Abstract
Background: SMN1 and SMN2 are causative and modifier genes, respectively, for spinal muscular atrophy (SMA). The incidence of SMN1 homozygous deletion in Japan is 1 in 20,000. However, the incidence of SMN2 homozygous deletion in Japan remains unknown. Methods: To clarify the incidence of homozygous SMN2 deletion in Japan, real-time polymerase chain reaction (PCR) was performed on dried blood spot (DBS) samples collected from newborns nationwide. Samples with positive or ambiguous results were retested using PCR-restriction fragment length polymorphism (PCR-RFLP) and nucleotide sequence analysis. Results: Of the 1000 DBS samples that were screened using real-time PCR, 51 were positive. Retesting using PCR-RFLP analysis identified 10 false results: six false positives and four false negatives. Therefore, there were 49 true positives among the 1000 samples. Notably, nucleotide sequence analysis revealed that the false negatives were caused by the cross-reactivity of SMN2 primers with SMN1 sequences. Conclusions: The incidence of homozygous SMN2 deletion in Japan is approximately 1 in 20 people. This incidence is much higher than that of homozygous SMN1 deletion and may reflect the vulnerability of the SMN2 region. Importantly, the results of the present study suggest that false negatives in the screening process were caused by cross-reactivity with non-target gene sequences.
1. Introduction
Spinal muscular atrophy (SMA) is a lower motor neuron disease (LMND) that is inherited in an autosomal recessive manner [1]. In 1995, Lefebvre et al. [2] identified SMN1 and SMN2 as SMA-related genes on chromosome 5q13. SMN1 and SMN2 are nearly identical genes that are located in the SMA locus. However, only SMN1 is a causative gene for SMA. More than 95% of SMA patients completely lack SMN1 (i.e., homozygous SMN1 deletion); the remaining patients may have point mutations or short deletions [2]. In contrast, more than 4% of control individuals lack SMN2 (i.e., homozygous SMN2 deletion) [2]. On the basis of genetic analysis, the incidence of SMA has been reported as 1 in 10,000–20,000 newborns, and the carrier frequency is 1 in 50–70 [3]. In Japan, the disease incidence is 1 in 20,000 [4,5].
The protein product of SMN1 is a full-length survival motor neuron protein (SMN), which has been known to exert a wide variety of cellular functions [6]. On the other hand, the protein products of SMN2 are the full-length SMN protein and an SMN protein lacking the exon 7 domain (Δ7-SMN). Full-length SMN is a minor product of SMN2, whereas Δ7-SMN is a major product; however, Δ7-SMN is unstable and non-functional [6].
Why is SMN2 considered a modifier gene for SMA? No homozygous deletion of SMN2 has been identified in SMA patients, supporting the hypothesis that the complete loss of SMN1 and SMN2 would result in embryonic or fetal lethality [6]. Thus, even small amounts of full-length SMN produced by SMN2 might be essential for embryonic or fetal survival in SMA patients. More importantly, SMA patients who have higher copy numbers of SMN2 exhibit ameliorated phenotypes [7]. SMN2 may therefore have a role in compensating for SMN1 loss (or deleterious mutation), at least to some degree.
The other roles of SMN2 in neurological diseases are largely unknown. Moulard et al. [8] identified homozygous SMN2 deletion in 36% of individuals with sporadic adult-onset LMND. Furthermore, Srivastava et al. [9] and Liping et al. [10] suggested that homozygous SMN2 deletion may cause distal muscle disorders. However, the relationship between homozygous SMN2 deletion and neurological disease remains a subject of long-standing debate.
Anthropologically, the frequency of SMN2 deletion is of great interest. Vorster et al. [11] reported a surprisingly high frequency of homozygous SMN2 deletion in South Africa. They identified homozygous SMN2 deletion in 12% of Black controls and 30% of Black patients with SMA-like symptoms. The authors suggested that these data may reflect architectural changes in the SMN region, including novel gene conversion or rearrangement events.
To clarify the relationship between homozygous SMN2 deletion and neurological diseases or the architectural stability of SMN2 in the SMA locus, it is essential to determine the incidence of homozygous SMN2 deletion in each ethnic group. Given that no epidemiological studies on homozygous SMN2 deletion were conducted in Japan until 2003, we planned to establish a screening system for homozygous SMN2 deletions. In 2023, we developed a real-time polymerase chain reaction (PCR) screening method to detect homozygous SMN2 deletions [12].
As a preliminary step to clarify the relationship between homozygous SMN2 deletion and neurological diseases in Japan, it is necessary to determine the frequency (or incidence) of homozygous SMN2 deletion in the Japanese population. In the present study, we used our real-time PCR method to screen 1000 dried blood spot (DBS) samples collected from newborns nationwide to estimate the incidence of homozygous SMN2 deletion in Japan. During the screening process, we encountered both false-positive and false-negative samples, which we also report in this article.
2. Materials and Methods
2.1. Residual DBS Samples
In the present study, 1000 residual DBS samples on filter paper (FTA Elute Cards; GE Healthcare, Boston, MA, USA) from newborn infants born all over Japan were used. The samples were randomly selected from residual samples that had been collected from 4157 newborn infants in our previous pilot study [13].
2.2. Real-Time PCR
A punched circle (1.2 mm in diameter) from the DBS card was placed directly into a PCR mixture [12]. The components of the real-time PCR mixture and their concentrations are listed in Table S1. The primer sequences were cenSMNex7forw (5′-TTT ATT TTC CTT ACA GGG TTT TA-3′) [7] and cenSMNint7rev (5′-GTG AAA GTA TGT TTC TTC CAC GCA-3′) [7]. The primer positions are shown in Figure 1.
