Abstract
Background: The family Buprestidae is one of the largest families in Coleoptera; however, the number of reported mitochondrial genomes for this family is limited. Methods: In this study, mitogenomes of Chrysobothris violacea, C. shirakii, Buprestis fairmairei, and Phaenops yin were sequenced, assembled, and annotated. The mitogenomes of Chrysobothris, Phaenops, and Buprestis are reported for the first time. Results: The mitogenomes of Chrysobothris violacea, C. shirakii, and Phaenops yin are complete, while the mitogenome of Buprestis fairmairei is partial, lacking trnV and 12S genes. The AT-skew of these four mitogenomes is positive (0.02–0.09). Among the protein-coding genes, the Ka/Ks ratio for cox1 is the lowest (0.05), and the nucleotide diversity for nd6 is the highest. Conclusions: The phylogenetic trees based on mitogenome sequences suggest that the target genus Chrysobothris is sister to Phaenops, and the target genus Buprestis is sister to (Melanophila + (Chrysobothris + Phaenops)) clade. The results of this study will provide mitogenomic data for further research on the mitogenome and phylogeny of Buprestidae.
1. Introduction
The Buprestidae is one of the largest families in Coleoptera, comprising six subfamilies and more than 15,000 species worldwide [1]. All the adult forms of Buprestidae are phytophagous insects, except Xyroscelis crocata (Gory & Laporte, 1839) feeding on the sap of Macrozamia communis [2]. Some species, such as Agrilus planipennis (Fairmaire, 1888), Agrilus mali (Matsumura, 1924), Lamprodila festiva (Linnaeus, 1767), and Melanophila acuminata (DeGeer, 1774), are known as forestry pests or invasive species [3,4,5,6,7]. Buprestid beetles are very widely distributed and can be found in almost all terrestrial and insular habitats where green plants exist. Generally, species with wood-boring habits are primarily distributed in temperate and subtropical regions, while those with leaf-mining habits are more common in moist tropical regions [8].
Although the classification system proposed by Bellamy [1] is widely used, high-level phylogenetic relationships from that system have come into question, including the monophyly of Buprestinae and Chrysochroinae [9]. The mitochondrial genome is an important molecular marker in animals. With the rapid development of sequencing technologies, the extraction, sequencing, and assembly of mitogenome have become much easier. To date, mitogenomes have been widely used for phylogenetic studies of insects [10,11,12,13]. The insect mitogenome, a double-stranded and closed molecule, contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a control region (CR) also called A + T-rich region [14]. The length of the complete mitogenome of Buprestidae ranges from 15 to 16 kbp [15]. Although 31 mitogenomes have been sequenced and annotated in Buprestidae, those of Buprestinae remain limited.
In the present study, the mitogenomes of Chrysobothris violacea (Kerremans, 1892), Chrysobothris shirakii (Miwa & Chûjô, 1935), Buprestis fairmairei (Théry, 1910), and Phaenops yin (Kubáň & Bílý, 2009) were sequenced, assembled, and annotated. The base composition, gene rearrangement, relative synonymous codon usage (RSCU), nucleotide diversity (Pi), and ratio of non-synonymous substitutions (Ka) to synonymous substitutions (Ks) were analyzed and compared. The phylogenetic relationships of subfamilies in Buprestidae were studied based on the mitogenomic data, including four new mitogenome sequences provided in this study.
2. Materials and Methods
2.1. Sampling and DNA Extraction
The specimen of Chrysobothris violacea was collected from Xinhua Village, Weideng Township, Weixi County, Yunnan Province, China on 1 August 2023. The specimens of Chrysobothris shirakii were collected from Yangjiayu, Yuguan Township, Qinhuangdao City, Hebei Province, China on 6 May 2024. The specimens of Buprestis fairmairei were collected from Baijixun Township, Weixi County, Yunnan Province, China on 29 Jun 2021. The specimens of Phaenops yin were collected from Dapingshan, Mianning City, Sichuan Province, China on 22 July 2023. The adults of Phaenops yin were discovered on the wood of burn areas, while some larvae were found under the bark. Specimens of adult form were stored in 95% alcohol and then stored at −20 °C when returned to the laboratory. All these specimens were identified based on the original descriptions [16,17,18,19]. In order to ensure sequencing quality, specimens were sequenced as soon as possible after collection and identification. All the studied specimens were deposited in Museum of China West Normal University. The tissues of thorax and legs were used for total DNA extraction, following the protocol of the Ezup Column Animal Genomic DNA Purification Kit (Shanghai, China). The Illumina libraries were constructed with an average insert size of 350 bp and then sequenced using Illumina platform with a short paired-end strategy (150 bp).
2.2. Mitogenome Assembly, Annotation, and Analysis
To obtain the target reads, Trimmomatic v. 0.36 [20] was used to remove low-quality bases and adapter sequences from the raw data. Subsequently, IDBA-UD v. 1.1.1 [21] was employed to assemble the target reads. The identification of open reading frames led to the discovery of 13 PCG genes. Additionally, the analysis of tRNA gene boundaries facilitated the recognition of two rRNA genes, as well as the A + T-rich region. Finally, the contigs were assembly using Geneious v. 11.0.2 [22]. The mitogenomic maps were drawn using Organellar Genome DRAW [23].
