Abstract
Members of the genus Capsicum are of great economic importance, including both wild forms and cultivars of peppers and chilies. The high number of potentially informative characteristics that can be identified through next-generation sequencing technologies gave a huge boost to evolutionary and comparative genomic research in higher plants. Here, we determined the complete nucleotide sequences of the plastomes of eight Capsicum species (eleven genotypes), representing the three main taxonomic groups in the genus and estimated molecular diversity. Comparative analyses highlighted a wide spectrum of variation, ranging from point mutations to small/medium size insertions/deletions (InDels), with accD, ndhB, rpl20, ycf1, and ycf2 being the most variable genes. The global pattern of sequence variation is consistent with the phylogenetic signal. Maximum-likelihood tree estimation revealed that Capsicum chacoense is sister to the baccatum complex. Divergence and positive selection analyses unveiled that protein-coding genes were generally well conserved, but we identified 25 positive signatures distributed in six genes involved in different essential plastid functions, suggesting positive selection during evolution of Capsicum plastomes. Finally, the identified sequence variation allowed us to develop simple PCR-based markers useful in future work to discriminate species belonging to different Capsicum complexes.
1. Introduction
In recent decades, plastid DNA (cpDNA) markers were used either to infer species-level phylogenetic and phylogeographic relationships in plants or to identify species via barcoding approaches [1,2,3]. Although cpDNA sequence divergence is often unable to provide adequate resolution of genetic differences at the intra-specific level because of its slow evolutionary rate, chloroplast DNA-based molecular markers, such as microsatellites and tandem repeats, are widely exploited to reveal inter-specific variation [4,5,6]. The progress of high-throughput sequencing technologies and the relatively simple assembling process of cpDNA gave a huge boost to genomic and phylogenetic studies [5,6,7,8]. As chloroplast genomes are characterized by a high number of potentially informative nucleotide sites, they became an efficient and cost-effective option for evolutionary and comparative genomic research in higher plants [9,10,11,12,13,14].
The genus Capsicum (Solanaceae), native to South and Central America and the southern United States of America (USA), includes sweet (peppers) and hot (chillies) cultivars of great economic importance that are cultivated and consumed around the world as vegetables and spices rich in vitamins A and C [15,16,17]. Capsicum-specific starch fossils, found at seven sites from the Bahamas to southern Peru, dating 6000 years before first contact with Europeans, clearly demonstrate that members of the genus Capsicum were extensively cultivated initially in the Americas and, after Columbus, were dispersed around the World [18]. Capsicum species exhibit flowers with stellate or rotate corollas characterized by diverse patterns of pigmentation and fleshy berries, which differ in shape, size, and color. In addition, plants of the genus Capsicum show an entire cup-shaped calyx, a unique trait among Solanaceae flowers and only shared with flowers of the genus Lycianthes [15].
Taxonomic studies of the genus, based on morphological, cytogenetic, biochemical, and ethnobotanical data, grouped Capsicum species into three main complexes: annuum (CA), baccatum (CB), and pubescens (CP). The CA complex includes wild and domesticated species of C. annuum, Capsicum chinense, Capsicum frutescens, and Capsicum galapagoense; the CB complex contains C. baccatum, Capsicum praetermissum, and Capsicum tovarii, whereas the CP complex comprises C. pubescens, Capsicum eximium, and Capsicum cardenasii [19,20]. Capsicum chacoense is considered a bridge species that could be included in either the CA or CB complex [19]. Although this classification is commonly accepted, establishing the genetic relationships within and between Capsicum species is still debated. Many studies were performed using different nuclear and plastid molecular markers to gain a better understanding of the genetic relationships within Capsicum and to assess genetic diversity in populations or core collections [5,15,16,21,22,23].
Next-generation sequencing (NGS) technologies provided a significant advancement in high-throughput data generation; however, the simultaneous analysis of a large number of genotypes is still challenging. Indeed, the whole chloroplast genome sequences of few Capsicum species have been released so far into the public domain [24,25,26,27,28,29,30]. The availability of a larger set of complete plastomes would allow for a better understanding of nuclear and cytoplasmic genome co-evolution, and would favor the development of more powerful methods for taxonomic barcoding and phylogenetic studies, as well as of novel biotechnological approaches for breeding purposes [31,32,33].
Here, we determined the complete nucleotide sequences of plastomes of eleven genotypes belonging to eight Capsicum species representing the three main taxonomic complexes, and performed a genome-wide analysis of molecular diversity among Capsicum plastomes. In addition to the assembly and annotation of plastomes, our aims were to (i) measure global patterns of sequence variations and establish the relationships among sequenced species; (ii) evaluate the extent of sequence similarity between plastomes; (iii) investigate any significant characteristics suggesting plastome rearrangements in Capsicum; (iv) derive estimates for molecular evolution of plastid protein-coding genes within Capsicum; (vi) and identify divergent regions suitable for the development of simple PCR-based molecular markers as a baseline to discriminate among Capsicum species.
2. Materials and Methods
2.1. Plant Material
A panel of eleven genotypes representing three complexes of the genus Capsicum was sampled for chloroplast isolation, cpDNA extraction, and sequencing. For the CA complex, we sampled three C. annuum genotypes (ann1, ann2, and ann3) and one genotype each for the species C. chinense (chi), C. frutescens (fru) and C. galapagoense (gal). For the CB complex, we sampled C. baccatum subsp. baccatum (bac.b), C. baccatum subsp. pendulum (bac.p), and C. praetermissum (pra). Finally, we also included a genotype from the CP complex, namely C. pubescens (pub), and a C. chacoense genotype (cha) that, depending on the classification schemes, is included in either CA or in CB (Table 1).

Table 1.
Plastome features of the eleven Capsicum genotypes.
Seeds that were provided by the Centre for Genetic Resources (Wageningen, The Netherlands) and Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK, Gatersleben, Germany) were germinated in the presence of 3 mM gibberellic acid. After germination, seedlings were transferred into pots and cultivated in a greenhouse under controlled conditions.
2.2. Chloroplast Isolation and DNA Extraction
Plants were kept in the dark for 48 h before harvesting to reduce starch contamination. A pool of about 15–25 g of fresh leaves collected from different individuals were used for chloroplast isolation with discontinuous sucrose gradients according to Kemble [34]. Purified chloroplasts were lysed with a detergent and the resulting cpDNA was dissociated from the proteins using proteinase K and phenol/chloroform treatments following the procedure described in Scotti et al. [35].
2.3. Chloroplast DNA Sequencing and Genome Assembly
Genomic libraries of C. baccatum subsp. baccatum, C. frutescens, C. praetermissum, and C. pubescens were sequenced on an Illumina HiSeq 2500 using the Illumina TruSeq DNA (Illumina, San Diego, CA, USA) sample preparation kit with 2 × 101 paired-end runs. The remaining plastomes were sequenced on an Illumina MiSeq platform with 2 × 251 paired-end runs after library preparation with an Illumina Nextera XT sample preparation kit.
