Results of Chromosomal Microarray Need to Always Be Checked by (Molecular) Cytogenetics—Even If They Seem to Be Simple Deletions
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Molecular Cytogenetics
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CMA | chromosome microarray |
| CNVs | copy number variations |
| DAPI | 4′,6-diamidino-2-phenylindole |
| FISH | fluorescence in situ hybridization |
| MCB | multicolor banding |
| NGS | next-generation sequencing |
| OGM | optical genomic mapping |
| pcp | partial chromosome paint |
| SNPs | single-nucleotide polymorphisms |
| sSMC | small supernumerary marker chromosome |
| wcp | whole chromosome paint |
References
- Wang, Y.; Liehr, T. The need for a concert of cytogenomic methods in chromosomic research and diagnostics. Genes 2025, 16, 533. [Google Scholar] [CrossRef] [PubMed]
- Dharmadhikari, A.V.; Markowitz, A.L.; Han, J.; Estrine, D.B.; Xu, D.; Ma, K.; Fong, C.; Fernandez, B.A.; Deardorff, M.A.; Schmidt, R.J.; et al. Optical genome mapping improves clinical interpretation of constitutional copy number gains and reduces their VUS burden. Genet. Med. 2025, in press. [Google Scholar] [CrossRef]
- Bionano. Available online: https://ir.bionano.com/news-releases/news-release-details/international-consortium-optical-genome-mapping-publishes-expert#:~:text=The%20consensus%20from%20a%20global,tests%20based%20on%20traditional%20methods (accessed on 23 May 2025).
- Coughlin, C.R., 2nd; Scharer, G.H.; Shaikh, T.H. Clinical impact of copy number variation analysis using high-resolution microarray technologies: Advantages, limitations and concerns. Genome Med. 2012, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.W.; Bi, W. Novel applications of array comparative genomic hybridization in molecular diagnostics. Expert Rev. Mol. Diagn. 2018, 18, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Molecular cytogenetics in the era of chromosomics and cytogenomic approaches. Front. Genet. 2021, 12, 720507. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, L.G.; Beaudet, A.L.; Brothman, A.R.; Hirsch, B.; Levy, B.; Martin, C.L.; Mascarello, J.T.; Rao, K.W.; Working Group of the Laboratory Quality Assurance Committee of the American College of Medical Genetics. Microarray analysis for constitutional cytogenetic abnormalities. Genet. Med. 2007, 9, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef] [PubMed]
- 3Billion. Available online: https://3billion.io/blog/microarray-vs-wes (accessed on 23 May 2025).
- Agilent. Available online: https://www.agilent.com/en/solutions/inherited-disease/postnatal?&utm_source=bing&utm_medium=cpc&utm_campaign=B_PS_NBr_HRG_EMEA_B_E_P&utm_term=chromosomal%20microarray&utm_content=Postnatal_P&gclid=09c7c545d1e913fc8014a7d3d94445fe&gclsrc=3p.ds (accessed on 23 May 2025).
- Dr Lalpathlabs. Available online: https://www.lalpathlabs.com/images/our-department/cytogenetics/knowledge-centre/Microarrays.pdf (accessed on 23 May 2025).
- Illumina. Available online: https://www.illumina.com/areas-of-interest/genetic-disease/rare-disease-genomics/cma-constitutional-cytogenetics.html (accessed on 23 May 2025).
- PerkinElmer. Available online: https://perkinelmer-appliedgenomics.com/home/products/molecular-cytogenetics/cytogenetic-microarrays/ (accessed on 23 May 2025).
- Soysal, Y.; Balci, S.; Hekimler, K.; Liehr, T.; Ewers, E.; Schoumans, J.; Bui, T.H.; Içduygu, F.M.; Kosyakova, N.; Imirzalioğlu, N. Characterization of double ring chromosome 4 mosaicism associated with bilateral hip dislocation, cortical dysgenesis, and epilepsy. Am. J. Med. Genet. A 2009, 149, 2782–2787. [Google Scholar] [CrossRef] [PubMed]
- Weise, A.; Mrasek, K.; Fickelscher, I.; Claussen, U.; Cheung, S.W.; Cai, W.W.; Liehr, T.; Kosyakova, N. Molecular definition of high-resolution multicolor banding probes: First within the human DNA sequence anchored FISH banding probe set. J. Histochem. Cytochem. 2008, 56, 487–493. [Google Scholar] [CrossRef] [PubMed]
- BACPAC Resources Center (BPRC). Available online: https://bacpacresources.org (accessed on 23 May 2025).