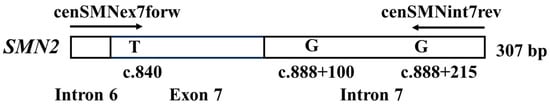
Figure 1.
Primer positions used in the real-time polymerase chain reaction analysis. The forward primer (cenSMNex7forw) binds to the intron 6/exon 7 boundary, including the SMN2-specific nucleotide, c.840T. The reverse primer (cenSMNint7rev) binds to the intron 7 sequence, including the SMN2-specific nucleotide, c.888+215G. The third SMN2-specific nucleotide, G at c.888+100, exists in intron 7. When false-positive results caused by cross-reactivity with the SMN1 sequence were obtained, the amplified products included the SMN1-specific nucleotide, A at c.888+100. This figure is reproduced from our previous report [12].
The PCR conditions were as follows: (1) initial denaturation at 94 °C for 7 min; (2) 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min; and (3) a final extension at 72 °C for 7 min.
2.3. PCR-Restriction Fragment Length Polymorphism (RFLP)
To distinguish SMN2 from SMN1, PCR-RFLP was performed according to the method reported by van der Steege et al. [14], with some modifications. The DNA sources in this study were DBS samples. The components of the PCR mixture and their concentrations are listed in Table S2. The primer sequences were R111 (5′-AGA CTA TCA ACT TAA TTT CTG ATCA-3′) [2] and X7-Dra (5′-CCT TCC TTC TTT TTG ATT TTG TTT-3′) [14]. The primer positions and restriction enzyme site are shown in Figure 2.
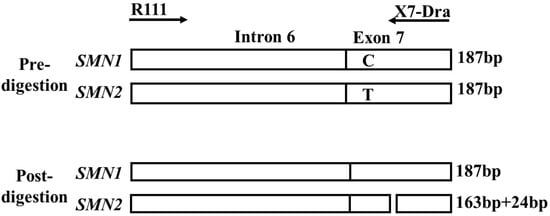
Figure 2.
Primer positions used in the polymerase chain reaction (PCR)-restriction fragment length polymorphism analysis. The forward primer (R111) bound to intron 6 of SMN1 and SMN2. The reverse mismatched primer (X7-Dra) bound to a region near c.840. The nucleotide at c.840 was G in SMN1 and T in SMN2. A restriction enzyme, DraI, cut the PCR-amplified fragment of SMN2 at c.840, creating two fragments (163 and 24 bp). This figure is a slightly modified version of a figure from our previous report [12].
The PCR conditions were as follows: (1) an initial denaturation at 94 °C for 7 min; (2) 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 1 min; and (3) a final extension at 72 °C for 7 min. Thereafter, the amplified products were digested by restriction enzyme DraI at 37 °C for 12 h.
2.4. Nucleotide Sequencing
The real-time PCR products that had been amplified for 45 cycles in the screening procedures were purified using ISOSPIN PCR Product (NIPPON GENE Co., Ltd., Tokyo, Japan). The purified products were then submitted for sequencing. All sequencing analyses were performed by FASMAC Co., Ltd. (Atsugi, Japan).
The sequencing reactions were performed using an Applied Biosystems Big Dye Terminator V3.1 cycle-sequencing kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). Reaction products were analyzed using an Applied Biosystems 3730xl DNA Analyzer (Thermo Fisher Scientific Inc.).
In the current study, we selected a direct sequence for sequencing DNA bases using products amplified directly by PCR. This eliminated the need for steps such as DNA cloning and plasmid purification and allowed us to directly read specific regions of the genome, which is useful when searching for genetic mutations.
2.5. Statistical Analyses
We used Excel software (version number 2025) (Microsoft Corp., Redmond, WA, USA) for the summary statistics. The cycle threshold (Ct) values of real-time PCR are presented as the mean ± standard deviation (SD). The mean Ct values of the SMN2 deletion and SMN2 retention groups were compared using Welch’s t-test. A p-value < 0.05 was considered significant.
3. Results
3.1. Detection of Homozygous SMN2 Deletion Using Real-Time PCR
We subjected 1000 DBS samples to real-time PCR, which is a suitable screening method because it can analyze many samples at once. The Ct value (mean ± SD) of 1000 samples was 32.7 ± 2.1.
The real-time PCR results were classified into three groups based on the Ct values (Figure 3): Group A, DBS samples with Ct values ≥ mean + 2 SD; Group B, DBS samples with Ct values ≥ mean + 1 SD and <mean + 2 SD; Group C, DBS samples with Ct values < mean + 1 SD.
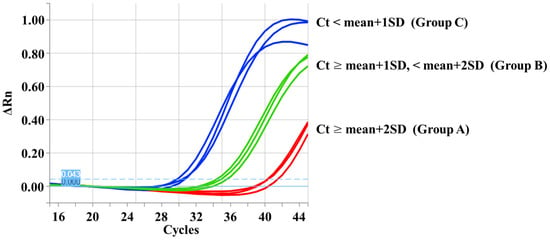
Figure 3.
Real-time polymerase chain reaction amplification curves. The curves of representative cases from each group are shown.
Group A samples (n = 51) were most likely to be positive cases for homozygous SMN2 deletion, Group B samples (n = 46) were ambiguous cases on the border between positive and negative results for homozygous SMN2 deletion., and Group C samples (n = 903) were most likely to be negative cases for homozygous SMN2 deletion.