The mitogenomic annotation was conducted using online tool Mitos WebServer [24]. The predicted secondary structures of 22 tRNA genes were visualized using VARNA v. 3.9 [25] based on the result of annotation completed by Mitos. The base composition of mitogenome sequences and the relative synonymous codon usage (RSCU) of 13 PCG genes were calculated and analyzed using MEGA v. 10.0.2 [26]. The AT-skew and GC-skew of four mitogenome sequences were calculated based on the formula proposed by Perna and Kocher [27]. For 13 PCG genes, the values of nucleotide diversity (Pi), nonsynonymous substitution (Ka), synonymous substitution (Ks), and Ka/Ks were calculated using DnaSP v. 5 [28]. In these calculations, the mitogenome of Chalcophora japonica (Gory, 1840) was used as the reference.
2.3. Phylogenetic Analyses
A total of 35 species (Table 1) were used for phylogenetic analyses of Buprestidae, including four mitogenome sequences provided in the present study. Both maximum-likelihood (ML) and Bayesian inference (BI) trees were reconstructed using two data sets (PCGs and PCGs + rRNAs). The sequences of PCGs and rRNAs were aligned using MAFFT v. 7.313 and trimmed using trimAl v. 1.2 [29], which are integrated in PhyloSuite v. 1.2.2 [30]. The sequences of each species were concatenated using ‘concatenate sequence’ integrated in PhyloSuite. The best-fit models of the ML and BI methods were computed using ModelFinder [31]. The ML and BI trees were reconstructed using IQ-TREE v. 1.6.8 [32] and MrBayes v. 3.2.6 [33], respectively. The ML analyses were conducted with the following parameters: the number of bootstraps: 5000; replicates: 1000; and minimum correlation coefficient: 0.9. The BI analyses were performed with the following parameters: 5,000,000 generations, sampling every 100th generation, 2 independent runs, and a burn-in fraction of 0.25. In ML analysis, the nodal support was estimated through 2000 ultrafast bootstrap resampling. The Markov chain Monte Carlo chains were executed independently after excluding constant sites from the sequence alignment. The analyses were terminated when both runs achieved satisfactory convergence. A consensus tree was generated from the post-burn-in trees after discarding the first 25% of sampled trees from each run. The original trees were edited and visualized using Figtree v. 1.4.3.

Table 1.
Information on the Buprestidae species and outgroup taxa used for phylogeny.
Table 1.
Information on the Buprestidae species and outgroup taxa used for phylogeny.
| Taxa | Accession No. | Genome Size (bp) | A + T% | AT-Skew | References |
|---|---|---|---|---|---|
| Agrilus sichuanus Jendek, 2011 | OK189519 | 16,521 | 71.73 | 0.12 | [34] |
| Agrilus adelphinus Kerremans, 1895 | NC071932 | 15,732 | 71.35 | 0.10 | [35] |
| Agrilus mali Matsumura, 1924 | MN894890 | 16,204 | 74.46 | 0.08 | [36] |
| Agrilus planipennis Fairmaire, 1888 | KT363854 | 15,942 | 71.90 | 0.12 | [37] |
| Agrilus discalis Saunders, 1873 | ON644870 | 15,784 | 74.59 | 0.11 | [38] |
| Agrilus zanthoxylumi Li, 1989 | OQ197496 | 16,320 | 74.70 | 0.08 | [39] |
| Coraebus diminutus Gebhardt, 1928 | OK189521 | 15,499 | 68.42 | 0.12 | [34] |
| Coraebus cloueti Théry, 1895 | OK189520 | 15,514 | 69.27 | 0.11 | [34] |
| Coraebus cavifrons Descarpentries and Villier, 1967 | MK913589 | 15,686 | 69.79 | 0.12 | [40] |
| Meliboeus sinae Obenberger, 1935 | OK189522 | 16,108 | 72.42 | 0.11 | [34] |
| Sambus femoralis Kerremans, 1892 | OK349489 | 15,367 | 73.23 | 0.12 | [34] |
| Sambus kanssuensis Ganglbauer, 1890 | OQ784265 | 15,411 | 72.4 | 0.10 | [38] |
| Habroloma sp. | OQ784266 | 16,273 | 73.99 | 0.11 | [38] |
| Endelus continentalis Obenberger, 1944 | OL702762 | 16,246 | 75.60 | 0.13 | [38] |
| Cantonius szechuanensis Obenberger, 1958 | OQ784264 | 15,927 | 73.09 | 0.11 | [38] |
| Trachys auricollis Saunder, 1873 | MH638286 | 16,429 | 71.05 | 0.10 | [41] |
| Trachys troglodytiformis Obenberger, 1918 | KX087357 | 16,316 | 74.62 | 0.10 | Unpublished |
| Trachys variolaris Saunders, 1873 | MN178497 | 16,771 | 72.11 | 0.11 | [42] |