High-quality reads were aligned onto the reference C. annuum cpDNA (NC_018552.1) with the BWA software [36] (version 0.7.12; Heng Li and Richard Durbin, Wellcome Trust Sanger Institute, Cambridge, UK). The Picard software (version 1.131; Broad Institute of MIT, Cambridge, MA, USA) was used to collect metrics (mean and standard deviation) on insert size distribution of each paired-end library. The Velvet software (version 1.2.10; Daniel R. Zerbino, Wellcome Trust Sanger Institute, Cambridge, UK) [37] was used for de novo assembly with the following settings: -ins_length XX, -ins_length_sd YY, scaffolding yes, and -exp_cov 100. Values XX and YY were obtained from the Picard analysis. Kmer length was set to 95 for all samples with the exception of C. chinense chi (kmer = 121) and pra C. praetermissum (kmer = 89).
2.4. Genome Annotation and Analysis of Nucleotide Variability
Genome annotation was carried out using the web tool DOGMA [38]. Gene structures were manually curated using Nicotiana tabacum (NC_001879.2) and Solanum lycopersicum (NC_007898.3) structural annotations as references. The circular C. pubescens plastome map was drawn using the online webtool OGDRAW—Draw Organelle Genome Maps [39]. Newly assembled chloroplast genomes and the plastome of Capsicum lycianthoides (NC_026551) were subjected to multiple alignment using the ClustalW program [40].
Genetic variability among pepper cpDNAs was investigated using different bioinformatic tools. Single-nucleotide variants (SNVs) were identified using the SNP-sites software [41] (Wellcome Trust Sanger Institute, Cambridge, UK). Such a tool extracted single-nucleotide polymorphisms (SNPs) from a multiple-sequence alignment using the cpDNA of C. lycianthoides as the reference sequence. Microsatellites (simple sequence repeats (SSRs)) were identified running the MIcroSAtellite (MISA) identification tool (http://pgrc.ipk-gatersleben.de/misa/) using the unit_size/min_repeats parameters as follows: 1/8, 2/6, 3/5, 4/5, 5/5, 6/5. Tandem repeats were identified using the Tandem Repeat Finder web tool accessible at https://tandem.bu.edu/trf/trf.basic.submit.html. Only perfect repeats were considered for downstream analysis. To visualize the overall differences among plastomes, we built pairwise alignments among eleven Capsicum plastomes by running mVISTA (Stanford University, Stanford, CA, USA) in LAGAN (Limited Area Global Alignment of Nucleotides) mode [42] and using the annotation of C. lycianthoides (NC_026551) as a reference. Aligned plastomes were also used to perform sliding window analysis using the DnaSP software [43] (University of Barcelona, Barcelona, Spain).
Seven regions, namely trnH-psbA, matK, rps16, trnL intron, atpB-rbcL, rbcL, and ndhF, that are traditionally used in previous phylogenetic studies based on Sanger sequencing methods [15,16,18,19,20] were used to infer a maximum-likelihood (ML) phylogenetic tree. The regions were extracted from each plastome using a custom perl script; then, a concatemer per genotype was generated, and a multiple-sequence alignment was built and manually inspected using the Bioedit software (Tom Hall, Ibis Therapeutics, Carlsbad, ON, Canada). An ML tree with 10,000 rapid bootstrap inferences, a generalized time reversible (GTR) substitution matrix and Gamma model of rate heterogeneity was inferred using the RAxML (The Exelixis Lab 2013, Scientific Computing Group, Heidelberg, Germany) program [44]. The RAxML results were visualized with the FigTree software, v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The same approach was used to infer a second tree based on the alignment of complete plastid sequences of the same genotypes.
Capsicum chloroplast genomes released into the public domain with accession numbers NC_028007.1/KR078312.1 (C. frutescens), NC_033525.1/KX913218.1 (C. chacoense), NC_030543.1/KU041709.1 (C. chinense), NC_033524.1/KX913216.1 (C. galapagoense), NC_018552.1/JX270811.1 (C. annuum), KR078313.1 (C. annuum), and KR078314.1 (C. baccatum var. baccatum) were downloaded from GenBank.
Pairwise global alignments between already publically available chloroplast sequences and plastomes that were sequenced and assembled in this study were performed using the European Molecular Biology Open Software Suite (EMBOSS) Stretcher tool. SNVs were identified using SNP-sites [41], while insertions/deletions (InDels) were manually scored.
2.5. Molecular Evolution Analysis on Protein-Coding Genes
The coding sequences of the 79 protein-coding genes present in all Capiscum plastomes and in C. lycianthoides (NC_026551) were extracted and fed into the Selecton web server [45] (http://selecton.tau.ac.il/) in order to investigate amino-acid sites under positive selection. The evolutionary model M8a (ωs = 1) was used. We considered a site under positive selection if the lower bound was >1 and the p-value was <0.01.
2.6. Primer Design and PCR Amplification
Primers for the development of ccsA-ndhD (forward (F): ACACATAGAAATTTGCGGGGTGC; reverse (R): TCGATGGCTTCCCTTGCATTACCA) and trnL-trnF (F: ATCGAAGAAATTCCCCGGCT; R: GCGCACATTACTTAGACGGGTT) molecular markers were designed from assembled plastomes by using the MacVector software (MacVector Inc., Apex, NC, USA). PCR amplifications were carried out, using Taq DNA polymerase according to the manufacturer’s instructions (Invitrogen, Paisley, UK), on 25 ng of total DNA or cpDNA of the following genotypes: C. baccatum subsp. baccatum (bac.b, Table 1), C. baccatum subsp. pendulum (bac.p, Table 1; bac.p2, CGN17015; bac.p3, CGN22181; bac.p4, CGN17174), C. praetermissum (pra, Table 1), C. pubescens (pub, Table 1; pub2, CGN22796; pub3, CAP1486), C. chacoense (cha, Table 1; cha2, CAP1445; cha3, CAP499; cha4, CAP501), C. annuum (ann2, Table 1; ann4, CGN17175; ann5, CGN21490; ann6, CGN24355; ann7, CGN23249), C. chinense (chi2, CGN17220; chi3, CGN23565; chi4, CGN17219), and C. frutescens (fru2, CGN22792; fru3, RCAT077650; fru4, CGN21546). The reaction conditions for all amplifications were as follows: denaturation at 94 °C for 3 min, then 30 cycles (94 °C, 30 s; annealing temperature, 30 s; 72 °C, 1 min/kb), followed by 5 min final extension at 72 °C.