3.2. Confirmation of the Real-Time PCR Results Using PCR-RFLP
Next, we retested the samples from Groups A and B (n = 51 + 46) using PCR-RFLP analysis. We did not retest the samples from Group C for two reasons: (1) there was a very small possibility that Group C included cases with homozygous SMN2 deletion, and (2) given the time it takes to perform PCR-RFLP, we considered that there were too many samples in this group (n = 903) to feasibly retest using this method.
PCR-RFLP is relatively cumbersome for gene detection because it includes PCR amplification, enzymatic digestion, and agarose gel electrophoresis, meaning that it takes several hours or even half a day from start to finish. However, this method is very robust and produces stable results because sufficient PCR product can be obtained for subsequent RFLP analysis regardless of the original DNA quality and quantity from the DBS samples. Moreover, when the PCR products are completely digested, RFLP analysis provides accurate information about the presence or absence of the target gene.
To retest the samples with positive (Group A) and ambiguous (Group B) results, we conducted PCR-RFLP analysis using a second punched circle (near the first one) from each DBS. Figure 4 displays a representative gel electrophoresis result showing the digestion products with SMN2 deletion and retention.
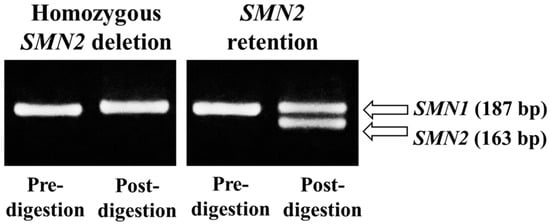
Figure 4.
Polymerase chain reaction (PCR)-restriction fragment length polymorphism. The restriction enzyme DraI cuts the PCR-amplified fragment of SMN2 exon 7 at c.840, creating two fragments (see Figure 2). The absence of 163-bp fragments indicates SMN2 deletion.
Table 1 summarizes the PCR-RFLP results in Groups A and B. Of the 51 DBS samples with Ct values ≥ mean + 2 SD (Group A), 45 had a homozygous SMN2 deletion, whereas 6 retained SMN2 alleles. Of the 46 samples with Ct values ≥ mean + 1 SD and <mean + 2 SD (Group B), 4 had a homozygous SMN2 deletion, whereas 42 retained SMN2 alleles. When Groups A and B were combined, 49 of the 97 samples had a homozygous SMN2 deletion, and 48 samples retained at least one copy of SMN2.

Table 1.
PCR-RFLP summary of positive and ambiguous samples (n = 97).
3.3. Nucleotide Sequencing Analysis of the Real-Time PCR Amplified Products
After confirming the presence or absence of homozygous SMN2 deletion using PCR-RFLP analysis, we conducted a nucleotide sequencing analysis using the real-time PCR-amplified products, which were obtained during the screening procedures.
All real-time PCR-amplified products could be subjected to nucleotide sequencing analysis. However, we only conducted a nucleotide sequencing analysis of the positive (Group A) and ambiguous (Group B) samples. We did not include the Group C samples in this analysis because of time and budget constraints. Even so, this nucleotide sequencing analysis clarified the cause of false-negative results and confirmed that the false-positive samples indeed retained SMN2.
In this analysis, we identified two kinds of amplified products: Type I amplified products (with SMN1 sequences) and Type II amplified products (with SMN2 sequences) (Figure 5). The presence of Type I amplified products indicated that the SMN2 primers had non-specifically amplified the SMN1 sequence from samples with at least one copy of SMN1. The presence of Type II amplified products indicated that the SMN2-specific primers had specifically amplified the SMN2 sequence from samples with at least one copy of SMN2. Of note, no mixed data (containing both types of amplified products) were obtained.
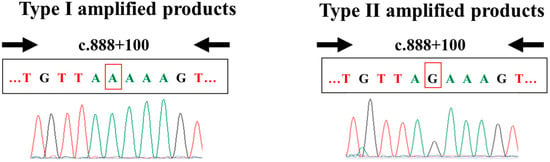
Figure 5.
Type I and Type II amplified products. The primer set in this experiment generated two types of amplification products: Type I and Type II. (Left) Type I amplified products were amplified products that contained an SMN1-specific nucleotide: A at c.888+100. When true-positive or false-negative results (i.e., the absence of SMN2) were obtained, we only observed Type I products. (Right) Type II amplified products were amplified products that contained an SMN2-specific nucleotide: G at c.888+100. When true-negative or false-positive results (i.e., the presence of SMN2) were obtained, we only observed Type II products.
Table 2 summarizes the nucleotide sequencing results in Groups A and B. Of the 51 DBS samples with Ct values ≥ mean + 2 SD (Group A), 45 had a homozygous SMN2 deletion (Type I), whereas 6 retained SMN2 alleles (Type II). Of the 46 samples with Ct values ≥ mean + 1 SD and <mean + 2 SD (Group B), 4 had a homozygous SMN2 deletion (Type I), whereas 42 retained SMN2 alleles (Type II). When Groups A and B were combined, 49 of the 97 samples had a homozygous SMN2 deletion, and 48 samples retained at least one copy of SMN2. These results were entirely consistent with the PCR-RFLP data (Table 1).

Table 2.
Nucleotide sequencing analysis summary of positive and ambiguous samples (n = 97).
