of DNA metabolism and repair. One significant question is whether all are expressed under normal growing conditions, or whether the redundancy of homologs includes non-transcribed genes. The rfa3 operon has been shown to be upregulated in response to UV [31,32], but absolute expression of the RPA genes has not yet been established. We sought to establish the basal level of transcription of the genes/operons in wild-type Hbt. salinarum as well as regulation under DNA-damaging conditions. Levels of the third genes in the two multigene operons, ral and yhcR, were not measured. Because of the high level of identity between rfa5 and rfa6, these were measured together using one primer pair.
Absolute expression was determined for each gene target during early log-phase normal growth. These values are presented in Table 4, with the housekeeping gene eef2 as control. rfa5/6 and rfa2 were expressed at the lowest, but still detectable, levels. The rfa3 operon (rfa3 and rfa8) was the most highly expressed, with levels comparable to eef2.

Table 4.
Absolute mRNA levels of RPA genes.
Interestingly, rfa7 levels were 5-fold higher than rfa2 levels despite the co-transcription. This suggests a potential second promoter for rfa7 and a function distinct from rfa2.
3.4.2. DNA-Damaging Treatment Results in Differential Expression
It has previously been reported from microarray analysis that rfa3 operon expression increased following IR and UV treatment [14,22,32]. We measured absolute levels of mRNA, as above, three hours after separate treatment with UV-C and mitomycin C. The rfa3 operon genes were modestly, but not statistically significantly, upregulated following UV treatment, and similarly following mitomycin C treatment (Table 5). Much more striking was the seven-fold upregulation of rfa2, but not rfa7, following both UV and mitomycin C treatment. A modest upregulation of rfa5/6 following mitomycin C treatment was only slightly below the 95% confidence level, and may indicate a response in transcription to the crosslinking damage, suggesting a role for these homologs in that specific repair pathway.

Table 5.
Absolute levels and relative in expression of RPA genes 3 hours following treatment with UV-C or mitomycin C (MMC). Relative change to the untreated RPA genes based on expression values normalized to the corresponding maintenance gene eef2 expression.
The upregulation of rfa5 and rfa6 following treatment with mitomycin C is in line with the sensitivity of the corresponding deletion strains to this damaging agent. This suggests a specific role in the repair pathway for the damage inflicted by mitomycin C, and provides a reason for the retention of these genes in the genome. The upregulation of rfa2, but not rfa7, following both UV-C and mitomycin C treatment supports the independent roles of these co-transcribed genes in the damage response.
4. Discussion
In this report, we have successfully utilized an established method to individually replace wild-type genes encoding RPA homologs of the haloarchaeon Hbt. salinarum with engineered deletions. Although each deletion was constructed to remove only the coding region of each gene, it is possible that polar effects were exerted on remaining genes within the operon. However, we consider this unlikely, as deletion of each individual gene had a unique phenotype. For example, in the rfa3 operon, deletion of rfa3 and rfa8 distinctly affected radiation survival, whereas deletion of ral did not. Similarly, in the rfa2 operon, rfa7 and yhcR deletions affected mitomycin C survival, and a deletion of rfa2 could not be recovered. Given the ease of generating these deletions, it is unlikely that second-site suppressors have masked the essentiality of these genes. Polarity effects as well as essentiality questions could be addressed by providing the deleted gene in trans on a multicopy plasmid, which should completely restore the wild-type phenotype of each individual mutation. In the case of an essential gene, with the wild-type gene supplied in trans during the “pop-in” phase of deletion construction, equal numbers of deletion and wild-type alleles should be recovered during the “pop-out” phase. However, such complementation experiments have been hampered by the dearth of selectable markers available historically in Hbt. salinarum. The widely used selection for novobiocin resistance is complicated by spontaneous resistance as well as detrimental growth effects in the minimal medium required for maintenance of the deletion plasmid.