| Catoxantha luodiana (Yang and Xie, 1993) | PP211020 | 15,594 | 68.68 | 0.12 | [43] |
| Anthaxia chinensis Kerremans, 1989 | MW929326 | 15,881 | 73.61 | 0.09 | [44] |
| Coomaniella dentata Song, 2021 | OL694144 | 16,179 | 76.59 | 0.01 | [45] |
| Coomaniella copipes Jendek and Pham, 2013 | OL694145 | 16,196 | 74.47 | 0.03 | [45] |
| Melanophila acuminata (De Geer, 1774) | MW287594 | 15,853 | 75.66 | 0.02 | [46] |
| Nipponobuprestis guangxiensis Peng, 1995 | PP133641 | 15,775 | 65.33 | 0.14 | [43] |
| Chalcophora japonica (Gory, 1840) | OP388437 | 15,759 | 67.97 | 0.13 | [15] |
| Chrysochroa fulgidissima (Schönherr, 1817) | EU826485 | 15,592 | 69.92 | 0.15 | [47] |
| Dicerca corrugata Fairmaire, 1902 | OL753086 | 16,276 | 71.76 | 0.09 | [45] |
| Chrysochroa opulenta (Gory, 1832) | PP211021 | 15,587 | 67.16 | 0.16 | [43] |
| Acmaeodera sp. | FJ613420 | 16,217 | 68.41 | 0.11 | [48] |
| Ptosima chinensis Marseul, 1867 | OP388449 | 16,115 | 67.00 | 0.13 | [15] |
| Julodis variolaris (Pallas, 1771) | OP390084 | 16,227 | 70.43 | 0.12 | [15] |
| Chrysobothris shirakii Miwa and Chûjô, 1935 | PV339621 | 15,789 | 78.54 | 0.02 | This study |
| Chrysobothris violacea Kerremans, 1892 | PV339622 | 15,961 | 79.29 | 0.02 | This study |
| Phaenops yin Kubáň and Bílý, 2009 | PV339623 | 16,051 | 76.51 | 0.04 | This study |
| Buprestisfairmairei Théry, 1910 | PV339624 | 13,390 | 73.42 | 0.09 | This study |
| Dryops ernesti Gozis, 1886 | KX035147 | 15,672 | 72.98 | 0.07 | Unpublished |
| Heterocerus parallelus Gebler, 1830 | KX087297 | 15,845 | 74.03 | 0.13 | Unpublished |
3. Results
3.1. Genome Organization and Base Composition
This study sequenced, assembled, and annotated the mitogenomes of Chrysobothris shirakii (GenBank No. PV339621), Chrysobothris violacea (No. PV339622), Phaenops yin (No. PV339623), and Buprestis fairmairei (No. PV339624). The average read coverage of Chrysobothris shirakii, C. violacea, Phaenops yin, and Buprestis fairmairei are 116.4 X, 82.2 X, 211.9 X, and 56 X, respectively. The number of paired-end reads is 15,220,144 (C. shirakii); 13,640,446 (C. violacea); 22,951,006 (P. yin); and 16,068,162, respectively. The lengths of these mitogenome sequences range from 15,789 bp to 16,051 bp, with the mitogenome of Buprestis fairmairei being 13,390 bp in size and encoding 35 genes, lacking the trnV and 12S genes (Table 2). Except for the partial mitogenome of Buprestis fairmairei, the mitogenomes of the other three species are complete, circular, and double-stranded molecules. Each mitogenome contains 37 genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and an A+T-rich region (control region, CR). Among the 37 genes, 14 (including nad1, nad4L, nad4, nad5, trnQ, trnV, trnL1, trnP, trnH, trnF, trnY, trnC, rrnL, and rrnS) genes are encoded on the N-strand, while the remaining 23 genes are encoded on the J-strand (Figures S1–S4).
These four mitogenome sequences all exhibit a high A + T bias, with Chrysobothris shirakii and C. violacea having an A + T content of 79.29%, higher than that of Phaenops yin (76.51%) and Buprestis fairmairei (73.42%). The AT-skew of these four mitogenomes is positive (0.02–0.09), while the GC-skew is negative (−0.2–−0.1). The gene intergenic spacers range from 1 to 30 bp in length, with the longest spacer located between the trnC and trnY of Phaenops yin. There are a total of 50 overlapping regions, with lengths ranging from 1 to 8 bp.

Table 2.
The mitogenomes of Chrysobothris shirakii, Chrysobothris violacea, Phaenops yin, and Buprestis fairmairei.
Table 2.
The mitogenomes of Chrysobothris shirakii, Chrysobothris violacea, Phaenops yin, and Buprestis fairmairei.
| Gene | Strand | Position From | To | Start Codons | Stop Condons | Intergenic Nucleotides |
|---|---|---|---|---|---|---|
| trnI | J | 1/1/1/1 | 65/65/66/64 | −3/−3/−3/−1 | ||
| trnQ | N | 63/63/64/64 | 131/131/132/132 | −1/0/−1/−1 | ||
| trnM | J | 131/132/132/132 | 199/200/200/199 | 0/0/0/0 | ||
| nad2 | J | 200/201/201/200 | 1225/1226/1223/1225 | ATT/ATT/ATT/ATT | TAA/TAA/TAA/TAA | 8/8/3/8 |