Collectively, our findings indicate that the false-negative results of the real-time PCR screening were caused by relatively high amplification caused by the cross-reactivity of the SMN2 primers with the SMN1 sequence. Additionally, the false-positive results of the real-time PCR screening were caused by relatively low amplification, which can be caused by low amounts of DNA in the sample and/or the presence of PCR-inhibiting compounds such as heparin. However, heparin was not used during sample preparation in the present study. The most plausible cause of false-positive results was therefore a low amount of DNA in the DBS samples.
3.4. Sensitivity and Specificity of the Screening, and Incidence of Homozygous SMN2 Deletion
In the present study, all DBS samples with Ct values < mean + 1 SD (Group C) were assumed to retain SMN2. Table 3 shows the sensitivity and specificity of real-time PCR in the 1000 samples in this study using the assumed data from Group C combined with the confirmed data from Groups A and B (Table 1). Although the 903 samples from Group C were not retested using PCR-RFLP, it is considered highly unlikely that this group contained cases of homozygous SMN2 deletions. We thus determined a sensitivity of 0.918 (45/49), specificity of 0.994 (945/951), positive predictive value of 0.882 (45/51), and negative predictive value of 0.996 (945/949).

Table 3.
Screening assay summary of all samples (n = 1000).
Using these data, we defined two new groups based on the absence/presence of SMN2: homozygous SMN2 deletion and SMN2 retention. The Ct values for the homozygous SMN2 deletion and SMN2 retention groups were 39.2 ± 1.9 (n = 49) and 32.4 ± 1.5 (n = 951), respectively. The Ct values were significantly different between the two groups (p < 0.01). However, in the actual screening situation, the Ct values for both groups had a small overlap caused by false results, as described in the previous sections.
Finally, using the real-time PCR results and the subsequent close examination of ambiguous cases using PCR-RFLP and nucleotide sequencing, we determined the incidence of individuals with homozygous SMN2 deletion in Japan as 49/1000 (4.9%) (Table 4).

Table 4.
Frequency of homozygous SMN2 deletion.
4. Discussion
4.1. Establishment of a Screening System for Homozygous SMN2 Deletion
Our previous study was performed to develop a methodology for detecting homozygous SMN2 deletion using real-time PCR [12]. To develop a methodology, we first needed to identify positive samples (i.e., those with homozygous SMN2 deletion). We therefore tested 300 newborn screening (NBS) samples using PCR-RFLP. However, the PCR-RFLP method is relatively time-consuming and laborious, and it is unsuitable for screening many samples. It was therefore necessary to establish a simple, rapid, and easy method using real-time PCR.
The present study was performed to screen more samples (1000 samples) using the real-time PCR method that was developed in our previous study. Because we faced problems (especially false results) in the screening process using real-time PCR, we retested some samples using PCR-RFLP and used nucleotide sequence analysis to identify the cause of false results (see Section 4.3). Finally, we estimated the incidence of homozygous SMN2 deletion in Japan as approximately 5% in the general population (see Section 4.2).
In our series of studies, we chose the direct PCR method using intercalating fluorescent technology because of its speed, simplicity, and low cost. Our screening system included no DNA extraction step (speed and simplicity), no fluorescent probes (simplicity and low cost), or no reference gene analysis (simplicity and low cost).
To avoid punch-to-punch variability, DNA should have been extracted from DBS in the first step of the analysis. However, this is time-consuming and requires many punched DBS circles. The direct PCR method allows us to skip the step of DNA extraction from DBS, thus saving DBS samples and completing the screening assay quickly.
To co-amplify the reference gene, we should have used the fluorescent probe method (5′ nuclease assay), but fluorescent probes are expensive. The intercalating fluorescent technology in our studies was unable to detect two or more different genes specifically, but intercalating fluorescent dyes are inexpensive. In addition, if one only aims to screen for the presence or absence of a target gene, introducing a reference gene may not be necessary, because screen-positive cases (i.e., target-gene-absent cases) or ambiguous cases should be retested, which was demonstrated in this study.
4.2. Incidence of Homozygous SMN2 Deletion in Japan
We screened DBS samples from newborns in Japan for homozygous SMN2 deletion using a real-time PCR method with an intercalating fluorescent dye. In our study, 951 of the 1000 DBS samples carried at least one copy of SMN2, whereas 49 samples had a homozygous SMN2 deletion. Our findings, therefore, indicate that the incidence of homozygous SMN2 deletion in Japan is 49 in 1000, or 4.9% (Table 4). The incidence of homozygous SMN2 deletion in Japan is similar to that in other Asian countries, slightly lower than that in European countries, and much lower than that in African countries (see Section 4.5).
4.3. False Results Identified in the Present Study
In the current study, Ct values ≥ mean + 2 SD were considered positive for homozygous SMN2 deletion, whereas Ct values < mean + 2 SD were considered negative for homozygous SMN2 deletion. To detect false-negative and -positive results, we also performed PCR-RFLP analysis for all samples with Ct values > mean + 1 SD. PCR-RFLP analysis identified six false-positive samples and four false-negative samples out of 1000 DBS samples. Subsequent nucleotide sequencing analysis demonstrated that false-negative results were caused by cross-reactivity with the SMN1 sequence.
Our real-time PCR method used specific primers to detect the target gene. However, although both the sense and antisense primers used in the present study were designed specifically for the target gene [7], false negatives still occurred. To prevent false negatives, future studies may consider the use of locked nucleic acid-modified primers [15]. In previously reported real-time PCR methods using specific probes, locked nucleic acid-modified probes have been used to increase the stringency of probe hybridization [16].
False positives are highly dependent on the condition of the DBS sample, including the coexistence of anticoagulants such as heparin [5,12]. To improve the accuracy of the screening system, attention should therefore be paid to sample preparation.