We were able to delete all of the RPA homologs apart from rfa2, one of two eukaryotic-type large subunit homologs. This could potentially be due to a severe growth defect present in Δrfa2 cells, but that is unlikely given the high number of clones that were screened. However, we were able to successfully engineer an rfa2 deletion in a strain with constitutively high rfa3 operon expression, suggesting that high levels of one RPA may be able to compensate for the loss of another if the homology is sufficiently high. This was demonstrated in the related halophile Hfx. volcanii, although in that organism the rfa1 homolog was found to be essential. Growth arrest following rfa1 downregulation was partially suppressed by overexpressing the rfa3 homolog despite the lack of homology between the two proteins [17]. It should be noted, however, that this was only discovered through deliberate overexpression, and such a suppressor was not isolated during construction of the deletion strains in Hfx. volcanii. In our suspected rfa2 deletion strain, however, although rfa2 expression was lower than in the wild-type, there was a detectable signal despite no amplification of the wild-type allele using primers outside of the transcript region. This suggests that the transcript is originating elsewhere in the genome through a possible rearrangement, underscoring its essentiality.
Our recovery of an otherwise wild-type strain carrying the rfa1 deletion allele in Hbt. salinarum demonstrates its non-essential nature. Rfa1 appears to be important in more than just repair of damage, as the strain carrying this deletion demonstrated a severe growth defect. Despite the parallels in gene organization and high identity of the homologs, the roles that these proteins play in these two related organisms have diverged, underscoring the multiple roles that RPAs play in normal growth as well as in response to genomic stressors. Although there was no change in expression in this study under UV or mitomycin C treatment, this gene has increased expression after H2O2 treatment, suggesting a role in oxidative damage repair [33].
The expression of two bacterial-type SSB homologs (rfa5 and rfa6) is particularly interesting given the identification of a bacterial-type SSB in human cells [9]. The human SSB homolog has been shown to be one of the first proteins to localize to sites of DNA double-strand breaks (DSB) and is an important player in the DNA damage response. In Hbt. salinarum, these homologs do not appear to play a major role in the repair of DSB induced by ionizing radiation, or in the repair of UV-induced damage, based on the lack of sensitivity to these agents in the deletion strains. However, they likely play some role in the repair of interstrand crosslinks induced by mitomycin C, as evidenced by sensitivity in the deletion strains and induction of expression. The conservation of these homologs (with 81% identity) on the minichromosomes, and across the Haloarchaea, would indicate their importance to the cell under certain conditions encountered on a regular basis.
Of note is the significant upregulation of rfa2 expression, but not of the cotranscribed gene rfa7, following UV and mitomycin C treatment. This gene has also been demonstrated to be upregulated following gamma irradiation and H2O2 stress; in both cases, the accompanying genes also appear to not be regulated [33,34] The Rfa2 protein is suspected to be the primary RPA involved at the replication fork in Hbt. salinarum given the proposed essentiality of the gene. It may be surprising, then, that the expression levels rise so dramatically following DNA damage given the plethora of other proteins available to be involved in those repair pathways. However, in Deinococcus radiodurans, despite the presence of the IR-induced novel SSB DdrB, the replicative SSB is also induced following DNA damage [25,35]. Like SSB, Rfa2 may be a critical component of all replication and repair protein complexes.
The phosphoesterase component, yhcR, of the rfa2 operon was assumed to be uninvolved in the repair pathways based on the IR and UV survival phenotype of the deletion strain. The drastic effect of deleting this gene on mitomycin C survival, but not from deletion of the co-transcribed rfa7, indicates an unanticipated relationship with repair of crosslinking damage. Phosphesterases from many genera of bacteria and archaea are part of a superfamily involved in non-homologous end-joining [36]. These results may indicate a similar function for the YhcR protein in Hbt. salinarum.
In most bacterial and eukaryotic organisms, a multifunctional RPA/SSB carries out the essential roles of DNA replication and repair. The euryarchaeota have evolved a unique set of proteins to carry out these functions [8] and this broad repertoire of RPA homologs may be a major determinant of the high tolerance of these organisms to genomic stress. The evolutionary conservation of the multiple homologs within the Haloarchaea indicates a possible adaptation unique to the saline environment. The difference in response of the various RPA deletions in Hbt. salinarum suggests a unique distribution of those functions and provides an opportunity to investigate the individual activities in separate proteins. A genetic analysis using strains containing multiple individual deletions would establish epistatic relationships as well as delineate the minimum repertoire of RPA genes necessary for carrying out life in Hbt. salinarum.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/4/223/s1, Figure S1: Verification of deletion of rfa2 in LH154, Figure S2: Survival of additional Hbt.salinarum deletion strains to UV-C damage, Table S1: Primers used for deletion creation and verification, Table S2: Transcript primers used for transcript visualization of the rfa2 operon. Table S3: qPCR primers and Taqman™ probes used in this study.
