From Model to Crop: Roles of Macroautophagy in Arabidopsis and Legumes
Abstract
1. Introduction
2. Molecular Mechanisms of Autophagy in Arabidopsis and Leguminous Plants
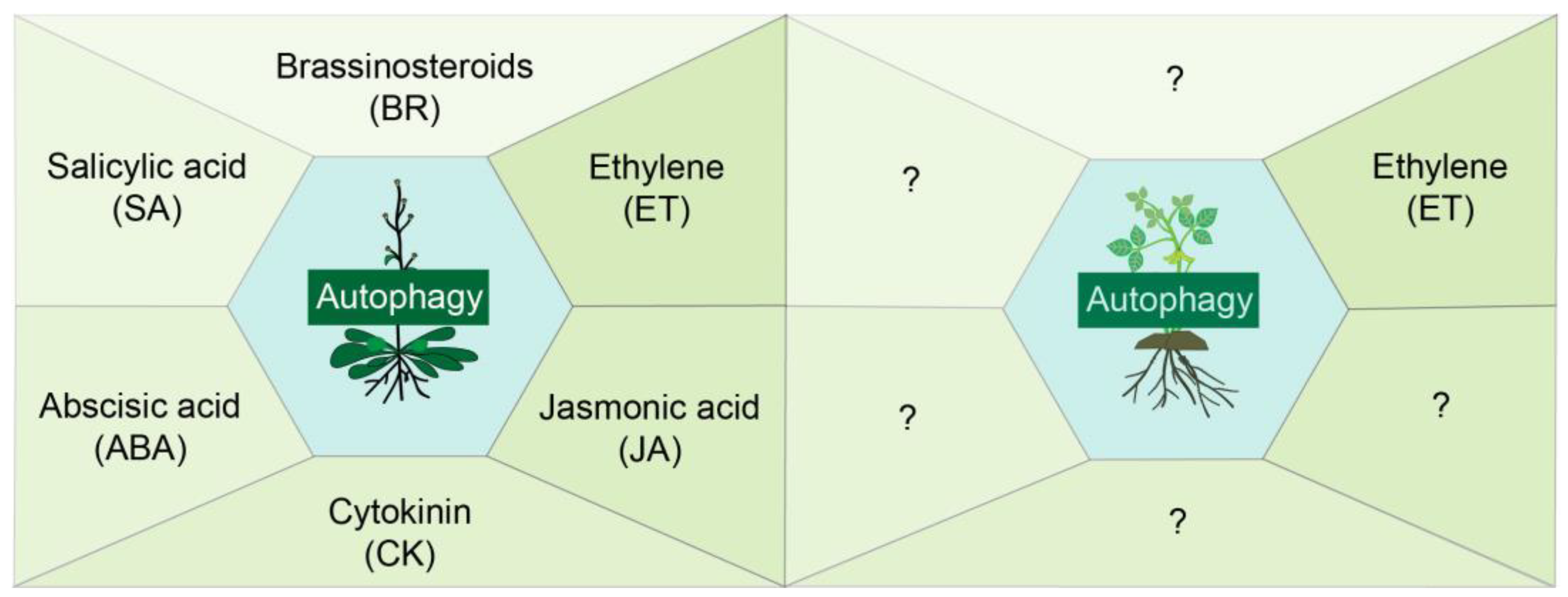
2.1. Phytohormones and Autophagy in Arabidopsis and Legumes
2.2. Transcription Factors and Epigenetic Regulation of Autophagy in Arabidopsis and Legumes
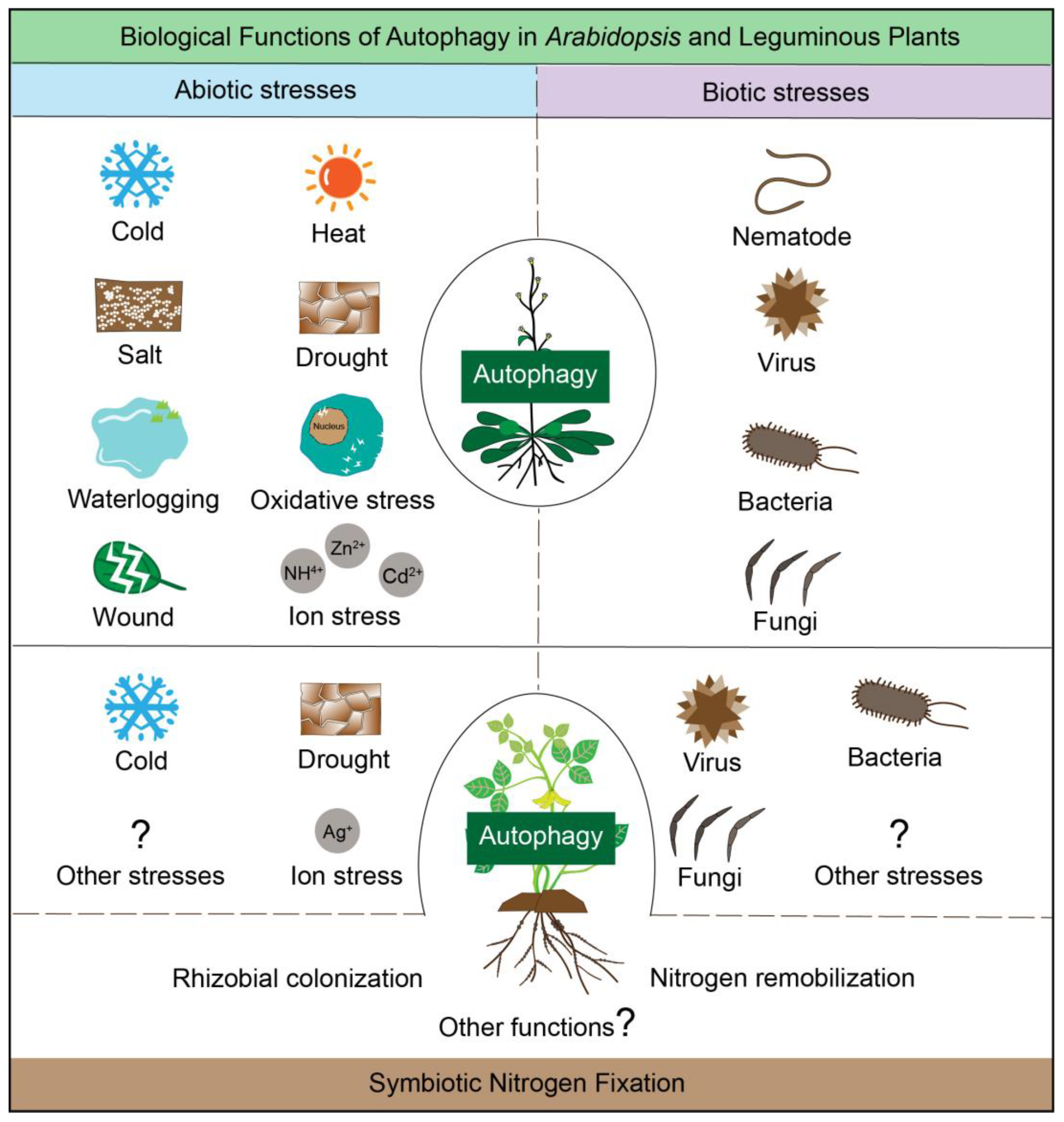
3. Biological Functions of Autophagy in Arabidopsis and Leguminous Plants
3.1. Biological Functions of Autophagy in Arabidopsis
3.2. Autophagy in Abiotic Stress Tolerance of Legumes
3.3. Autophagy in Biotic Stress Defense of Legumes
3.4. Autophagy in Seed Development and Symbiotic Nitrogen Fixation of Legumes
4. Genome-Wide Identification of ATG8 Gene Family in Peanut
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhong, Y.; Tian, J.; Li, X.; Liao, H. Cooperative interactions between nitrogen fixation and phosphorus nutrition in legumes. New Phytol. 2023, 237, 734–745. [Google Scholar] [CrossRef]
- Jan, N.; Rather, A.M.; John, R.; Chaturvedi, P.; Ghatak, A.; Weckwerth, W.; Zargar, S.M.; Mir, R.A.; Khan, M.A.; Mir, R.R. Proteomics for abiotic stresses in legumes: Present status and future directions. Crit. Rev. Biotechnol. 2023, 43, 171–190. [Google Scholar] [CrossRef]
- Lu, Z.X.; He, J.F.; Zhang, Y.C.; Bing, D.J. Composition, physicochemical properties of pea protein and its application in functional foods. Crit. Rev. Food Sci. Nutr. 2020, 60, 2593–2605. [Google Scholar] [CrossRef]
- Yadav, R.K.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Asati, R.; Patel, V.; Sikarwar, R.S.; Payasi, D.K. Breeding and Genomic Approaches towards Development of Fusarium Wilt Resistance in Chickpea. Life 2023, 13, 988. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Farooq, A.; Sharma, V.; Sofi, S.A.; Bashir, S.; Rai, P.K.; Sofi, P.A.; Deveshwar, P.; Zargar, S.M. A comprehensive multi-omics approach for understanding common bean’s (Phaseolus vulgaris L.) response to phosphorus stress. Plant Sci. 2025, 359, 112632. [Google Scholar] [CrossRef]