4.4. Possible Pitfalls of Neonatal SMN1 Deletion Screening
For the early detection of SMA, NBS programs to detect SMN1 deletion (SMA-NBS) are being implemented around the world [17], and real-time PCR analyses are used in many SMA-NBS laboratories [18].
A systematic review by Cooper et al. [18] noted that most previous SMA-NBS studies reported no false-negative or -positive cases (i.e., they had a sensitivity and specificity of 100%). The authors suggested that some false negatives may have been missed because of a lack of follow-up for the negative results. When a screening test determines that SMN1 is absent (i.e., the case is positive for SMA), retesting is usually performed to avoid false-positive cases. However, when a screening test determines that SMN1 is present (i.e., the case is negative for SMA), retesting is usually skipped, meaning that false-negative samples containing SMN1 may be overlooked, although this is very rare.
Importantly, the pitfalls of screening tests that were identified in the present study, particularly regarding false-negative results, may also exist in SMA-NBS.
4.5. Anthropological Importance of SMN2 Deletion
A large-scale gene duplication in primates gave rise to a variety of paralogous genes [19]. Human SMN2 is one such paralogous gene; it was created by the duplication of an SMN1-including segment approximately three million years ago [19]. The difference between human SMN1 and SMN2, which are both approximately 20 kb in size, was initially thought to be only 5 bp [2] but was later reported to be at least 22 bp [20]. Nonetheless, SMN1 and SMN2 are almost identical genes. This high homology makes the SMA locus on the chromosome unstable, which leads to genomic instability and a predisposition to gene deletions, duplications, and conversions between both SMN1 and SMN2 [20].
Table 5 shows the frequencies of homozygous SMN2 deletion around the world, demonstrating that more than 10% of Sub-Saharan individuals have a complete absence of SMN2. Notably, Sub-Saharan individuals are likely to have three or more copies of SMN1, and the carrier frequency of SMA is much lower in this region than in other areas [11,21]. Furthermore, a high copy number of SMN1 in conjunction with a lack of SMN2 is relatively common in Black South African individuals, whereas a low copy number of SMN1 is relatively common in European/Asian individuals. These observations may be explained by the two following scenarios:


Table 5.
Frequencies of homozygous SMN2 deletion around the world *.
Table 5.
Frequencies of homozygous SMN2 deletion around the world *.
| Country/ Ethnicity | Controls | NeuromuscularDisorders | References | |||
|---|---|---|---|---|---|---|
| SMN2-Deletion/ Sample Number | Frequency | SMN2-Deletion/ Sample Number | Frequency | |||
| European | ||||||
| 1 | UK | (-) | (-) | 16/154 | 10.4% | [22] |
| 2 | USA and Canada | 4/54 | 7.4% | (-) | (-) | [23] |
| 3 | France | 8/90 | 8.9% | (-) | (-) | [24] |
| France | 15/167 | 9.0% | 16/167 | 9.6% | [25] | |
| 4 | France | 52/621 | 8.4% | 54/600 | 9.0% | [26] |
| 5 | Sweeden | 37/502 | 7.4% | 29/502 | 5.8% | [26] |
| 6 | Germany | 9/100 | 9.0% | (-) | (-) | [27] |
| 7 | The Netherlands | 78/984 | 7.9% | 62/847 | 7.3% | [28] |
| African | ||||||
| 8 | Sub-Saharan (Mali) | 150/613 | 24.5% | (-) | (-) | [21] |
| 9 | Sub-Saharan (Nigeria) | 33/120 | 27.5% | (-) | (-) | [21] |
| 10 | Sub-Saharan (Kenya) | 23/120 | 19.2% | (-) | (-) | [21] |
| 11 | Black South African | 15/122 | 12.3% | 60/122 | 49.2% | [11] |
| Asian | ||||||
| 12 | Vietnam | 2/52 | 3.9% | (-) | (-) | [29] |
| 13 | Chinese | 89/1712 | 5.29% | (-) | (-) | [30] |
| 14 | Taiwan | 30/520 | 5.8% | (-) | (-) | [31] |
| 15 | Taiwan | 5147/107,611 | 4.8% | (-) | (-) | [32] |
| 16 | Korea | 2/100 | 2.0% | 5/25 | 20.0% | [33] |
| 17 | Korea | 49/1581 | 3.1% | (-) | (-) | [34] |
| 18 | Japan | 16/300 | 5.3% | (-) | (-) | [12] |
| 19 | Japan | 19/399 | 4.8% | 41/537 | 7.6% | [35] |
| 20 | Japan | 49/1000 | 4.9% | (-) | (-) | This study |
* Homozygous deletion of SMN2 exon 7, specifically; (-): no data reported.
- (1)
- Vorster et al. [11] noted that primates have only one copy of SMN1, suggesting that the SMN region in early humans might have consisted of only the SMN1 gene. At a later stage, the hypervariable nature of the SMN region might have resulted in multiple copies of SMN1. This duplicated SMN1 might have then diverged into SMN2 as a result of mutations. Thus, a high copy number of SMN1 and a lack of SMN2 may reflect a state before the divergence of SMN2 from SMN1.
- (2)
- Sangaré et al. [23] posited that the population that migrated out of Africa to Asia/Europe may have had a lower SMN1 copy number by chance, or might have randomly drifted in this direction after the migration. Black South African individuals may therefore represent the descendants who retained genetic diversity, whereas European/Asian individuals represent the descendants with reduced genetic diversity caused by the bottleneck phenomenon. Thus, the bottleneck phenomenon might be the driving force underlying the differences in SMA-related genotypes between Black South African and European/Asian individuals.