3. Results
3.1. Chloroplast Genome Size and Organization
Sequencing of the eleven Capsicum genotypes produced 5,634,814–404,910,769 base pairs (bp) of high-quality plastid reads with per-base mean coverage ranging from 26 to 2581. A combination of de novo and reference-guided assembly with the C. annuum chloroplast genome (NC_018552.1) as a reference was used to obtain the complete plastome for all genotypes. Genome sizes ranged from 156,836 bp in C. frutescens to 157,390 in C. pubescens (Table 1).
As expected, all Capsicum genotypes exhibited the typical quadripartite structure of angiosperms, including a pair of inverted repeats (IRs), ranging from 25,751 bp to 25,910 bp in size, separated by two single-copy regions, a large single copy (LSC, 87,288 bp min–87,688 bp max) and a small single copy (SSC, 17,860 bp min–17,973 bp max). A slight variation in guanine/cytosine (GC) content among genotypes was observed (Table 1). Each of the eleven plastomes, similar to other Solanaceae, contains 113 genes, including 79 protein-coding, four ribosomal RNA, and 30 transfer RNA (tRNA) genes. Seventeen genes, located in IR regions, were duplicated (Figure 1).
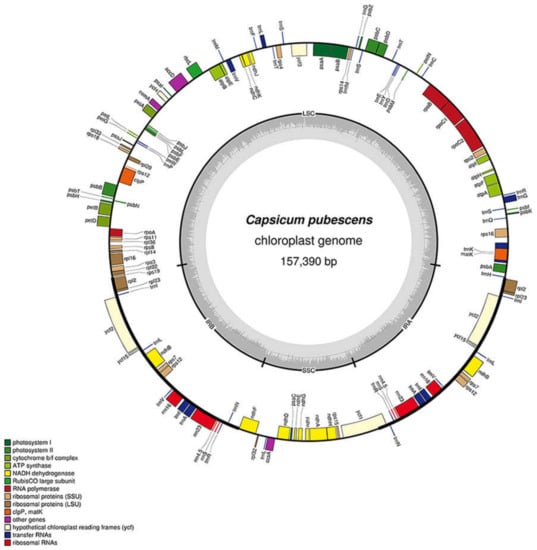
Figure 1.
Map of the Capsicum pubescens chloroplast genome. Genes inside of the outer circle are transcribed in the clockwise direction, while those outside are transcribed in the counterclockwise direction. Different color codes represent genes belonging to various functional groups. The circle inside GC content graph marks the 50% threshold. The inverted repeat, large single-copy, and small single-copy regions are denoted by IR, LSC, and SSC, respectively.
A detailed view of the IR–SSC/LSC junctions of the plastomes under investigation is provided in the Supplementary Materials (Figure S1). In all genotypes, the LSC/IRb and SSC/IRa junctions are in the rps19 and ycf1 genes, respectively, while the IRb/SSC and IRa/LSC ones are in the intergenic trnN–GUU/ndhF and rpl2/trnH–GUG regions. The junction position, however, slightly varies among different genotypes.
The eleven plastomes were deposited in GenBank under accession numbers: MH559320–MH559330.
3.2. Sequence Variation within Capsicum Genotypes
All comparative analyses across pepper genotypes were carried out using C. lycianthoides (NC_026551) as a reference genome. Sliding window analysis of the multiple-sequence alignment including the eleven Capsicum plastomes and C. lycianthoides showed high sequence similarity and indicated the trnN–GUU/ndhF intergenic region as a polymorphic hotspot (Figure 2).
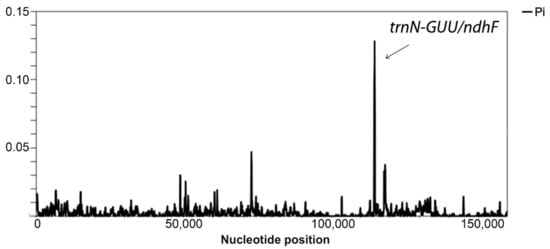
Figure 2.
Sliding window analysis of the multiple plastome sequence alignment within the Capsicum genus. The region with high nucleotide variability (Pi > 0.05), corresponding to the IR/SSC junction, is indicated. Window length = 200 base pairs (bp); step size = 50 bp.
Similarly, VISTA-based identity plots revealed moderate sequence divergences among the genotypes under investigation. Indeed, nucleotide differentiation mainly affects intergenic/non coding regions, as well as single-copy regions (Supplementary Figure S2). In comparison with C. lycianthoides, all pepper genotypes showed a large deletion (over 500 bp) in the intergenic region between ndhF and rpl32 genes (below 50% identity).
Variations SNPs, tandem repeats (TRs), and SSRs were assessed among the sequenced Capsicum genotypes. A range of 1152–1271 SNPs was detected among the eleven plastomes. The distribution of these variations in different regions was slight different among species and well conserved within genotypes belonging to the CA complex (Supplementary Figure S3). In nine cases, two alternative alleles, compared to the reference, were discovered (Supplementary Materials Table S1). The CB complex (including bac.b, bac.p, and pra) showed the highest SNP variations in intergenic (642–644), exon (497–500), and intron (125–130) regions, whereas the CA complex (ann1, ann2, ann3, chi, fru, and gal) was characterized by 581–600, 462–470, and 105–111 SNPs in intergenic, exon, and intron regions, respectively. Intermediate values were detected for C. chacoense and C. pubescens (Supplementary Materials, Figure S3). Although the total number of detected SNPs seems almost equally distributed between intergenic and exon regions, normalization of SNP number per kb highlighted higher values in intergenic (13.99–15.51) compared to exon (6.09–6.58) regions (data not shown). SNP distribution within LSC, SSC, and IRb regions is also shown in the Supplementary Materials (Figure S3).
We identified 92 SSRs, of which 65 were polymorphic among the eleven Capsicum species, including mononucleotide, dinucleotide, trinucleotide, and tetranucleotide repeats (Supplementary Materials Figure S4A and Table S2). No pentanucleotide or hexanucleotide repeats were observed. The mononucleotide repeat (adenosine/thymine (A/T)) was the most common type of microsatellite in pepper plastomes, whereas the tetranucleotide unit, repeated four times, was typical of the CA complex (Supplementary Materials Table S2). The distribution of SSRs showed that these loci were primarily located in intergenic regions and in the LSC, whilst the distribution in exon and intron regions and/or in the SSC and IRb was comparable (Supplementary Materials Figure S4B).
A total of 58 perfect tandem repeats (TRs) were identified, of which 51 are characterized by a period size of 9–30 bp, six have a period size ranging from 30 to 60 bp, and one is longer than 100 bp (Supplementary Materials Table S3). They are mostly located in intergenic regions (50), seven are in coding regions of accD, rpl33, ycf2, ndhD, and ycf1, and one was in the intron of the rps16 gene. Most of them are located in the LSC region (Supplementary Materials Figure S5 and Table S3). Thirty-two of the 58 TRs are polymorphic within the eleven Capsicum plastomes, while three of them are located in the coding regions of accD and ycf1. A tandem repeat of 30 nucleotides located in the ycf1 coding region is exclusive to the CB complex.