- Mrasek, K.; Heller, A.; Rubtsov, N.; Trifonov, V.; Starke, H.; Claussen, U.; Liehr, T. Detailed Hylobates lar karyotype defined by 25-color FISH and multicolor banding. Int. J. Mol. Med. 2003, 12, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Claussen, U. Current developments in human molecular cytogenetic techniques. Curr. Mol. Med. 2002, 2, 283–297. [Google Scholar] [CrossRef] [PubMed]
- De Gregori, M.; Pramparo, T.; Memo, L.; Gimelli, G.; Messa, J.; Rocchi, M.; Patricelli, M.G.; Ciccone, R.; Giorda, R.; Zuffardi, O. Direct duplication 12p11.21-p13.31 mediated by segmental duplications: A new recurrent rearrangement? Hum. Genet. 2005, 118, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Eussen, B.H.; van de Laar, I.; Douben, H.; van Kempen, L.; Hochstenbach, R.; De Man, S.A.; Van Opstal, D.; de Klein, A.; Poddighe, P.J. A familial inverted duplication 2q33-q34 identified and delineated by multiple cytogenetic techniques. Eur. J. Med. Genet. 2007, 50, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Neill, N.J.; Ballif, B.C.; Lamb, A.N.; Parikh, S.; Ravnan, J.B.; Schultz, R.A.; Torchia, B.S.; Rosenfeld, J.A.; Shaffer, L.G. Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH. Genome Res. 2011, 21, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Debost-Legrand, A.; Capri, Y.; Gouas, L.; Pebrel-Richard, C.; Veronese, L.; Tchirkov, A.; Haoud, K.; Boespflug-Tanguy, O.; Goumy, C.; Vago, P. De novo unbalanced translocation 2;4 characterized by metaphase CGH and array CGH in a child with mental retardation and dysmorphic features. Pathol. Biol. 2011, 59, 309–313. [Google Scholar] [CrossRef] [PubMed]
- González-Rodríguez, M.T.A.; Brukman-Jiménez, S.A.; Cuero-Quezada, I.; Corona-Rivera, J.R.; Corona-Rivera, A.; Serafín-Saucedo, G.; Aguirre-Salas, L.M.; Bobadilla-Morales, L. Identification of a small supernumerary marker chromosome in a Turner syndrome patient with karyotype mos 46,X,+mar/45,X. Genes 2023, 14, 253. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Yamamoto-Shimojima, K.; Yanagishita, T.; Ondo, Y.; Nishi, E.; Okamoto, N.; Yamamoto, T. Complex chromosomal rearrangements of human chromosome 21 in a patient manifesting clinical features partially overlapped with that of Down syndrome. Hum. Genet. 2020, 139, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- PubMed Searching for Array CGH Unbalanced Insertion Deletion. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=array%20cgh%20unbalanced%20insertion%20deletion&sort=date (accessed on 23 May 2025).
- PubMed Searching for Array CGH Unbalanced Translocation Deletion. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=array+cgh+unbalanced+translocation+deletion&sort=date (accessed on 23 May 2025).
- PubMed Searching for Array CGH Chromoanasynthesis Deletion. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=array+cgh+chromoanasynthesis+deletion&sort=date (accessed on 23 May 2025).