4.6. Perspectives from SMN2 Studies
Studies using animal models indicate that the loss of both SMN1 and SMN2 may lead to fetal lethality [36,37], indicating that the presence of SMN2 is critical for the survival of SMN1-deficient fetuses. This concept guided us to the idea that SMN2 loss may be related to disease development in patients with some genetic defects.
In recent years, there has been debate regarding the relationship between homozygous SMN2 deletion and motor neuron disease. Echaniz-Laguna and Lee [38] concluded that homozygous SMN2 deletion is a risk factor for developing amyotrophic lateral sclerosis (ALS), whereas Corcia [26] suggested that it is a preventive factor against ALS progression in some ethnicities. However, Gamez [39] reported that homozygous SMN2 deletion in ALS patients is not associated with survival or a decline in respiratory function.
More recently, Ishihara et al. [35] suggested that a decrease in the SMN2 copy number may adversely affect the onset and prognosis of motor neuron diseases, including ALS and LMND, in the Japanese population. These authors reported that the SMN2 copy number affects the survival time of patients with ALS (median survival: 0 copies, 34 months; 1 copy, 39 months; 2 copies, 44 months; 3 copies, 54 months; log-rank test, p < 0.05). Furthermore, SMN2-null cases (i.e., homozygous SMN2 deletion or a loss of SMN2) were significantly more common in the LMND group (12.0%) than in the control group (4.8%) (odds ratio = 2.73, 95% confidence interval = 1.06–6.98, p < 0.05).
However, these discussions were all based on retrospective cohort studies. In addition, the discussions did not include any advanced molecular genetic studies after the discovery of SMN1 and SMN2. Thus, prospective cohort studies incorporating the latest findings from molecular genetic research may provide more definitive conclusions about the relationship between homozygous SMN2 deletion and motor neuron disease.
4.7. Limitations
In the present study, DBS samples with Ct values < mean + 1 SD were considered to carry at least one copy of SMN2, and further testing using PCR-RFLP or nucleotide sequence analysis was not performed. Therefore, the possibility of unrecognized false-negative results could not be completely excluded. However, such cases are expected to be very rare, and we consider them unlikely to contradict our statistical conclusions. Furthermore, time and labor constraints mean that confirmatory testing usually cannot be performed on all DBS samples; our limitations are therefore likely to be widely acceptable.
4.8. Conclusions
The present study of screening for homozygous SMN2 deletion using real-time PCR revealed two important findings. First, the incidence of homozygous SMN2 deletion in Japan is approximately 5%. Second, cross-reactive amplification with SMN1 may result in false negatives in SMN2 deletion screening. Importantly, this finding also suggests that cross-reactivity with SMN2 may result in false-negative results in SMA-NBS, which screen for SMN1 deletion.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16060712/s1, Table S1: Real-time PCR reaction mixture; Table S2: PCR-RFLP mixtures.
Author Contributions
Conceptualization, Y.B. and H.N.; methodology, M.S. and H.N.; validation, Y.N. and H.N.; investigation, M.S. and H.N.; resources, Y.N., R.B. and H.A.; data curation, M.S., Y.B., S.-I.W., M.N., J.O. and H.N.; writing—original draft preparation, M.S. and H.N.; writing—review and editing, Y.B. and H.N. visualization; M.S. and H.N.; supervision, H.A. and H.N.; project administration, Y.B. and H.N.; funding acquisition, H.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Japan Society for the Promotion of Science: grant number JP23K07279.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Kobe University Graduate School of Medicine (reference B230027, approved on 14 June 2023) and the Ethics Committee of Kobe Gakuin University (reference 23-02, approved on 16 October 2023).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Although the residual DBS samples used in this study were anonymized, the study was made public through an opt-out procedure.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
We are grateful to Hirokuni Negishi for his advice on improving the manuscript.
Conflicts of Interest
R.B., H.A., and H.N. report personal compensation from Biogen Japan, Chugai Pharmaceutical Co., and Novartis Japan. H.N. also reports a consulting fee from Sekisui Medical Co. These companies had no role in the choice of research project; design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. All other co-authors declare no competing interests.