Among the annotated 79 protein-coding genes, 26 have perfectly conserved sequences and 48 have point mutations within the coding sequence, while five genes, namely accD, ndhB, rpl20, ycf1, and ycf2 are the most variable. The latter differ in gene length because of several insertions/deletions, also evident at the amino-acid level (Figure 3).
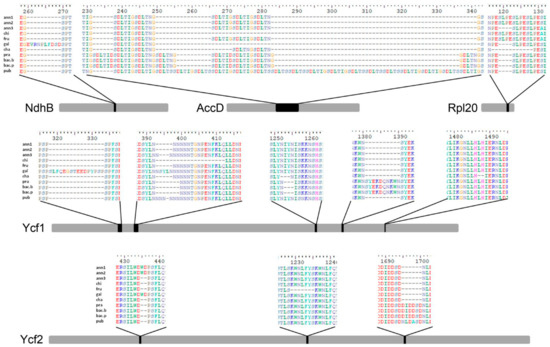
Figure 3.
Schematic representation of the five most variable genes (ndhB, accD, rpl20, ycf1 and ycf2) in the plastomes under investigation. Gray bars represent the multiple-sequence alignment (MSA) for each gene and are scaled according to the MSA length. Black boxes indicate highly variable regions in the MSA. Above each box, a snapshot of the MSA along with alignment positions is reported.
In order to further evaluate within-species nucleotide variability, already publically available Capsicum chloroplast genomes belonging to the same species whose plastomes were sequenced in this work were downloaded from GenBank for comparative genomics. Species-specific pairwise global alignments showed nucleotide variability to be in the range of 0.1% to 0.3% (Supplementary Materials Table S4). Generally, nucleotide variability is in non-coding regions and affects A/T stretches, the number of tandem repeat units, and DNA low-complexity regions (data not shown).
3.3. Phylogenetic Reconstruction and Molecular Evolution
In order to reconstruct the phylogeny of Capsicum and to verify the evolutionary significance of SNP, SSR, and TR variation, a phylogenetic tree was inferred from plastid concatemers of seven regions (trnH-psbA, matK, rps16, trnL intron, atpB-rbcL, rbcL, and ndhF) from the eleven newly assembled pepper plastomes and C. lycianthoides (as an outgroup). The phylogenetic tree inferred from maximum-likelihood analysis has strong bootstrap supports for all nodes with the exception of the placement of C. galapagoense with respect to other species of the CA complex, consisting of C. annuum, C. frutescens, and C. chinense. Nevertheless, the branch subtending the inclusion of C. galapagoense in the C. annuum clade is strongly supported. The C. chacoense genotype examined here is sister to the CB complex with strong support. Finally, C. pubescens (a member of the CP complex) is sister to both the C. annuum and C. baccatum lineages. The phylogenetic tree was compared with SNP variability in exon, intron, and intergenic regions (Supplementary Materials Figure S6), and with SSR and TR variation (Supplementary Materials Figure S7). In all cases, we found that the observed variability reflects the pattern of phylogenetic relationship resulting from the maximum-likelihood analysis. Based on this correspondence, we repeated the phylogenetic analysis using the alignment from the complete plastome sequences: the resulting ML tree (Figure 4) has the same topology as that based on concatemers of the seven plastid regions, but a stronger bootstrap supports all nodes.
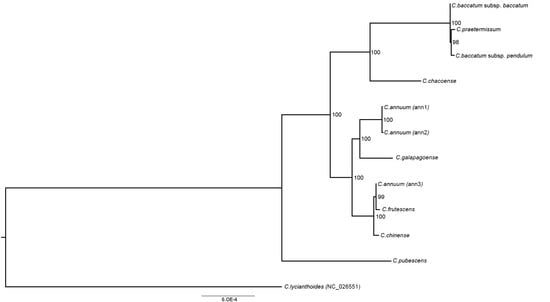
Figure 4.
Phylogenetic tree of Capsicum genotypes. Phylogram of the best maximum-likelihood (ML) tree as determined using the RAxML software from the complete plastome dataset. Numbers associated with branches are ML bootstrap support values.
Gene divergence analysis, based on Selecton, showed that protein-coding genes are generally well conserved among Capsicum species. The most divergent genes are rpl20 and rpl32, followed by rpl36, clpP, and accD (Figure 5A). This analysis also evidenced high divergent branch length for most genes in the outgroup C. lycianthoides. For example, in the accD gene, in addition to C. lycianthoides, the species that exhibited highly divergent branches are those belonging to the CB and CP complexes. Furthermore, we also investigated the positive selection of protein-coding genes, and identified 25 putative positive signatures distributed in six out of 79 protein-coding genes (matK, rbcL, accD, rpl20, petD, and rpl32) (Figure 5B).

Figure 5.
Results of molecular evolution analysis of plastid genes within the Capsicum genus. (A) Estimation of protein-coding gene divergence by the average branch length ± standard deviation for each gene tree; (B) number of putative sites under positive selection.
3.4. Chloroplast-Specific Molecular Markers for Capsicum spp.
Comparative analyses allowed us to identify divergent regions potentially useful for an in-depth molecular characterization of the Capsicum genus. Among them, we selected those suitable for the development of simple PCR-based molecular markers able to discriminate among different complexes. In Table 2, some examples of three types of potential chloroplast molecular markers in pepper spp. are reported.

Table 2.
Examples of chloroplast molecular markers (single-nucleotide polymorphisms, SNPs; simple sequence repeats, SSRs; tandem repeats, TRs) identified in this study using the accession NC_026551 of C. lycianthoides as a reference.
Based on SNP variations, the selected coding (psbA, atpI, rps2, rpoB, and atpB) and non-coding (rps16 intron) regions discriminated species or complexes through the loss or gain of restriction sites, making them useful for developing cleaved amplified polymorphic sequence (CAPS) markers. In particular, the SNPs present in psbA and atpI cause the gain and loss of a HpyCH4III and Hpy188III restriction sites and were specific to the CB complex; those in the rps16 intron and rps2 result in the loss of EcoRI and Sau3AI restriction sites in C. chacoense, and the loss of HpyCH4V and AluI sites in rpoB and atpB for the CP and CA complexes, respectively.
The rpl20–rps12 intergenic region showed the highest variability in the SSR, discriminating all three species complexes. By contrast, an SSR detected in the ycf3 intron is specific to C. chacoense, and microsatellites present in the psbK–psbI and atpB–rbcL intergenic regions univocally characterize C. pubescens. The rpl32–trnL intergenic region includes an SSR discriminating C. galapagoense from the remaining species of the CA complex.