- Quinonez, S.C.; Gelehrter, T.D.; Uhlmann, W.R. A Marfan syndrome-like phenotype caused by a neocentromeric supernumerary ring chromosome 15. Am. J. Med. Genet. A 2017, 173, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Small Supernumerary Marker Chromosomes, Basics; Epubli: Berlin, Germany, 2023. [Google Scholar]
- Conlin, L.K.; Kramer, W.; Hutchinson, A.L.; Li, X.; Riethman, H.; Hakonarson, H.; Mulley, J.C.; Scheffer, I.E.; Berkovic, S.F.; Hosain, S.A.; et al. Molecular analysis of ring chromosome 20 syndrome reveals two distinct groups of patients. J. Med. Genet. 2011, 48, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, J.; van Haeringen, A.; Hansson, K.B.; Lankester, A.; Smit, M.J.; Belfroid, R.D.; Bakker, E.; Rosenberg, C.; Tanke, H.J.; Szuhai, K. Ring chromosome formation as a novel escape mechanism in patients with inverted duplication and terminal deletion. Eur. J. Hum. Genet. 2007, 15, 548–555. [Google Scholar] [CrossRef] [PubMed]

| Source/Company (Alphabetically) | Statement (Cited as Verbatim Quote) | Summary as Statements for CMA | Classification of Statements |
|---|---|---|---|
| 3 billion [9] | Microarray is particularly useful for diagnosing conditions related to large chromosomal abnormalities, such as deletions, duplications, and unbalanced rearrangements. It remains a cost-effective and fast option for identifying known CNVs and SNPs in the genome. | CMA can detect chromosomal aberrations accurately CMA is more cost efficient than other approaches CMA is covering whole genome | No: CMA can detect CNVs with high accuracy, but not the nature of the chromosomal aberration No: Specialists are needed as in (molecular) cytogenetics for experiments and interpretation; machines and consumables are more expensive than in banding cytogenetics and FISH No: CMA can only cover parts of genome depending on the applied probes on the array; heterochromatic parts, euchromatic parts near heterochromatic ones and telomere-near parts are underrepresented 1 |
| Agilent [10] | Testing for diseases that are genetic in origin, caused by aberrations associated with developmental delay, intellectual disability, and congenital anomalies, can vastly impact quality of life. Whether you are looking for single gene or chromosomal anomalies, chromosomal microarrays and NGS application solutions and streamlined workflows enable accurate