- Wang, N.; Wang, T.; Chen, Y.; Wang, M.; Lu, Q.; Wang, K.; Dou, Z.; Chi, Z.; Qiu, W.; Dai, J.; et al. Microbiome convergence enables siderophore-secreting-rhizobacteria to improve iron nutrition and yield of peanut intercropped with maize. Nat. Commun. 2024, 15, 839. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, H.; Zhu, X.; Li, Y.; Zhang, B.; Tadege, M.; Wu, S.; Qi, Z.; Xia, Z. Functional Genomics: From Soybean to Legume. Int. J. Mol. Sci. 2025, 26, 6323. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Su, Y.; Jiang, S.; Liang, W.; Lou, Z.; Frugier, F.; Xu, P.; Murray, J.D. Applying conventional and cell-type-specific CRISPR/Cas9 genome editing in legume plants. aBIOTECH 2025, 6, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Nzepang, D.T.; Cissoko, M.; Gully, D.; Hocher, V.; Rami, J.F.; Fall, S.; Fonceka, D.; Svistoonoff, S. Transcriptomic analysis reveals genetic factors underlying impaired symbiotic nitrogen fixation in lines derived from crosses between cultivated peanut (Arachis hypogaea L.) and its wild ancestors. BMC Genom. 2025, 26, 556. [Google Scholar] [CrossRef]
- Wu, N.; Feng, Y.; Jiang, T.; Li, Z.; Liu, Y.; Yuan, M. Genome-wide study and expression analysis of soybean ERF transcription factors and overexpression of GmERF205 enhances drought resistance in soybean. BMC Genom. 2025, 26, 726. [Google Scholar] [CrossRef]
- Martinez, M.A.; Montechiarini, N.H.; Gosparini, C.O.; Oppedijk, B.; van Duijn, B. Chlorophyll Fluorescence, Oxygen Consumption Rates and Germination of Green Soybean Seeds Produced Under Heat-Drought Stress. Plant Direct 2025, 9, e70100. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, X.; Wei, X.; Li, X.; Sun, J.; Dong, S. The impact of drought stress under different soil matrices on physiological characteristics of soybean seedlings. AoB Plants 2025, 17, plaf026. [Google Scholar] [CrossRef]
- Yu, L.; Gao, Y.; Yang, Q.; Liu, J.; Liu, T.; Mo, W.; Liu, H.; Tian, Z.; Li, L. Large-scale pairing identifies a soybean phytocytokine-receptor module conferring disease resistance. Nat. Plants 2025, 11, 1739–1747. [Google Scholar] [CrossRef]
- Tian, M.; Sun, Y.; Zhang, G.; Xu, Y.; Zhu, J.; Huang, W.; Wang, Y.; Zhang, B.; Li, Z.; Lin, S.; et al. Metabolomics navigates natural variation in pathogen-induced secondary metabolism across soybean cultivar populations. Proc. Natl. Acad. Sci. USA 2025, 122, e2505532122. [Google Scholar] [CrossRef]