Acknowledgments
This work was supported by a grant from the Henry M. Jackson Foundation for the Advancement of Military Medicine and a graduate fellowship to P.E.G. from Idaho NASA EPSCoR and the Idaho Space Grant Consortium. J.J.E. was supported by the Biomedical Engineering Graduate Program of the South Dakota School of Mines and Technology. Reiko Scalarone provided excellent technical assistance.
Author Contributions
P.E.G., J.J.E., and L.C.D. conceived and designed the experiments and wrote the paper; P.E.G., J.J.E., and J.M. performed the experiments; and all authors analyzed the data.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Awate, S.; Brosh, R.M., Jr. Interactive roles of DNA helicases and translocases with the single-stranded DNA binding protein RPA in nucleic acid metabolism. Int. J. Mol. Sci. 2017, 18, 1233. [Google Scholar] [CrossRef] [PubMed]
- Komori, K.; Ishino, Y. Replication Protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 2001, 276, 25654–25660. [Google Scholar] [CrossRef] [PubMed]
- Shereda, R.D.; Bernstein, D.A.; Keck, J.L. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 2007, 282, 19247–19258. [Google Scholar] [CrossRef] [PubMed]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef] [PubMed]
- Plate, I.; Hallwyl, S.C.; Shi, I.; Krejci, L.; Muller, C.; Albertsen, L.; Sung, P.; Mortensen, U.H. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 2008, 283, 29077–29085. [Google Scholar] [CrossRef] [PubMed]
- Ashton, N.W.; Loo, D.; Paquet, N.; O’Byrne, K.J.; Richard, D.J. Novel insight into the composition of human single-stranded DNA-binding protein 1 (hSSB1)-containing protein complexes. BMC Mol. Biol. 2016, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lin, L.J.; Sriratana, P.; Coleman, K.; Ha, T.; Spies, M.; Cann, I.K. Engineering of functional Replication Protein A homologs based on insights into the evolution of oligonucleotide/oligosaccharide-binding folds. J. Bacteriol. 2008, 190, 5766–5780. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.B.; McKinney, M.C.; Guzman, C.E.; Sriratana, B.; Fitz-Gibbon, S.; Ha, T.; Cann, I.K. The euryarchaeota, nature’s medium for engineering of single-stranded DNA-binding proteins. J. Biol. Chem. 2005, 280, 15325–15339. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.J.; Bolderson, E.; Cubeddu, L.; Wadsworth, R.I.; Savage, K.; Sharma, G.G.; Nicolette, M.L.; Tsvetanov, S.; McIlwraith, M.J.; Pandita, R.K.; et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 2008, 453, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Norais, C.A.; Chitteni-Pattu, S.; Wood, E.A.; Inman, R.B.; Cox, M.M. Ddrb protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J. Biol. Chem. 2009, 284, 21402–21411. [Google Scholar] [CrossRef] [PubMed]
- Wold, M.S. Replication Protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.B.; Murphy, M.C.; White, B.A.; Mackie, R.I.; Ha, T.; Cann, I.K. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase bi. J. Biol. Chem. 2004, 279, 6315–6326. [Google Scholar] [CrossRef] [PubMed]
- Kamekura, M. Diversity of extremely halophilic bacteria. Extremophiles 1998, 2, 289–295. [Google Scholar] [CrossRef] [PubMed]
- DeVeaux, L.C.; Muller, J.A.; Smith, J.; Petrisko, J.; Wells, D.P.; DasSarma, S. Extremely radiation-resistant mutants of a halophilic archaeon with increased single-stranded DNA-binding protein (RPA) gene expression. Radiat. Res. 2007, 168, 507–514. [Google Scholar] [PubMed]
- Webb, K.M.; Yu, J.; Robinson, C.K.; Noboru, T.; Lee, Y.C.; DiRuggiero, J. Effects of intracellular Mn on the radiation resistance of the halophilic archaeon Halobacterium salinarum. Extremophiles 2013, 17, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Karan, R.; DasSarma, P.; Balcer-Kubiczek, E.; Weng, R.R.; Liao, C.C.; Goodlett, D.R.; Ng, W.V.; Dassarma, S. Bioengineering radioresistance by overproduction of RPA, a mammalian-type single-stranded DNA-binding protein, in a halophilic archaeon. Appl. Microbiol. Biotechnol. 2014, 98, 1737–1747. [Google Scholar] [CrossRef] [PubMed]