Compared with other potential molecular markers, tandem repeats showed lower discriminatory power among species complexes. In fact, the TRs present in the ccsA–ndhD intergenic and ycf1 coding regions distinguished the CB complex, whereas TRs within the trnS–rps14 and ycf2–trnI regions differentiated C. pubescens and species belonging to the CA complex, respectively.
A preliminary experimental validation in representative species confirmed the presence of the tandem repeat detected in the ccsA–ndhD intergenic region in genotypes of the CB complex, and the insertion in the C. pubescens plastome of a sequence of 98 bp in length in the trnL–trnF region that was previously undescribed (Figure 6). The validation of other candidate markers listed in Table 2 is currently underway, and it will be the main objective of a future study.
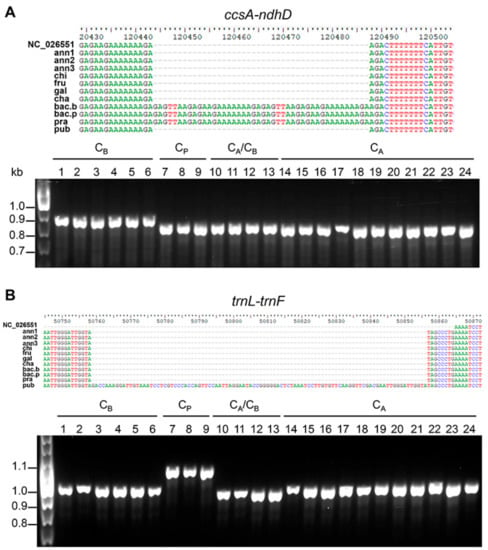
Figure 6.
Examples of chloroplast molecular markers developed in this study. PCR markers based on the presence of perfect tandem repeats and insertions/deletions (InDels) able to discriminate CB (A) and CP (B) complexes. PCR results from representative genotypes in each complex are shown. CB = C. baccatum; CP = C. pubescens; CA = C. annuum; 1 = bac.b; 2 = bac.p; 3 = bac.p2; 4 = bac.p3; 5 = bac.p4; 6 = pra; 7 = pub; 8 = pub2; 9 = pub3; 10 = cha; 11 = cha2; 12 = cha3; 13 = cha4; 14 = ann2; 15 = ann4; 16 = ann5; 17 = ann6; 18 = ann7; 19 = chi2; 20 = chi3; 21 = chi4; 22 = fru2; 23 = fru3; 24 = fru4.
4. Discussion
Until the ‘80s, the sequencing of single chloroplast genes and/or of non-coding regions was widely used for phylogenetic studies with the purpose of clarifying inter- and intra-species relationships and investigating plastid DNA diversity [31,46]. Improvements in protocols for chloroplast isolation and cpDNA extraction, coupled with the evolution and spread of NGS techniques, made complete plastid genome sequencing affordable [32,47]. This promptly allowed the extension of gene-based phylogenetics to phylogenomics and going beyond traditional molecular marker-based barcoding approaches. Indeed, the number of plastid genome sequences released into the public domain for land plant species is growing at an unprecedented rate (see https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles/). Several projects were recently undertaken with the aim of obtaining multiple complete chloroplast genomes and providing basic information for comparative analysis [14,48].
At present, ten cpDNA sequences are available in Genbank for Capsicum species. With the present study, we contributed to enriching the cpDNA sequence space available for Capsicum by releasing into the public domain the plastomes of eleven genotypes. Based on this resource, we performed the first large-scale genome-wide analysis of molecular diversity of Capsicum species belonging to the three main taxonomic complexes. Mapping of reads ranges from 36 to 2581 per-base coverage across chloroplast genomes. Although we recorded a 72-fold difference in mean sequence depth per base among plastomes, this coverage was nevertheless sufficient to assemble all full-length genomes. The plastomes were fairly well conserved in terms of size, gene arrangement, and gene number, and comparable with those of Capsicum species available at GenBank. In order to evaluate within-species nucleotide variability, we compared already publically available Capsicum plastomes with those produced in this work. Pairwise sequence comparisons showed that sequences belonging to the same species are almost identical, even if a variable number of SNPs and InDels was identified. A subset of the InDel events we observed fall within mono-nucleotide repetitive stretches (mainly A/T), which are known to be prone to sequencing errors. The remaining InDels affect tandem repeats and, to a lesser extent, microsatellite or low-complexity regions. It cannot be excluded that these InDels may be due to errors in the assembly procedure. Indeed, the assembly of large tandem-repeat arrays remains intractable especially if the sequencing technique is based on short reads [49].
Although comparative analysis of genomic sequences, which included C. lycianthoides as a reference genome, revealed high sequence similarity among the eleven pepper plastomes, a wide spectrum of variations, ranging from point mutations to small/medium-sized InDels, was observed in 67% of the genes. The differences in the most variable genes (e.g., accD, ndhB, rpl20, ycf1, and ycf2) were due to InDels. In particular, accD and rpl20 showed the highest variability between species as already observed [24] upon comparing the C. annuum plastome with those of other Solanaceae. Compared to previous results [24], we found both intra- and inter-specific variability in the CA complex. In addition, we detected a large insertion in the trnL–trnF intergenic region of C. pubescens, while ycf1 and ycf2 were characterized in most genotypes by various InDels.
Single-nucleotide polymorphisms, when normalized per kb, resulted in a comparable number among the eleven plastomes and mostly localized in intergenic regions as expected, since coding regions are, in general, more conserved than non-coding regions [50]. Simple sequence repeats or microsatellites are locus-specific and multi-allelic markers that were extensively involved in a variety of applications including cultivar identification [51], genetic diversity assessment [52], molecular evolution [53], etc. In the present study, SSRs were mainly localized in intergenic regions and in the LSC. This finding is in agreement with previous results for species belonging to Solanaceae, Poaceae, and Arecaceae [5,30,54,55]; indeed, the low number of SSRs within IRs is due to its duplicative nature that implies copy-correction activity [54]. Mono-(p1), di-(p2), and trinucleotide (p3) SSR types were detected in all genotypes under investigation. In particular, the majority involved p1 SSRs, representing 82–87% of the total, whilst tetranucleotides (p4) were only present in species belonging to the CA complex (1.6% of the total). This pattern of distribution was in accordance with previous results on four cultivated species of the Capsicum genus, reporting a frequency of 80% for mononucleotides, while tetranucleotides were the least frequent type [5]. The analysis of tandem repeats revealed that their period size was mostly between nine and 30 nucleotides and only one period sequence was longer than 100 nucleotides.
The strong bootstrap support of the ML tree based on the concatamers of seven plastid regions strengthens that it can represent a reliable phylogenetic framework for the assessment of repetitive element evolution in Capsicum species. When this phylogenetic tree was compared with variability derived from SSRs and TRs, the species grouping obtained by the ML analysis reflected the observed variability in repetitive sequences. While the CA complex is relatively homogeneous in terms of variation in SNPs and repeats, C. chacoense displays a different pattern of variation compared with genotypes in the CB complex, particularly for SNP and SSR variation.