- Buntic, A.; Jelusic, A.; Milic, M.; Dimitrijevic, S.; Buzurovic, U.; Milinkovic, M.; Knezevic, M. Antimicrobial peptides (AMP)-producing Bacillus spp. for the management of Fusarium infection and alfalfa growth promotion. Pest. Manag. Sci. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Silva Volpato, N.D.; Gomez, F.M.; Gimenez, V.D.; Ciampitti, I.A. A global dataset on mungbean for managing seed yield and quality. Sci. Data 2025, 12, 658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zou, Y.; Zhou, W.; Li, C.; Zuo, L.; Miao, L.; Cui, X. Current status and obstacles of narrowing yield gaps of four major crops. J. Sci. Food Agric. 2025, 105, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, R.; Zhu, L. Chaperone-Mediated Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 435–452. [Google Scholar] [CrossRef] [PubMed]
- Plott, S.; Dagdas, Y.F.; Ibl, V. Microautophagy in cereal grains: Protein storage or degradation? Trends Plant Sci. 2025, 30, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, O.K. N-degron-mediated ATG8 isoform switching controls plant thermotolerance. Autophagy 2025, 21, 2528–2530. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, X.; Otegui, M.S. Plant autophagy: New flavors on the menu. Curr. Opin. Plant Biol. 2018, 46, 113–121. [Google Scholar] [CrossRef]
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 2014, 19, 905–921. [Google Scholar] [CrossRef]
- Wang, Q.; Hou, S. The emerging roles of ATG1/ATG13 kinase complex in plants. J. Plant Physiol. 2022, 271, 153653. [Google Scholar] [CrossRef]
- Sawa-Makarska, J.; Baumann, V.; Coudevylle, N.; von Bulow, S.; Nogellova, V.; Abert, C.; Schuschnig, M.; Graef, M.; Hummer, G.; Martens, S. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 2020, 369, eaaz7714. [Google Scholar] [CrossRef]
- Cook, A.S.I.; Chen, M.; Nguyen, T.N.; Cabezudo, A.C.; Khuu, G.; Rao, S.; Garcia, S.N.; Yang, M.; Iavarone, A.T.; Ren, X.; et al. Structural pathway for PI3-kinase regulation by VPS15 in autophagy. Science 2025, 388, eadl3787. [Google Scholar] [CrossRef]
- Popelka, H.; Klionsky, D.J. When an underdog becomes a major player: The role of protein structural disorder in the Atg8 conjugation system. Autophagy 2024, 20, 2338–2345. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Li, B.; Jiang, L. Autophagosome biogenesis and organelle homeostasis in plant cells. Plant Cell 2024, 36, 3009–3024. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. A tissue-specific rescue strategy reveals the local roles of autophagy in leaves and seeds for resource allocation. Plant Physiol. 2024, 197, kiae647. [Google Scholar] [CrossRef]