| trnW | J | 1234/1235/1227/1234 | 1301/1303/1296/1300 | −8/−8/−8/−8 | ||
| trnC | N | 1294/1296/1289/1293 | 1357/1357/1356/1354 | 7/0/30/0 | ||
| trnY | N | 1365/1358/1387/1355 | 1429/1421/1452/1419 | 1/1/1/1 | ||
| cox1 | J | 1431/1423/1454/1421 | 2961/2953/2987/2951 | */*/* | T(AA)/T(AA)/T(AA)/T(AA) | 0/0/0/0 |
| trnL2 | J | 2962/2954/2988/2952 | 3026/3018/3054/3016 | 0/0/0/0 | ||
| cox2 | J | 3027/3019/3055/3017 | 3711/3703/3744/3698 | ATA/ATA/ATA/ATA | T(AA)/T(AA)/TAA/T(AA) | 0/0/4/0 |
| trnK | J | 3712/3704/3749/3699 | 3781/3774/3818/3768 | 0/-−/0/0 | ||
| trnD | J | 3782/3774/3819/3769 | 3847/3838/3886/3833 | 0/0/0/0 | ||
| atp8 | J | 3848/3839/3887/3834 | 4003/3994/4048/3989 | ATT/ATA/ATT/ATA | TAA/TAA/TAA/TAA | −7/−7/−7/−7 |
| atp6 | J | 3997/3988/4042/3983 | 4671/4662/4716/4657 | ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA | −1/−1/−1/−1 |
| cox3 | J | 4671/4662/4716/4657 | 5454/5445/5504/5445 | ATG/ATG/ATG/ATG | T(AA)/T(AA)/TAA/TAA | 0/0/3/−1 |
| trnG | J | 5455/5446/5508/5445 | 5518/5508/5572/5508 | 0/0/0/0 | ||
| nad3 | J | 5519/5509/5573/5509 | 5872/5862/5926/5862 | ATT/ATT/ATT/ATT | TAG/TAG/TAA/TAG | −2/−2/2/−2 |
| trnA | J | 5871/5861/5929/5861 | 5933/5922/5993/5923 | −1/−1/2/−1 | ||
| trnR | J | 5933/5922/5996/5923 | 5999/5988/6061/5988 | −3/−3/−1/−1 | ||
| trnN | J | 5997/5986/6061/5988 | 6062/6051/6127/6051 | 0/0/0/0 | ||
| trnS1 | J | 6063/6052/6128/6052 | 6130/6118/6194/6118 | 0/0/0/0 | ||
| trnE | J | 6131/6119/6195/6119 | 6196/6183/6259/6182 | −2/−2/−2/−2 | ||
| trnF | N | 6195/6182/6258/6181 | 6258/6245/6322/6247 | 0/0/0/−1 | ||
| nad5 | N | 6259/6246/6323/6247 | 7981/7968/8045/7965 | ATT/ATT/ATT/ATA | T(AA)/T(AA)/T(AA)/TAA | 0/0/0/0 |
| trnH | N | 7982/7969/8046/7966 | 8045/8033/8109/8029 | 0/0/0/0 | ||
| nad4 | N | 8046/8034/8110/8030 | 9381/9369/9445/9365 | ATG/ATG/ATG/ATG | T(AA)/T(AA)/T(AA)/T(AA) | −7/−7/−7/−7 |
| nad4L | N | 9375/9363/9439/9359 | 9665/9653/9729/9649 | ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAG | 2/2/2/2 |
| trnT | J | 9668/9656/9732/9652 | 9733/9719/9795/9715 | −1/−1/0/0 | ||
| trnP | N | 9733/9719/9796/9716 | 9797/9783/9861/9782 | 1/1/2/1 | ||
| nad6 | J | 9799/9785/9864/9784 | 10,302/10,288/10,370/10,293 | ATT/ATA/ATA/ATC | TAA/TAA/TAA/TAA | −1/−1/−1/1 |
| cytb | J | 10,302/10,288/10,370/10,293 | 11,441/11,427/11,512/11,435 | ATG/ATG/ATG/ATG | TAG/TAA/TAG/TAG | −2/−2/−2/−2 |
| trnS2 | J | 11,440/11,426/11,511/11,434 | 11,507/11,492/11,578/11,501 | 20/27/26/28 | ||
| nad1 | N | 11,528/11,520/11,605/11,530 | 12,481/12,473/12,555/12,480 | TTG/TTG/TTG/TTG | TAA/TAA/TAA/TAA | 1/1/1/1 |
| trnL1 | N | 12,483/12,475/12,557/12,482 | 12,547/12,539/12,621/12,545 | 0/0/0/0 | ||
| rrnL | N | 12,548/12,540/12,622/12,546 | 13,847/13,844/13,931/13,390 | 0/0/0/ | ||
| trnV | N | 13,848/13,845/13,932/* | 13,917/13,914/14,001/* | 0/0/0/ | ||
| rrnS | N | 13,918/13,915/14,002/* | 14,652/14,699/14,780/* | 0/0/0/ | ||
| CR | 14,653/14,700/14,781/* | 15,798/15,961/16,051/* |
The order of four mitogenomes is Chrysobothris shirakii, C. violacea, Phaenops yin, and Buprestis fairmairei. * represents missing data.
3.2. Protein-Coding Genes, Codon Usage, and Nucleotide Diversity
The sequence lengths of the PCGs of Chrysobothris shirakii and C. violacea are both 11,159 bp, encoding 3710 amino acid residues; the sequence length of the PCGs in Phaenops yin is 11,171 bp, encoding 3715 amino acid residues; and the sequence length of Buprestis fairmairei is 11,163 bp, encoding 3710 amino acid residues. Among them, the PCGs length and amino acid residue count of Phaenops yin are slightly higher than those of the other three species.
Except for the initiation codon of the nad1 gene, which is TTG, the initiation codons of most PCGs are ATN. The initiation codon of the cox1 gene could not be determined and may use a non-canonical initiation codon. For 13 PCGs, there are three types of termination codons: TAA, TAG, and T, with the single T as a termination codon being complemented by the 3′A residue on the mRNA. In these four mitogenome, L1, F, and I are three most frequently used amino acids (Figure 1).