- Skowyra, A.; MacNeill, S.A. Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 2012, 40, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Capes, M.D.; DasSarma, P.; DasSarma, S. The core and unique proteins of haloarchaea. BMC Genom. 2012, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Grey, V.L.; Fitt, P.S. An improved synthetic growth medium for Halobacterium cutirubrum. Can. J. Microbiol. 1976, 22, 440–442. [Google Scholar] [CrossRef] [PubMed]
- DasSarma, S.; Fleischmann, E. Halophiles. In Archaea: A Laboratory Manual; Robb, F.T., Place, A.R., Eds.; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1995; p. 280. [Google Scholar]
- Miller, J. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: New York, NY, USA, 1972. [Google Scholar]
- Gygli, P.E.; DeVeaux, L.C. Adaptation of the Halobacterium salinarum ssp. NRC-1 gene deletion system for modification of chromosomal loci. J. Microbiol. Methods 2014, 99, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Berman, M.L.; Enquist, L.W. Experiments with Gene Fusions; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1984; p. 303. [Google Scholar]
- Peck, R.F.; DasSarma, S.; Krebs, M.P. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 2000, 35, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J.S.; DeVeaux, L.C. The essential role of the Deinococcus radiodurans SSB gene in cell survival and radiation tolerance. PLoS ONE 2013, 8, e71651. [Google Scholar] [CrossRef] [PubMed]
- Boubriak, I.; Ng, W.L.; DasSarma, P.; DasSarma, S.; Crowley, D.J.; McCready, S.J. Transcriptional responses to biologically relevant doses of UV-B radiation in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2008, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Meth. 2012, 9, 671. [Google Scholar] [CrossRef]
- Ng, W.V.; Kennedy, S.P.; Mahairas, G.G.; Berquist, B.; Pan, M.; Shukla, H.D.; Lasky, S.R.; Baliga, N.S.; Thorsson, V.; Sbrogna, J.; et al. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 2000, 97, 12176–12181. [Google Scholar] [CrossRef] [PubMed]
- Gygli, P.E.; Prajapati, S.; DeVeaux, L.C.; DasSarma, S.; DasSarma, P.; Mestari, M.A.; Wells, D.P. Resistance of the extreme halophile Halobacterium sp. NRC-1 to multiple stresses. AIP Conf. Proc. 2009, 1099, 993–996. [Google Scholar]
- McCready, S.; Marcello, L. Repair of UV damage in Halobacterium salinarum. Biochem. Soc. Trans. 2003, 31, 694–698. [Google Scholar] [CrossRef] [PubMed]
- McCready, S.; Muller, J.A.; Boubriak, I.; Berquist, B.R.; Ng, W.L.; DasSarma, S. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2005, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Van, P.T.; Busch, C.R.; Robinson, C.K.; Pan, M.; Pang, W.L.; Reiss, D.J.; DiRuggiero, J.; Baliga, N.S. Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 2010, 6, 393. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.; Kish, A.; Pan, M.; Kaur, A.; Reiss, D.J.; King, N.; Hohmann, L.; DiRuggiero, J.; Baliga, N.S. An integrated systems approach for understanding cellular responses to gamma radiation. Mol. Syst. Biol. 2006, 2, 47. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Earl, A.M.; Howell, H.A.; Park, M.J.; Eisen, J.A.; Peterson, S.N.; Battista, J.R. Analysis of Deinococcus radiodurans’s transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 2004, 168, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.A.; Smith, P.; Shuman, S. Structure of bacterial Ligd 3′-phosphoesterase unveils a DNA repair superfamily. Proc. Natl. Acad. Sci. USA 2010, 107, 12822–12827. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).