Both the phylogenetic reconstructions based on the concatamers of seven plastid regions and on complete plastid sequence (Figure 3) correspond well to previous reports on the relationships among Capsicum species and complexes. However, the stronger bootstrap support of the latter tree allows the confirmation of the taxonomic placement of some critical species as the inclusion of C. galapagoense in the CA complex and of C. praetermissum in the CB complex. In particular, the C. chacoense genotype examined here can be unequivocally assigned as a sister to the CB complex, accordingly with the results of Walsh and Hoot [20], and ruling out the previous hypothesis by Ince, Karaca, and Onus [19], who postulated C. chacoense as a sort of bridge placement between the CA and CB complexes. Nevertheless, C. chacoense is the basal species in the clade, including the CB complex; thus, it is expected also to share some plesiomorphic traits with the CA complex.
We identified 25 putative positive signatures distributed in six protein-coding genes. Overall, the genes with higher divergence rate also showed higher positive selection. These genes were involved in different essential functions such as the Calvin cycle (rbcL), cytochrome b6f (petD), RNA maturation (matK), ribosomal proteins (rpl20, rpl32), and fatty-acid biosynthesis (accD). The genes under positive selection may be related to a recent increase in diversification rate following adaptation to novel ecological conditions [56,57]. In particular, as it was also found in other plant lineages, we detected both highly divergent branches and accelerated rates of evolution in the accD gene, a plastid-encoded subunit of the acetyl-coenzyme A (CoA) carboxylase [58]. accD was found to affect plant fitness and leaf longevity [59,60] and might have been involved in the adaptation to specific ecological niches during Capsicum diversification.
The sequence variations identified here were used to develop simple PCR-based markers useful to distinguish species belonging to different complexes. Single-nucleotide polymorphism SNP analysis led us to identify variation in the gain and loss of restriction sites allowing the development of CAPS markers, allowing the discrimination of different complexes. In particular, SNPs present in psbA and atpI were specific for the CB complex, whereas SNPs in atpB allowed discrimination among species belonging to the CA complex. The use of SSRs in intergenic regions as molecular markers was widely suggested, since these regions evolve faster than coding sequences [31,55]. Among them, we identified the variation in the atpB–rbcL region, already reported by Walsh and Hoot [20], that elucidated relationships between Capsicum spp., thereby contributing to their taxonomic grouping. We selected and tested, in a representative sample of Capsicum species, a TR in the ccsA–ndhD region that clearly enabled us to discriminate species belonging to the CB complex. Furthermore, in the trnL–trnF intergenic region, we identified and tested an insertion of 98 bp in C. pubescens different from that of 225 bp found in C. annuum by Jo et al. [24]. Moreover, Jarret [61] evaluated the feasibility of using this locus for DNA barcoding within the C. annuum complex and demonstrated its ability to differentiate among the examined species. Generally, plant DNA barcoding often showed their limit in species discrimination, especially for closely related taxa, making unrealistic the possibility of using a universal set of markers for species identification in higher plants. This limitation is further exacerbated for application at the intra-specific level. There is an increasing interest in expanding the genetic diversity in the production chain, as well as in the genetic traceability of foods with specific metabolic traits. While traditional barcoding often struggles to reliably differentiate within Capsicum complexes, full plastome sequences proved powerful to distinguish each cultivar, by virtue of global patterns of sequence variations. Indeed, thanks to the use of the full plastome barcode as the method of choice for plant identification, we envisage a growing use of full-length plastomes in the identification and traceability of pepper varieties.
5. Conclusions
The present study reports the complete plastomes of eleven genotypes belonging to the three main taxonomic species complexes of Capsicum. This sequence resource was exploited for the genome-wide analysis of molecular diversity within the Capsicum genus. Comparative analysis revealed a wide spectrum of variations, some of which were found at both the inter- and intra-specific level. Our phylogenetic reconstruction corresponds well to previous reports on the relationships among Capsicum species and complexes, but contributes to the taxonomic placement of some critical species. In particular, the C. chacoense genotype examined here can be unequivocally assigned as a sister to the CB complex. Furthermore, we identified 25 putative positive signatures distributed in six protein-coding genes involved in different essential functions of chloroplasts and probably related to the recent increase in diversification rate following adaptation to novel ecological conditions. Finally, the sequence variations allowed us to develop simple PCR-based markers that can be helpful to distinguish species belonging to different complexes.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/10/503/s1, Figure S1: Comparison of plastome junctions (LSC/IRb, SSC/IRa, IRb/SSC, and IRa/LSC) among pepper species. Numbers indicate the lengths of intergenic spacers (IGSs), genes, and spacers around IR/LSC and IR/SSC junctions; Figure S2: Comparison of eleven Capsicum plastome sequences using the VISTA software and the accession NC_026551 of C. lycianthoides as a reference. Blue and red regions correspond to coding and non-coding regions, respectively. The Y-axis represents percent similarity ranging from 50–100%; Figure S3: Distribution of single-nucleotide polymorphisms (SNPs) in the eleven Capsicum plastomes using the accession NC_026551 of C. lycianthoides as a reference. Number and SNP distribution among different regions: exon, intron, intergenic region, large single-copy region (LSC), small single-copy region (SSC), and inverted repeat b (IRb). The number of SNPs (left bar) does not correspond to SNP distribution (right bar) due to overlap of several genes on opposite strands; Figure S4: Distribution of simple sequence repeats (SSRs) in the eleven Capsicum plastomes and in the accession NC_026551 of C. lycianthoides used as an outgroup species. (A) Total number of SSRs reported as SSR type. (B) Number and SSR distribution among different regions: exon, intron, intergenic region, large single-copy region (LSC), small single-copy region (SSC), and inverted repeat b (IRb). P1 = mono-, p2 = di-, p3 = tri-, p4 = tetranucleotide; Figure S5: Distribution of perfect tandem repeats (TRs) in the eleven Capsicum plastomes and in the accession NC_026551 of C. lycianthoides used as an outgroup species. TR distribution among different regions: exon, intergenic region, large single-copy region (LSC), small single-copy region (SSC), and inverted repeat b (IRb); Figure S6: Molecular phylogenetic analysis using maximum-likelihood method and SNP variation across exon, intron, and intergenic regions among eleven Capsicum plastomes. (A) Phylogenetic tree inferred from maximum-likelihood analysis of seven combined plastid regions (RAxML maximum-likelihood bootstrap above nodes). Heat maps represent SNP variability in (B) exon, (C) intron, and (D) intergenic regions compared with the C. lycianthoides plastome (NC_026551) used as a reference. Yellow corresponds to the reference allele; red and blue correspond to alternative alleles. The arrows indicate the anticlockwise genome orientation; Figure S7: Molecular phylogenetic analysis using maximum-likelihood method and SSR and TR size variation among eleven Capsicum plastomes. (A) Phylogenetic tree inferred from maximum-likelihood analysis of seven combined plastid regions (RAxML maximum-likelihood bootstrap above nodes). Heat maps represent differences in SSR size (B) and in the number of copies of perfect tandem repeats (C) compared with C. lycianthoides plastome (NC_026551) used as a reference. Heat map colors range from green through yellow to red, where green and red indicate an SSR size greater or lesser than the reference, and a higher and lower number of copies than the reference. The arrows indicate the anticlockwise genome orientation; Table S1: Single-nucleotide polymorphisms (SNPs) in the eleven Capsicum plastomes identified using the SNP-sites tool [41] using the accession NC_026551 of C. lycianthoides as a reference. “0” indicates the reference allele, “1” or “2” indicates the alternative allele; Table S2: Simple sequence repeats (SSRs) in the eleven Capsicum plastomes using the accession NC_026551 of C. lycianthoides as a reference. SSR size, location, and distribution among different regions: exon, intron, and intergenic regions are reported. SSRs were identified using the MISA (MIcroSAtellite) identification tool (http://pgrc.ipk-gatersleben.de/misa/); Table S3: Perfect tandem repeats (TRs) in the eleven Capsicum plastomes using the accession NC_026551 of C. lycianthoides as reference. TR period size, copy number, location, and distribution among different regions: exon, intron, and intergenic regions are reported. TRs were identified using the Tandem Repeats Finder tool (https://tandem.bu.edu/trf/trf.basic.submit.html); Table S4: Summary of within-species nucleotide variability assessed starting from pairwise global alignments.