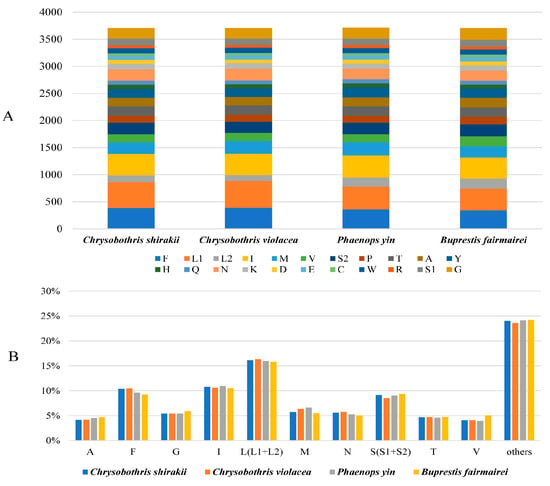
Figure 1.
Numbers of different amino acids in the four new mitogenome sequences (A) and the percentages of the top eleven amino acids (B). The stop codons are not included in PCGs.
A comparative analysis of the Ka/Ks ratios (Figure 2) of the four mitogenomes showed that the Ka/Ks value of the PCGs are all less than 1, indicating that these genes are under purifying selection. Among them, the ratio for cox1 is the lowest (0.05). The Pi values of the PCGs in these four mitogenomes range from 0.149 (cox1) to 0.291 (nd6), with the highest nucleotide diversity level found in the nd6 gene (Pi = 0.291), followed by atp8 (Pi = 0.266).
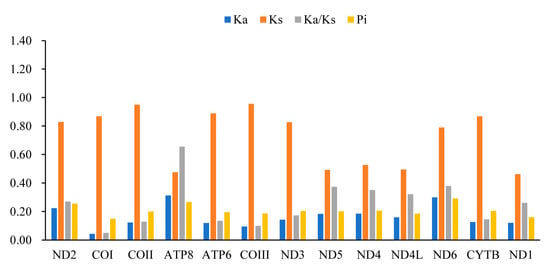
Figure 2.
The Ka/Ks and Pi of PCGs in the four newly sequenced Buprestidinae mitogenomes. Pi: the values of nucleotide diversity; Ka: nonsynonymous substitution; and Ks: synonymous substitution.
A comparative analysis of the codon usage frequencies (RSCU) for these four species showed that the most frequently used codons are UUU, UUA, and AUU (Figure 3).
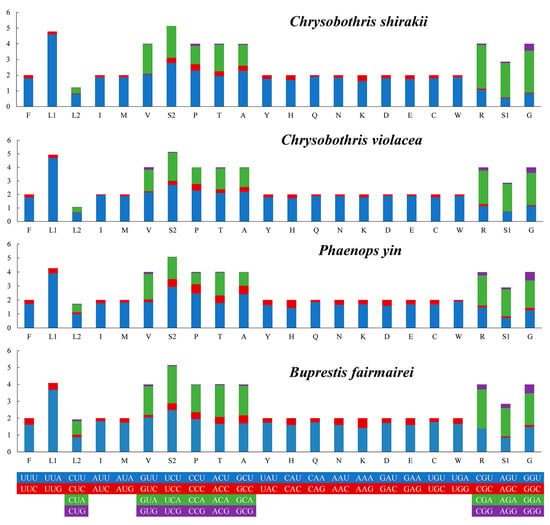
Figure 3.
Relative synonymous codon usage (RSCU) of the four new mitogenome sequences. The codon families are displayed on the x-axis, with the codon usage frequency represented on the y-axis.
3.3. Ribosomal and Transfer RNA Genes
Two rRNA genes are located between the CR region and trnL1, separated by trnV. The sequence length of the rRNA in the mitogenomes of the three species (Chrysobothris shirakii, C. violacea, and Phaenops yin) ranges from 2035 to 2090 bp, while the 12S is missing in Buprestis fairmairei. The 16S rRNA length in these four mitogenomes ranges from 845 to 1305 bp. The A + T content of the rRNAs in the three mitogenomes (Chrysobothris shirakii, C. violacea, and Phaenops yin) ranges from 79.85% (Phaenops yin) to 82.63% (C. violacea).
The tRNA sequence lengths in the four mitogenomes range from 1377 bp (Chrysobothris violacea) to 1457 bp (C. shirakii), with individual gene lengths varying from 62 bp (trnA) to 71 bp (trnK). Eight tRNA genes are located on the N strand, while the remaining 14 genes are located on the J strand. Among the 22 tRNA genes, except for trnS1, which lacks the DHU arm and cannot form a cloverleaf structure, the other 21 tRNA genes are able to form typical cloverleaf structures (Figures S5–S8).
3.4. Phylogenetic Relationships
Based on the mitogenomes of the four species in this study and 32 previously known mitogenomes of Buprestidae, phylogenetic trees (ML and BI trees) of the Buprestidae family were constructed using the concatenated sequences of PCGs + RNAs. The results showed that the BI and ML trees have the same topological structure, with the phylogenetic relationships of the subfamilies as follows: (Chrysochroniae + ((Julodinae + Polycestinae) + Buprestinae) + Agrilinae) (Figure 4 and Figure 5). The four target species cluster together with other species of Buprestinae, with high support values (ML = 98, BI = 1). In Buprestinae, the phylogenetic relationships of the genus are as follows: ((Dicerca + Coomaniella) + (Buprestis + (Anthaxia + (Melanophila + (Chrysobothris + Phaenops)))). For the target genera, Chrysobothris is sister to Phaenops, and Buprestis is sister to (Melanophila + (Chrysobothris + Phaenops)) clade.