- Kim, S.H.; Kwon, C.; Lee, J.H.; Chung, T. Genes for plant autophagy: Functions and interactions. Mol. Cells 2012, 34, 413–423. [Google Scholar] [CrossRef]
- Wada, S.; Hayashida, Y.; Izumi, M.; Kurusu, T.; Hanamata, S.; Kanno, K.; Kojima, S.; Yamaya, T.; Kuchitsu, K.; Makino, A.; et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 2015, 168, 60–73. [Google Scholar] [CrossRef]
- Sera, Y.; Hanamata, S.; Sakamoto, S.; Ono, S.; Kaneko, K.; Mitsui, Y.; Koyano, T.; Fujita, N.; Sasou, A.; Masumura, T.; et al. Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci. Rep. 2019, 9, 18544. [Google Scholar] [CrossRef]
- Zhen, X.; Xu, F.; Zhang, W.; Li, N.; Li, X. Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS ONE 2019, 14, e0223011. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Shi, M.; Luo, X.; Huang, Y.; Zeng, H.; Liu, Y.; Huang, Y.; Xu, P.; Qian, Y.; et al. Duplicated OsATG9 Genes Antagonise Autophagy to Balance Growth and Drought Tolerance in Rice. Plant Cell Environ. 2025, 48, 6397–6401. [Google Scholar] [CrossRef]
- Liu, M.; Ma, L.; Tang, Y.; Yang, W.; Yang, Y.; Xi, J.; Wang, X.; Zhu, W.; Xue, J.; Zhang, X.; et al. Maize Autophagy-Related Protein ZmATG3 Confers Tolerance to Multiple Abiotic Stresses. Plants 2024, 13, 1637. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, Z.; Liu, M.; Wang, Z.; Zhang, X.; Guo, X.; Zhang, J.; Hu, J.; Zhu, W.; Li, Q.; et al. Manipulation of the central autophagy component ZmATG8c affects thermotolerance in maize. Plant Physiol. 2025, 199, kiaf299. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, F.; Meng, C.; Si, J.; Zhang, W.; Sun, J.; Wang, Z. Identification of the ZmATG18 subfamily genes in maize and the role of ZmATG18a in drought stress. BMC Genom. 2025, 26, 777. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.M.; Wu, N.N.; Ran, J.; Liu, J.Z. Silencing Autophagy-Related Gene 2 (ATG2) Results in Accelerated Senescence and Enhanced Immunity in Soybean. Int. J. Mol. Sci. 2021, 22, 11749. [Google Scholar] [CrossRef] [PubMed]
- Borek, S.; Stefaniak, S.; Nuc, K.; Wojtyla, L.; Ratajczak, E.; Sitkiewicz, E.; Malinowska, A.; Swiderska, B.; Wleklik, K.; Pietrowska-Borek, M. Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds. Int. J. Mol. Sci. 2023, 24, 11773. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Wang, W.; Luo, N.; Li, H.; Liu, J.; Wang, Y.; Ye, Y.; Zhu, H.; Li, F.; Yu, H.; et al. MtNAD1 associates with the autophagy complex to contribute to the degradation of immunity-related proteins in Medicago truncatula nodules. New Phytol. 2025, 245, 2186–2201. [Google Scholar] [CrossRef]