results. Make risk-informed decisions with confidence and take your clinical research further. | Chromosomal aberrations can be picked up accurately by CMA Clinical research based on CMA provide reliable risk-informed results | No: CNVs can be picked up by CMA; not the (type of) rearrangement No: Without (molecular) cytogenetics no reliable risk-informed results can be established 1 |
| Dr Lalpathlabs [11] | Most copy number variations can be interpreted based on gene content, size, inheritance, databases. | In most cases CMA is enough to solve a clinical case | No: in most cases CMA result needs to be checked by (molecular) cytogenetics 1 |
| Illumina [12] | SNP arrays for cytogenetics research The identification of structural chromosomal aberrations can provide insight into causative relationships with complex phenotypes. Chromosomal microarrays leverage the investigative power of SNP genotypes to detect imbalances in copy number and allelic homozygosity, which are commonly associated with genetic constitutional disorders. Chromosomal microarrays can detect variations that may be missed by other technologies. | Cytogenetic research can be based on CMA Disease causing structural chromosomal aberrations can be identified by CMA; sometimes only by CMA CMA is superior to other approaches in detection of CNVs | No: Cytogenetic research can be based on (molecular) cytogenetics and CMA Yes: if “chromosomal aberrations” is replaced by “CNVs” Yes: concerning resolution level between (molecular) cytogenetics and NGS 1 |
| PerkinElmer [13] | Cytogenomic microarrays offer a simple, reliable method for assessing chromosomal aberrations and their biological relevance at a higher resolution. | CMA is simple to do and interpret CMA has a high resolution CMA can detect chromosomal aberrations accurately CMA is more cost efficient than other approaches CMA is covering whole genome | No: Clinical Laboratory Geneticists are needed and inclusion of databases Yes: concerning resolution level between (molecular) cytogenetics and NGS No: CMA can detect CNVs with high accuracy, but not the nature of the chromosomal aberration No: Specialists are needed as in (molecular) cytogenetics for experiments and interpretation; machines and consumables are more expensive than in banding cytogenetics and FISH No: CMA can only cover parts of genome covered by the applied probes on the array; heterochromatic parts, euchromatic parts near heterochromatic ones and telomere-near parts are underrepresented 1 |