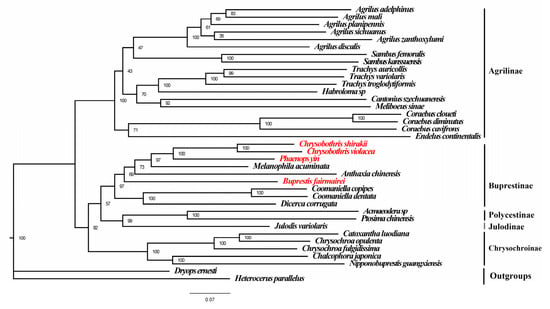
Figure 4.
Maximum likelihood tree of Buprestidae based on PCGs + rRNAs. Values at nodes are ML bootstrap values. Red names represent the mitogenome data provided in this study.
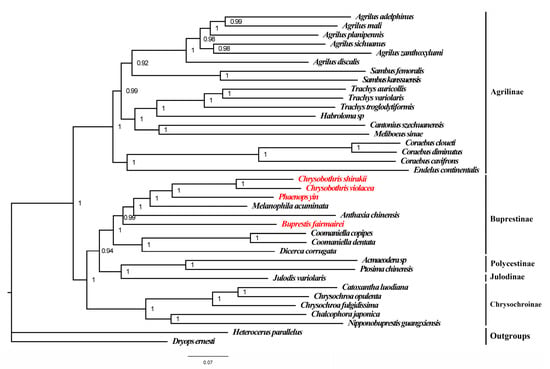
Figure 5.
Bayesian tree of Buprestidae based on PCGs + rRNAs. Values at nodes are posterior probability. Red words are the target species provided in this study. Red names represent the mitogenome data provided in this study.
4. Discussion
Compared to the known species of Buprestidae [1], the number of reported mitogenomes is very limited. Currently, only 35 mitogenomes (including four mitogenomes provided in this study) have been reported [43]. In this study, the three complete mitogenomes contain 37 typical genes and a CR region, and the partial mitogenome of Buprestis fairmairei. Currently, the mitogenome composition of Buprestidae is consistent with that of other insects [10,11,12,13,49]. All four mitogenomes exhibit a high AT-skew. The gene order of these four mitogenomes is also consistent, aligning with the results from other species of the Buprestidae. In PCGs, the Ka/Ks of cox1 is the lowest, suggesting that a large number of synonymous mutations have occurred in this gene, while cox1 exhibits the highest level of conservation. Based on the most frequently used codons (UUU, UUA, and AUU), it can be inferred that the mitogenome exhibits a significant AT bias, which is consistent with previous studies [36,37,39–41]. Although gene rearrangements have been reported in some insects [50], this phenomenon has not been observed within Buprestidae to date.
Although different taxonomists have established various classification systems, research on the phylogeny of higher taxa within the Buprestidae is relatively scarce [9,51,52]. Firstly, the morphological characteristics were used to construct phylogenetic relationships among subfamilies [52]; however, the results of that study were severely inconsistent with the current classification system of Buprestidae, which is why it is seldom mentioned. Then, morphological characteristics were used to investigate the phylogenetic relationships among genera within Chrysochroina [53]. Subsequently, four gene fragments from 137 species [9] and their results supported the monophyly of Agrilinae, Julodinae, Galbellinae, and Polycestinae but did not support the monophyly of Chrysochroinae and Buprestinae. The relationships among tribes within Agrilinae are also unresolved. In recent years, mitogenome-based phylogenetic analysis has also been applied to Buprestidae [36,39,40,41,43], and the study results indicate that the phylogenetic relationships among subfamilies are relatively stable. The phylogenetic relationship of subfamilies is (Chrysochroniae + ((Julodinae + Polycestinae) + Buprestinae) + Agrilinae), which is largely consistent with mitogenome-based phylogenetic trees. In Buprestinae clade, Coomaniella is close to Dicerca, which is consistent with previous studies [9,45]. The phylogenetic tree of this study is consistent with recent research findings, possibly because both used mitochondrial genome data and the species used are largely the same. However, this result is inconsistent with the topology of the phylogenetic tree constructed based on gene fragments [9], which may be due to differences in the species and molecular data used in previous studies.
In this study, the mitogenomes of the genera Buprestis, Chrysobothris, and Phaenops are reported for the first time. The results indicate that Chrysobothris is close to Phaenops, and Buprestis is close to the clade composed of Melanophila, Chrysobothris, and Phaenops. Relying solely on morphological traits or molecular data to study phylogenetic issues is unreliable. These phylogenetic questions will be addressed based more mitogenome or genome data combined with morphological characteristics in the future studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes16070828/s1, Figure S1. The mitogenome maps of Chrysobothris shirakii. Figure S2. The mitogenome maps of Chrysobothris violacea. Figure S3. The mitogenome maps of Phaenops yin. Figure S4. The mitogenome maps of Buprestis fairmairei. Figure S5. The secondary cloverleaf structure for the tRNA of Chrysobothris shirakii. Figure S6. The secondary cloverleaf structure for the tRNA of Chrysobothris violacea. Figure S7. The secondary cloverleaf structure for the tRNA of Phaenops yin. Figure S8. The secondary cloverleaf structure for the tRNA of Buprestis fairmairei.
Author Contributions
Conceptualization: Z.W. and A.S. Data curation: Y.L., B.O., and J.W. Formal analysis: Y.L. and Z.W. Funding acquisition: Z.W. Software: J.W. and B.O. Writing—original draft: Y.L. and Z.W. Writing—review and editing: Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was founded by the Sichuan Provincial Natural Science Foundation (2024NSFSC0076; 2025LHJJ0100).
Institutional Review Board Statement
No ethical statement was reported.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete mitogenome sequences are available in NCBI (PV339621, PV339622, PV339623, and PV339624).