Abbreviations
The following abbreviations are used in this manuscript:
| ALS | Amyotrophic lateral sclerosis. |
| Ct | Cycle threshold. |
| DBS | Dried blood spot. |
| LMND | Lower motor neuron disease. |
| NBS | Newborn screening. |
| PCR | Polymerase chain reaction. |
| RFLP | Restriction fragment length polymorphism. |
| SD | Standard deviation. |
| SMA | Spinal muscular atrophy. |
| SMA-NBS | Newborn screening programs to detect SMN1 deletion. |
| SMN | Survival motor neuron. |
References
- Arnold, W.D.; Kassar, D.; Kissel, J.T. Spinal muscular atrophy: Diagnosis and management in a new therapeutic era. Muscle Nerve 2015, 51, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Verhaart, I.E.C.; Robertson, A.; Wilson, I.J.; Aartsma-Rus, A.; Cameron, S.; Jones, C.C.; Cook, S.F.; Lochmüller, H. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—A literature review. Orphanet J. Rare Dis. 2017, 12, 124. [Google Scholar] [CrossRef]
- Ito, M.; Yamauchi, A.; Urano, M.; Kato, T.; Matsuo, M.; Nakashima, K.; Saito, K. Epidemiological investigation of spinal muscular atrophy in Japan. Brain Dev. 2022, 44, 2–16. [Google Scholar] [CrossRef]
- Noguchi, Y.; Bo, R.; Nishio, H.; Matsumoto, H.; Matsui, K.; Yano, Y.; Sugawara, M.; Ueda, G.; Wijaya, Y.O.S.; Niba, E.T.E.; et al. PCR-based screening of spinal muscular atrophy for newborn infants in Hyogo Prefecture, Japan. Genes 2022, 13, 2110. [Google Scholar] [CrossRef]
- Nishio, H.; Niba, E.T.E.; Saito, T.; Okamoto, K.; Takeshima, Y.; Awano, H. Spinal muscular atrophy: The past, present, and future of diagnosis and treatment. Int. J. Mol. Sci. 2023, 26, 11939. [Google Scholar] [CrossRef] [PubMed]
- Feldkötter, M.; Schwarzer, V.; Wirth, R.; Wienker, T.F.; Wirth, B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002, 70, 358–368. [Google Scholar] [CrossRef]
- Moulard, B.; Salachas, F.; Chassande, B.; Briolotti, V.; Meininger, V.; Malafosse, A.; Camu, W. Association between centromeric deletions of the SMN gene and sporadic adult-onset lower motor neuron disease. Ann. Neurol. 1998, 43, 640–644. [Google Scholar] [CrossRef]
- Srivastava, S.; Mukherjee, M.; Panigrahi, I.; Shanker Pandey, G.; Pradhan, S.; Mittal, B. SMN2-deletion in childhood-onset spinal muscular atrophy. Am. J. Med. Genet. 2001, 101, 198–202. [Google Scholar] [CrossRef]
- Liping, L.; Hongwei, M.; Lin, W. Homozygous survival motor neuron 2 gene deletion and sporadic lower motor neuron disease in children: Case report and literature review. J. Child. Neurol. 2013, 28, 509–516. [Google Scholar] [CrossRef]
- Vorster, E.; Essop, F.B.; Rodda, J.L.; Krause, A. Spinal muscular atrophy in the Black South African population: A matter of rearrangement? Front. Genet. 2020, 11, 54. [Google Scholar] [CrossRef]
- Bouike, Y.; Sakima, M.; Taninishi, Y.; Matsutani, T.; Noguchi, Y.; Bo, R.; Awano, H.; Nishio, H. Real-time PCR-based screening for homozygous SMN2 deletion using residual dried blood spots. Genes 2023, 14, 2159. [Google Scholar] [CrossRef]
- Shinohara, M.; Niba, E.T.E.; Wijaya, Y.O.S.; Takayama, I.; Mitsuishi, C.; Kumasaka, S.; Kondo, Y.; Takatera, A.; Hokuto, I.; Morioka, I.; et al. A novel system for spinal muscular atrophy screening in newborns: Japanese pilot study. Int. J. Neonatal Screen. 2019, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Van der Steege, G.; Grootscholten, P.M.; Van der Vlies, P.; Draaijers, T.G.; Osinga, J.; Cobben, J.M.; Scheffer, H.; Buys, C.H. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet 1995, 345, 985–986. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, C.; Teng, Y.; Zeng, S.; Chen, S.; Liang, D.; Li, Z.; Wu, L. Detection of spinal muscular atrophy using a duplexed real-time PCR approach with locked nucleic acid-modified primers. Ann. Lab. Med. 2021, 41, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.L.; Lee, F.K.; Yazdanpanah, G.K.; Staropoli, J.F.; Liu, M.; Carulli, J.P.; Sun, C.; Dobrowolski, S.F.; Hannon, W.H.; Vogt, R.F. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin. Chem. 2015, 61, 412–419. [Google Scholar] [CrossRef]
- Dangouloff, T.; Vrščaj, E.; Servais, L.; Osredkar, D.; SMA NBS World Study Group. Newborn screening programs for spinal muscular atrophy worldwide: Where we stand and where to go. Neuromuscul. Disord. 2021, 31, 574–582. [Google Scholar] [CrossRef]
- Cooper, K.; Nalbant, G.; Sutton, A.; Harnan, S.; Thokala, P.; Chilcott, J.; McNeill, A.; Bessey, A. Systematic review of newborn screening programmes for spinal muscular atrophy. Int. J. Neonatal Screen. 2024, 10, 49. [Google Scholar] [CrossRef]
- Rochette, C.F.; Gilbert, N.; Simard, L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001, 108, 255–266. [Google Scholar] [CrossRef]
- Blasco-Pérez, L.; Costa-Roger, M.; Leno-Colorado, J.; Bernal, S.; Alias, L.; Codina-Solà, M.; Martínez-Cruz, D.; Castiglioni, C.; Bertini, E.; Travaglini, L.; et al. Deep molecular characterization of milder spinal muscular atrophy patients carrying the c.859G>C variant in SMN2. Int. J. Mol. Sci. 2022, 23, 8289. [Google Scholar] [CrossRef]