Author Contributions
N.D., S.C., T.C., and N.S. conceived and designed the research. N.D., C.C., and V.D.C. performed bioinformatic analyses. R.T., L.S., and N.S. carried out wet-lab experiments. N.D., R.T., S.C., T.C., and N.S. contributed to data interpretation. N.D., R.T., and N.S. wrote the manuscript. S.C. and T.C. revised the manuscript. All authors read and approved the manuscript.
Funding
This research was partially funded by grants from the Italian Ministry of Research (MIUR) grant number: PON02_00395_3215002 “GenHORT”.
Acknowledgments
Technical assistance of G. Guarino and R. Nocerino (CNR-IBBR, Portici, Italy), and G. Festa and M. Salzano (CREA, Pontecagnano, Italy) with artwork and plant growth is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; de Pamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in Angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Olmstead, R.G.; Bohs, L.; Migid, H.A.; Santiago-Valentin, E.; Garcia, V.F.; Collier, S.M. A molecular phylogeny of the Solanaceae. Taxon 2008, 57, 1159–1181. [Google Scholar]
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F.; et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Powell, W.; Morgante, M.; Andre, C.; McNicol, J.W.; Machray, G.C.; Doyle, J.J.; Tingey, S.V.; Rafalski, J.A. Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr. Biol. 1995, 5, 1023–1029. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, Z.; Li, B.; Qin, C.; Wu, Z.; Trejo-Saavedra, D.L.; Luo, X.; Cui, J.; Rivera-Bustamante, R.F.; Li, S.; et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 2016, 6, 18919. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, S.; Ding, Y.; Xu, J.; Li, M.F.; Zhu, S.; Chen, N. Chloroplast genomic resource of Paris for species discrimination. Sci. Rep. 2017, 7, 3427. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Woeste, K.E.; Zhao, P. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front. Plant Sci. 2016, 7, 1955. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, R.; Li, X. Comparative analysis of the complete chloroplast genome of four known Ziziphus species. Genes 2017, 8, 340. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-P.; Huang, J.-P.; Wu, C.-S.; Hsu, C.-Y.; Chaw, S.-M. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2010, 2, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; De Paola, D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS ONE 2015, 10, e0120589. [Google Scholar] [CrossRef] [PubMed]
- Rousseau-Gueutin, M.; Bellot, S.; Martin, G.E.; Boutte, J.; Chelaifa, H.; Lima, O.; Michon-Coudouel, S.; Naquin, D.; Salmon, A.; Ainouche, K.; et al. The chloroplast genome of the hexaploid Spartina maritima (Poaceae, Chloridoideae): Comparative analyses and molecular dating. Mol. Phylogenet. Evol. 2015, 93, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Gu, C.; Tembrock, L.R.; Zhang, D.; Ge, S. Characterization of the whole chloroplast genome of Chikusichloa mutica and its comparison with other rice tribe (Oryzeae) species. PLoS ONE 2017, 12, e0177553. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Q.; Drew, B.T.; Yang, J.-B.; Gao, L.-M.; Li, D.-Z. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS ONE 2017, 12, e0178026. [Google Scholar] [CrossRef] [PubMed]
- Carrizo García, C.; Barfuss, M.H.J.; Sehr, E.M.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 2016, 118, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.A.; Ashrafi, H.; Reyes-Chin-Wo, S.; Yao, J.; Stoffel, K.; Truco, M.-J.; Kozik, A.; Michelmore, R.W.; Van Deynze, A. Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30 k unigene pepper genechip. PLoS ONE 2013, 8, e56200. [Google Scholar] [CrossRef] [PubMed]
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sánchez García, Y.; Jarret, R.; Daviña, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The Evolution of Chili Peppers (Capsicum—Solanaceae): A Cytogenetic Perspective, 2007; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2007; pp. 137–170. [Google Scholar]
- Perry, L.; Dickau, R.; Zarrillo, S.; Holst, I.; Pearsall, D.M.; Piperno, D.R.; Berman, M.J.; Cooke, R.G.; Rademaker, K.; Ranere, A.J.; et al. Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the americas. Science 2007, 315, 986–988. [Google Scholar] [CrossRef] [PubMed]
- Ince, A.G.; Karaca, M.; Onus, A.N. Genetic relationships within and between Capsicum species. Biochem. Genet. 2010, 48, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Walsh, B.M.; Hoot, S.B. Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: The chloroplast atpB-rbcL spacer region and nuclear waxy introns. Int. J. Plant Sci. 2001, 162, 1409–1418. [Google Scholar] [CrossRef]
- Rai, V.P.; Kumar, R.; Kumar, S.; Rai, A.; Kumar, S.; Singh, M.; Singh, S.P.; Rai, A.B.; Paliwal, R. Genetic diversity in Capsicum germplasm based on microsatellite and random amplified microsatellite polymorphism markers. Physiol. Mol. Biol. Plants 2013, 19, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Nicolaï, M.; Cantet, M.; Lefebvre, V.; Sage-Palloix, A.-M.; Palloix, A. Genotyping a large collection of pepper (Capsicum spp.) with SSR loci brings new evidence for the wild origin of cultivated C. annuum and the structuring of genetic diversity by human selection of cultivar types. Genet. Resour. Crop Evol. 2013, 60, 2375–2390. [Google Scholar] [CrossRef]
- Thul, S.T.; Darokar, M.P.; Shasany, A.K.; Khanuja, S.P.S. Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol. Biotechnol. 2012, 51, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.D.; Park, J.; Kim, J.; Song, W.; Hur, C.-G.; Lee, Y.-H.; Kang, B.-C. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 2011, 30, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Raveendar, S.; Na, Y.-W.; Lee, J.-R.; Shim, D.; Ma, K.-H.; Lee, S.-Y.; Chung, J.-W. The complete chloroplast genome of Capsicum annuum var. glabriusculum using illumina sequencing. Molecules 2015, 20, 13080–13088. [Google Scholar] [CrossRef] [PubMed]