Acknowledgments
We thank the collectors for their hard work in field collection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bellamy, C.L. A World Catalogue and Bibliography of the Jewel Beetles (Coleoptera: Buprestoidea), Pensoft Series Faunistica; Pensoft Publishers: Sofia, Bulgaria; Moscow, Russia, 2008; Volume 1–4, pp. 1–2684. [Google Scholar]
- Bellamy, C.L. Phylogenetic relationships of Xyroscelis (Coleoptera: Buprestidae). Invertebr. Syst. 1997, 11, 569–574. [Google Scholar] [CrossRef]
- Barr, W.F.; Linsley, E.G. Distributional and biological notes on the species of the subgenus Melanophila occurring in western North America (Coleoptera: Buprestidae). Pan-Pac. Entomol. 1947, 23, 162–166. [Google Scholar]
- Bozorov, A.T.; Luo, Z.H.; Li, X.S.; Zhang, D.Y. Agrilus mali Matsumara (Coleoptera: Buprestidae), a new invasive pest of wild apple in western China: DNA barcoding and life cycle. Ecol. Evol. 2019, 9, 1169–1172. [Google Scholar] [CrossRef]
- Bunescu, H.; Florian, T. The jewel beetle Lamprodila (Palmar) festiva Linné, 1767, a new invasive urban pest of Cupressaceae in Cluj area (Romania) (Coleoptera: Buprestidae). Fragm. entomol. 2019, 51, 241–246. [Google Scholar] [CrossRef]
- Chamorro, M.L.; Volkovitsh, M.G.; Poland, T.M.; Haack, R.A.; Lingafelter, S.W. Preimaginal stages of the emerald ash borer, Agrilus planipennis Fairmaire (Coleoptera: Buprestidae): An invasive pest on Ash trees (Fraxinus). PLoS ONE 2012, 7, e33185. [Google Scholar] [CrossRef]
- Orlova-Bienkowskaja, M.J.; Volkovitsh, M.G. Range expansion of Agrilus convexicollis in European Russia expedited by the invasion of the emerald ash borer, Agrilus planipennis (Coleoptera: Buprestidae). Biol. Invasions 2015, 17, 537–544. [Google Scholar] [CrossRef]
- Bellamy, C.L. The stunning world of jewel beetles. Wings Essays Invertebr. Conserv. 2003, 26, 13–17. [Google Scholar]
- Evans, A.M.; Mckenna, D.D.; Bellamy, C.L.; Farrell, B.D. Large-scale molecular phylogeny of metallic wood-boring beetles (Coleoptera: Buprestoidea) provides new insights into relationships and reveals multiple evolutionary origins of the larval leaf-mining habit. Syst. Entomol. 2015, 40, 385–400. [Google Scholar] [CrossRef]
- Xu, S.; Wu, Y.; Liu, Y.; Zhao, P.; Chen, Z.; Song, F.; Li, H.; Cai, W. Comparative mitogenomics and phylogenetic analyses of Pentatomoidea (Hemiptera: Heteroptera). Genes 2021, 12, 1306. [Google Scholar] [CrossRef]
- Wang, J.J.; Yang, M.F.; Dai, R.H.; Li, H.; Wang, X.Y. Characterization and phylogenetic implications of the complete mitochondrial genome of Idiocerinae (Hemiptera: Cicadellidae). Int. J. Biol. Macromol. 2018, 120, 2366–2372. [Google Scholar] [CrossRef]
- Yang, M.S.; Song, L.; Zhou, L.; Shi, Y.X.; Song, N.; Zhang, Y.L. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wen, Q.; Wang, T.F.; Ran, F.R.; Wang, M.; Fan, X.L.; Wei, S.J.; Li, Z.H.; Tan, J.L. Next-generation sequencing of four mitochondrial genomes of Dolichovespula (Hymenoptera: Vespidae) with a phylogenetic analysis and divergence time estimation of Vespidae. Animals 2022, 12, 3004. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.H.; Huang, X.Y.; Shi, A.M. First mitochondrial genome of subfamily Julodinae (Coleoptera, Buprestidae) with its phylogenetic implications. ZooKeys 2023, 1139, 165–182. [Google Scholar] [CrossRef]
- Abeille de Perrin, E. Rectification sur deux buprestides. Bull. Soc. Entomol. Fr. 1894, cxxx–cxxxix. [Google Scholar]
- Kubáň, V.; Bílý, S. Phaenops yin sp. nov. and P. yang sp. nov. from China (Coleoptera: Buprestidae:Buprestinae: Melanophilini). Folia Heyrovskyana, Series A 2009, 17, 111–125. [Google Scholar]
- Eschscholtz, J.F.; Rathke, H.; Predpriaetie (Ship). Zoologischer Atlas: Enthaltend Abbildungen Und Beschreibungen Neuer Thierarten, Während Des Flottcapitains v. Kotzebue Zweiter Reise um Die Welt, Auf der Russisch-Kaiserlichen Kriegsschlupp Predpriaetië in Den Jahren 1823–1826; Erstes Heft.; G. Reimer: Berlin, Germany, 1829; pp. 1–17. [Google Scholar]
- Peng, Z.L.; Shi, A.M.; Xu, H.X. An Iconography of Chinese Buprestidae (Coleoptera); The Straits Publishing House: Fuzhou, China, 2021; pp. 1–860. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.J.T.N.; Gimmel, M.L.; Narayanan Kutty, S.; Cockerill, T.D.; Khen, C.V.; Vogler, A.P. Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a bornean rainforest sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic. Acids. Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA second-ary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 3, 353–358. [Google Scholar] [CrossRef]