| Results of | ||
|---|---|---|
| Case | CMA | (Molecular) Cytogenetics |
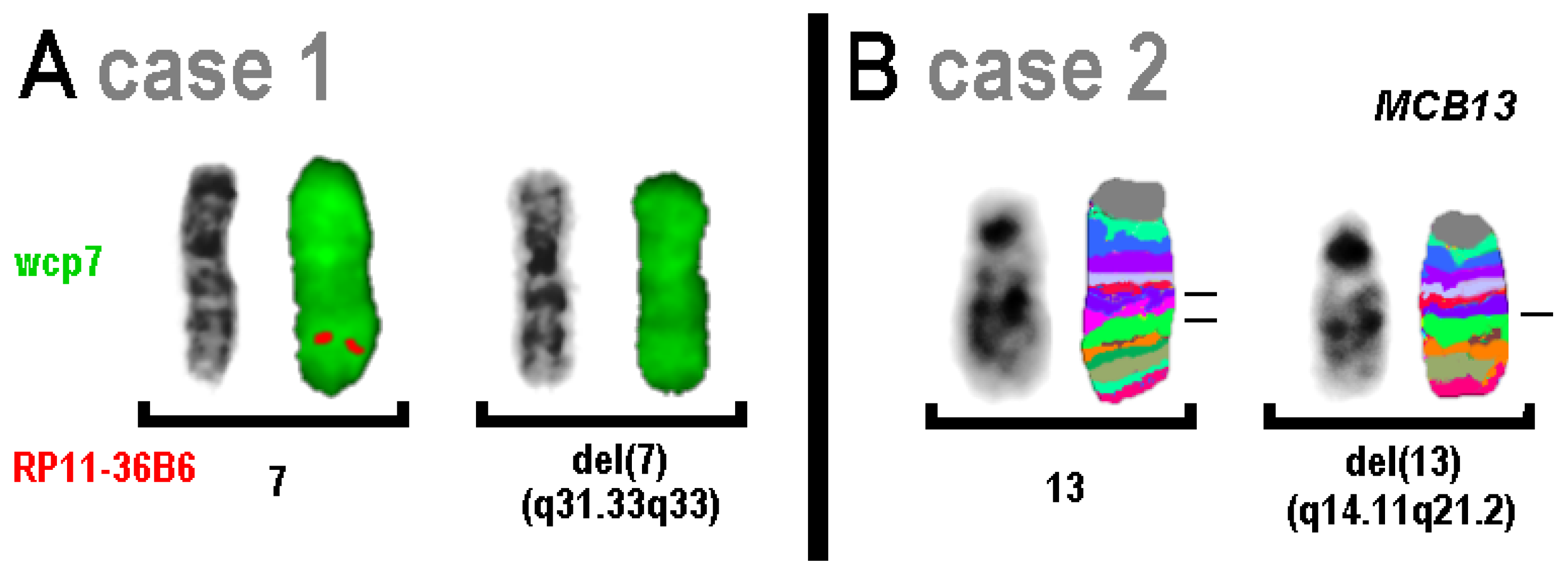
| 1 | arr[GRCh37] 7q31.33q33(124,810,332_135,770,066)x1 | 46,XX.ish del(7)(RP11-36B6-) (RP11-36B6: 130,474,967-130,476,483) |
| 2 | arr[GRCh37] 13q14.11q21.2(43,067,728_61,154,642)x1 | 46,XY,del(13)(q14.11q21.2) |
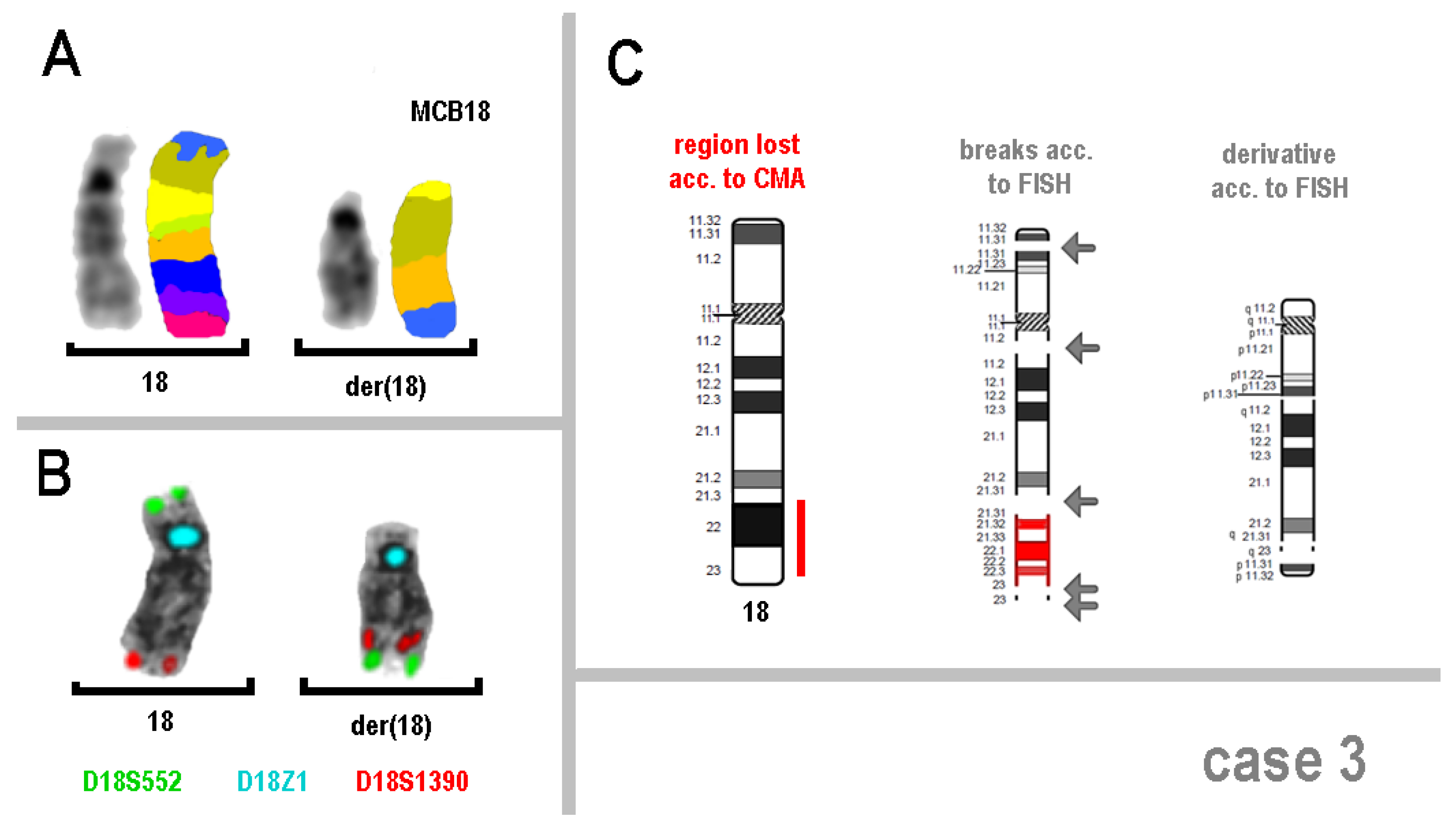
| 3 | arr[GRCh37] 18q21.32q23(56,390,211_77,541,179)x1 | 46,XY,der(18)(:q11.2->p13.31::q11.2->q21.32::q23->q23:p13.31->pter) |
| 4 | arr[GRCh37] 4p16.3(98,378_997,434)x1 | 46,XY,r(4).ish r(4)(D4S3359-,D4S2930+) (D4S3359: position not available) (D4S2930: 189,996,850-190,197,190) |
| 5 | arr[GRCh37] 15q26.1q26.3(91,659,385_102,531,389)x1 | 46,XX,r(15)(p11.2q26.1) |
| 6 | arr[GRCh37] 13q34(114,052,627_115,169,824)x1 | 46,XY,der(13)(13pter->13q34::15p12->15pter) |
| 7 | arr[GRCh37] 8q12.1(60,206,035_60,779,958)x1 | 46,XX,der(8); acc. to FISH der(8) = ins(8)(q24.3q21.13q12.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liehr, T.; Singer, S.; Mau-Holzmann, U.; Kankel, S.; Padutsch, N.; Person, L.; Daumiller, E.; Kornak, U. Results of Chromosomal Microarray Need to Always Be Checked by (Molecular) Cytogenetics—Even If They Seem to Be Simple Deletions. Genes 2025, 16, 714. https://doi.org/10.3390/genes16060714
Liehr T, Singer S, Mau-Holzmann U, Kankel S, Padutsch N, Person L, Daumiller E, Kornak U. Results of Chromosomal Microarray Need to Always Be Checked by (Molecular) Cytogenetics—Even If They Seem to Be Simple Deletions. Genes. 2025; 16(6):714. https://doi.org/10.3390/genes16060714
Chicago/Turabian StyleLiehr, Thomas, Sylke Singer, Ulrike Mau-Holzmann, Stefanie Kankel, Niklas Padutsch, Luisa Person, Eva Daumiller, and Uwe Kornak. 2025. "Results of Chromosomal Microarray Need to Always Be Checked by (Molecular) Cytogenetics—Even If They Seem to Be Simple Deletions" Genes 16, no. 6: 714. https://doi.org/10.3390/genes16060714
APA StyleLiehr, T., Singer, S., Mau-Holzmann, U., Kankel, S., Padutsch, N., Person, L., Daumiller, E., & Kornak, U. (2025). Results of Chromosomal Microarray Need to Always Be Checked by (Molecular) Cytogenetics—Even If They Seem to Be Simple Deletions. Genes, 16(6), 714. https://doi.org/10.3390/genes16060714