- Raveendar, S.; Jeon, Y.-A.; Lee, J.-R.; Lee, G.-A.; Lee, K.J.; Cho, G.-T.; Ma, K.-H.; Lee, S.-Y.; Chung, J.-W. The complete chloroplast genome sequence of Korean landrace “Subicho” pepper (Capsicum annuum var. annuum). Plant Breed. Biotechnol. 2015, 3, 88–94. [Google Scholar] [CrossRef]
- Shim, D.; Raveendar, S.; Lee, J.-R.; Lee, G.-A.; Ro, N.-Y.; Jeon, Y.-A.; Cho, G.-T.; Lee, H.-S.; Ma, K.-H.; Chung, J.-W. The complete chloroplast genome of Capsicum frutescens (Solanaceae). Appl. Plant Sci. 2016, 4, 1600002. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Lee, J.; Lee, S.-C.; Yang, T.-J.; Yoon, J.B. The complete chloroplast genome sequence of Capsicum chinense jacq. (Solanaceae). Mitochondrial DNA B Resour. 2016, 1, 164–165. [Google Scholar] [CrossRef]
- Zeng, F.-C.; Gao, C.-W.; Gao, L.-Z. The complete chloroplast genome sequence of American bird pepper (Capsicum annuum var. glabriusculum). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Raveendar, S.; Lee, K.J.; Shin, M.-J.; Cho, G.-T.; Lee, J.-R.; Ma, K.-H.; Lee, G.-A.; Chung, J.-W. Complete chloroplast genome sequencing and genetic relationship analysis of Capsicum chinense jacq. Plant Breed. Biotechnol. 2017, 5, 261–268. [Google Scholar] [CrossRef]
- Rogalski, M.; do Nascimento Vieira, L.; Fraga, H.P.; Guerra, M.P. Plastid genomics in horticultural species: Importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 2015, 6, 586. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Kemble, R.J. A rapid, single leaf, nucleic acid assay for determining the cytoplasmic organelle complement of rapeseed and related Brassica species. Theor. Appl. Genet. 1987, 73, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Scotti, N.; Cardi, T.; Marechal Drouard, L. Mitochondrial DNA and RNA isolation from small amounts of potato tissue. Plant Mol. Biol. Rep. 2001, 19, 67. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with dogma. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellar genome draw (ogdraw): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using clustalw and clustalx. Curr. Protoc. Bioinform. 2003. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. Snp-sites: Rapid efficient extraction of SNPs from multi-fasta alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, A.; Foong, J.; Brudno, M.; Dubchak, I. Genomevista—An integrated software package for whole-genome alignment and visualization. Bioinformatics 2014, 30, 2654–2655. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Doron-Faigenboim, A.; Erez, E.; Martz, E.; Bacharach, E.; Pupko, T. Selecton 2007: Advanced models for detecting positive and purifying selection using a bayesian inference approach. Nucleic Acids Res. 2007, 35, W506–W511. [Google Scholar] [CrossRef] [PubMed]
- Kelchner, S.A. The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann. Mo. Bot. Gard. 2000, 87, 482–498. [Google Scholar] [CrossRef]
- Twyford, A.D.; Ness, R.W. Strategies for complete plastid genome sequencing. Mol. Ecol. Resour. 2017, 17, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The complete plastome sequences of four orchid species: Insights into the evolution of the orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 2017, 8, 715. [Google Scholar] [CrossRef] [PubMed]
- Weissensteiner, M.H.; Pang, A.W.C.; Bunikis, I.; Höijer, I.; Vinnere-Petterson, O.; Suh, A.; Wolf, J.B.W. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res. 2017, 27, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Rongwen, J.; Akkaya, M.; Bhagwat, A.; Lavi, U.; Cregan, P. The use of microsatellite DNA markers for soybean genotype identification. Theor. Appl. Genet. 1995, 90, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.B.; Linares, A.R.; Cavalli-Sforza, L.L.; Feldman, M.W. An evaluation of genetic distances for use with microsatellite loci. Genetics 1995, 139, 463–471. [Google Scholar] [PubMed]
- Wang, H.-L.; Yang, J.; Boykin, L.M.; Zhao, Q.-Y.; Wang, Y.-J.; Liu, S.-S.; Wang, X.-W. Developing conversed microsatellite markers and their implications in evolutionary analysis of the Bemisia tabaci complex. Sci. Rep. 2014, 4, 6351. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, K.; Hodkinson, T.R.; Barth, S. New chloroplast microsatellite markers suitable for assessing genetic diversity of Lolium perenne and other related grass species. Ann. Bot. 2012, 110, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- De Santana Lopes, A.; Pacheco, T.G.; Nimz, T.; do Nascimento Vieira, L.; Guerra, M.P.; Nodari, R.O.; de Souza, E.M.; de Oliveira Pedrosa, F.; Rogalski, M. The complete plastome of macaw palm [Acrocomia aculeata (Jacq.) Lodd. ex Mart.] and extensive molecular analyses of the evolution of plastid genes in Arecaceae. Planta 2018, 247, 1011–1030. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Andrew, R.L.; Rieseberg, L.H. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 2014, 23, 4899–4911. [Google Scholar] [CrossRef] [PubMed]
- Piot, A.; Hackel, J.; Christin, P.-A.; Besnard, G. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 2018, 247, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-M.; Wang, J.; Feng, L.; Liu, S.; Pang, H.; Qi, L.; Li, J.; Sun, Y.; Qiao, W.; Zhang, L.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.; Clements, M.A.; Arroyo, M.T.; Leebens-Mack, J. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B 2015, 282, 20151553. [Google Scholar] [CrossRef] [PubMed]
- Kode, V.; Mudd, E.A.; Iamtham, S.; Day, A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005, 44, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Jarret, R.L. DNA barcoding in a crop genebank: The Capsicum annuum species complex. Open Biol. J. 2008, 1, 35–42. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).