- Rozas, J.; Sanchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhnam, S.; Largetm, B.; Lium, L.; Suchardm, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Wei, Z.H. The complete mitochondrial genomes of five Agrilinae (Coleoptera, Buprestidae) species and phylogenetic implications. ZooKeys 2022, 1092, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Gan, Y.J.; Wang, L.; Xu, Y.Y.; Wei, Z.H.; Shi, A.M. The larval, pupal and mitogenomic characteristics of Agrilus Adelphinus Kerremans, 1895 (Coleoptera, Buprestidae) from China. Zookeys 2023, 1174, 15–33. [Google Scholar] [CrossRef]
- Sun, H.Q.; Zhao, W.X.; Lin, R.Z.; Zhou, Z.F.; Huai, W.X.; Yao, Y.X. The conserved mitochondrial genome of the jewel beetle (Coleoptera: Buprestidae) and its phylogenetic implications for the suborder Polyphaga. Genomics 2020, 112, 3713–3721. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Quan, G.X.; Mittapalli, O.; Cusson, M.; Krell, P.J.; Doucet, D. The complete mitogenome of the Emerald Ash Borer (EAB), Agrilus planipennis (Insecta: Coleoptera: Buprestidae). Mitochondrial DNA B Resour. 2017, 2, 134–135. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Wei, Z.H.; Lu, J.W.; Shi, A.M. Mitogenomic analysis and phylogenetic relationships of Agrilinae: Insights into the evolutionary patterns of a diverse buprestid subfamily. PLoS ONE 2023, 18, e0291820. [Google Scholar] [CrossRef]
- Li, J.J.; Yan, B.; Song, H.T.; Yang, M.F. The complete mitochondrial genome of Agrilus zanthoxylumi Li, 1989 (Buprestidae: Agrilinae): Genome characterization and phylogenetic implications. J. Entomol. Res. Soc. 2025, 27, 121–138. [Google Scholar] [CrossRef]
- Cao, L.M.; Wang, X.Y. The complete mitochondrial genome of the jewel beetle Coraebus cavifrons (Coleoptera: Buprestidae). Mitochondrial DNA B Resour. 2019, 4, 2407–2408. [Google Scholar] [CrossRef]
- Xiao, L.F.; Zhang, S.D.; Long, C.P.; Guo, Q.Y.; Xu, J.S.; Dai, X.H.; Wang, J.G. Complete mitogenome of a leaf-mining buprestid beetle, Trachys auricollis, and its phylogenetic implications. Genes 2019, 10, 992. [Google Scholar] [CrossRef]
- Cao, L.M.; Wang, X.Y. The complete mitochondrial genome of the jewel beetle Trachys variolaris (Coleoptera: Buprestidae). Mitochondrial DNA B Resour. 2019, 4, 3042–3043. [Google Scholar] [CrossRef]
- Ouyang, B.W.; Huang, X.Y.; Gan, Y.J.; Wei, Z.H.; Shi, A.M. Three mitochondrial genomes of Chrysochroinae (Coleoptera, Buprestidae) and phylogenetic analyses. Genes 2024, 15, 1336. [Google Scholar] [CrossRef]
- Chen, B.; Wei, Z.H.; Shi, A.M. The complete mitochondrial genome of the jewel beetle, Anthaxia chinensis (Coleoptera: Buprestidae). Mitochondrial DNA B Resour. 2021, 6, 2962–2963. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Chen, B.; Wei, Z.H.; Shi, A.M. First report of complete mitochondrial genome in the tribes Coomaniellini and Dicercini (Coleoptera: Buprestidae) and phylogenetic implications. Genes 2022, 13, 1074. [Google Scholar] [CrossRef]
- Peng, X.J.; Liu, J.; Wang, Z.; Zhan, Q.Z. The complete mitochondrial genome of the pyrophi-lous jewel beetle Melanophila acuminata (Coleoptera: Buprestidae). Mitochondrial DNA B Resour. 2021, 6, 1059–1060. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.Y.; Jeong, H.C.; Kim, M.J.; Jeong, H.U.; Lee, S.H.; Kim, I. Complete mitogenome sequence of the jewel beetle, Chrysochroa fulgidissima (Coleoptera: Buprestidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2009, 20, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009, 58, 381–394. [Google Scholar] [CrossRef]
- Bai, Y.; Yang, K.; Ye, L.; Gao, X.Y. Complete Mitogenome and Phylogenetic Analyses of Galerita Orientalis Schmidt-Goebel, 1846 (Insecta: Coleoptera: Carabidae: Galeritini). Genes 2022, 13, 2199. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Vogler, A.P. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol. Phylogenet. Evol. 2012, 63, 299–304. [Google Scholar] [CrossRef]
- Deuve, T.; Cruaud, A.; Genson, G.; Rasplus, J.Y. Molecular systematics and evolutionary history of the genus Carabus (Col. Carabidae). Mol. Phylogenet. Evol. 2012, 65, 259–275. [Google Scholar] [CrossRef]
- Kolibáč, J. Classification and phylogeny of the Buprestoidea (Insecta: Coleoptera). Acta Mus. Moraviae Sci. biol. 2000, 85, 113–184. [Google Scholar]
- Hołyński, R.B. Taxonomic Structure of the Subtribe Chrysochroina Cast with Review of the Genus Chrysochroa Dej. (Coleoptera: Buprestidae); Gondwana: Warszawa, Poland, 2009; p. 379. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).