- Sangaré, M.; Hendrickson, B.; Sango, H.A.; Chen, K.; Nofziger, J.; Amara, A.; Dutra, A.; Schindler, A.B.; Guindo, A.; Traoré, M.; et al. Genetics of low spinal muscular atrophy carrier frequency in sub-Saharan Africa. Ann. Neurol. 2014, 75, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Morrison, K.E.; Al-Chalabi, A.; Bakker, M.; Leigh, P.N. Analysis of chromosome 5q13 genes in amyotrophic lateral sclerosis: Homozygous NAIP deletion in a sporadic case. Ann. Neurol. 1996, 39, 796–800. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, P.E.; Parsons, D.W.; Simard, L.R.; Rochette, C.; Ray, P.N.; Mendell, J.R.; Prior, T.W.; Burghes, A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997, 60, 1411–1422. [Google Scholar] [CrossRef]
- Gérard, B.; Ginet, N.; Matthijs, G.; Evrard, P.; Baumann, C.; Da Silva, F.; Gérard-Blanluet, M.; Mayer, M.; Grandchamp, B.; Elion, J. Genotype determination at the survival motor neuron locus in a normal population and SMA carriers using competitive PCR and primer extension. Hum. Mutat. 2000, 16, 253–263. [Google Scholar] [CrossRef]
- Corcia, P.; Camu, W.; Halimi, J.M.; Vourc’h, P.; Antar, C.; Vedrine, S.; Giraudeau, B.; de Toffol, B.; Andres, C.R.; French ALS Study Group. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology 2006, 67, 1147–1150. [Google Scholar] [CrossRef]
- Corcia, P.; Ingre, C.; Blasco, H.; Press, R.; Praline, J.; Antar, C.; Veyrat-Durebex, C.; Guettard, Y.O.; Camu, W.; Andersen, P.M.; et al. Homozygous SMN2 deletion is a protective factor in the Swedish ALS population. Eur. J. Hum. Genet. 2012, 20, 588–591. [Google Scholar] [CrossRef]
- Anhuf, D.; Eggermann, T.; Rudnik-Schöneborn, S.; Zerres, K. Determination of SMN1 and SMN2 copy number using TaqMan technology. Hum. Mutat. 2003, 22, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Blauw, H.M.; Barnes, C.P.; Van Vught, P.W.; Van Rheenen, W.; Verheul, M.; Cuppen, E.; Veldink, J.H.; Van den Berg, L.H. SMN1 gene duplications are associated with sporadic ALS. Neurology 2012, 78, 776–780. [Google Scholar] [CrossRef]
- Tran, V.K.; Sasongko, T.H.; Hong, D.D.; Hoan, N.T.; Dung, V.C.; Lee, M.J.; Gunadi Takeshima, Y.; Matsuo, M.; Nishio, H. SMN2 and NAIP gene dosages in Vietnamese patients with spinal muscular atrophy. Pediatr. Int. 2008, 50, 346–351. [Google Scholar] [CrossRef]
- Sheng-Yuan, Z.; Xiong, F.; Chen, Y.J.; Yan, T.Z.; Zeng, J.; Li, L.; Zhang, Y.N.; Chen, W.Q.; Bao, X.H.; Zhang, C.; et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur. J. Hum. Genet. 2010, 18, 978–984. [Google Scholar] [CrossRef]
- Kao, H.Y.; Su, Y.N.; Liao, H.K.; Liu, M.S.; Chen, Y.J. Determination of SMN1/SMN2 gene dosage by a quantitative genotyping platform combining capillary electrophoresis and MALDI-TOF mass spectrometry. Clin. Chem. 2006, 52, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.N.; Hung, C.C.; Lin, S.Y.; Chen, F.Y.; Chern, J.P.; Tsai, C.; Chang, T.S.; Yang, C.C.; Li, H.; Ho, H.N.; et al. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005–2009: A prospective population-based cohort study. PLoS ONE 2011, 6, 17067. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Lee, K.A.; Hong, J.M.; Suh, G.I.; Choi, Y.C. Homozygous SMN2 deletion is a major risk factor among twenty-five Korean sporadic amyotrophic lateral sclerosis patients. Yonsei Med. J. 2012, 53, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Yun, S.A.; Roh, E.Y.; Yoon, J.H.; Shin, S.; Ki, C.S. Carrier frequency of spinal muscular atrophy in a large-scale Korean population. Ann. Lab. Med. 2020, 40, 326–330. [Google Scholar] [CrossRef]
- Ishihara, T.; Koyama, A.; Atsuta, N.; Tada, M.; Toyoda, S.; Kashiwagi, K.; Hirokawa, S.; Hatano, Y.; Yokoseki, A.; Nakamura, R.; et al. SMN2 gene copy number affects the incidence and prognosis of motor neuron diseases in Japan. BMC Med. Genom. 2024, 17, 263. [Google Scholar] [CrossRef] [PubMed]
- Hsieh-Li, H.M.; Chang, J.G.; Jong, Y.J.; Wu, M.H.; Wang, N.M.; Tsai, C.H.; Li, H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000, 24, 66–70. [Google Scholar] [CrossRef]
- Monani, U.R.; Sendtner, M.; Coovert, D.D.; Parsons, D.W.; Andreassi, C.; Le, T.T.; Jablonka, S.; Schrank, B.; Rossoll, W.; Prior, T.W.; et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn−/− mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000, 9, 333–339. [Google Scholar] [CrossRef]
- Echaniz-Laguna, A.; Guiraud-Chaumeil, C.; Tranchant, C.; Reeber, A.; Melki, J.; Warter, J.M. Homozygous exon 7 deletion of the SMN centromeric gene (SMN2): A potential susceptibility factor for adult-onset lower motor neuron disease. J. Neurol. 2002, 249, 290–293. [Google Scholar] [CrossRef]
- Gamez, J.; Barceló, M.J.; Muñoz, X.; Carmona, F.; Cuscó, I.; Baiget, M.; Cervera, C.; Tizzano, E.F. Survival and respiratory decline are not related to homozygous SMN2 deletions in ALS patients. Neurology 2002, 59, 1456–1460. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).