- Agbemafle, W.; Wong, M.M.; Bassham, D.C. Transcriptional and post-translational regulation of plant autophagy. J. Exp. Bot. 2023, 74, 6006–6022. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.Y.; Wang, P.; Yin, Y.; Bassham, D.C. Interactions between autophagy and phytohormone signaling pathways in plants. FEBS Lett. 2022, 596, 2198–2214. [Google Scholar] [CrossRef]
- Yang, C.; Luo, M.; Zhuang, X.; Li, F.; Gao, C. Transcriptional and Epigenetic Regulation of Autophagy in Plants. Trends Genet. 2020, 36, 676–688. [Google Scholar] [CrossRef]
- Liao, C.Y.; Pu, Y.; Nolan, T.M.; Montes, C.; Guo, H.; Walley, J.W.; Yin, Y.; Bassham, D.C. Brassinosteroids modulate autophagy through phosphorylation of RAPTOR1B by the GSK3-like kinase BIN2 in Arabidopsis. Autophagy 2023, 19, 1293–1310. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009, 21, 2914–2927. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, G.; Wang, H.; Zhou, Z. The relationship between ethylene-induced autophagy and reactive oxygen species in Arabidopsis root cells during the early stages of waterlogging stress. PeerJ 2023, 11, e15404. [Google Scholar] [CrossRef]
- Laureano-Marin, A.M.; Aroca, A.; Perez-Perez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic Acid-Triggered Persulfidation of the Cys Protease ATG4 Mediates Regulation of Autophagy by Sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Acheampong, A.K.; Shanks, C.; Cheng, C.Y.; Schaller, G.E.; Dagdas, Y.; Kieber, J.J. EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc. Natl. Acad. Sci. USA 2020, 117, 27034–27043. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Yamamoto, H.; Hirakawa, T.; Uemura, A.; Pelayo, M.A.; Iimura, H.; Katagiri, N.; Takeda-Kamiya, N.; Kumaishi, K.; Shirakawa, M.; et al. Petal abscission is promoted by jasmonic acid-induced autophagy at Arabidopsis petal bases. Nat. Commun. 2024, 15, 1098. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Nang, M.P.; Oshima, K.; Ishibashi, Y.; Zheng, S.H.; Yuasa, T.; Iwaya-Inoue, M. The ethylene signal mediates induction of GmATG8i in soybean plants under starvation stress. Biosci. Biotechnol. Biochem. 2011, 75, 1408–1412. [Google Scholar] [CrossRef]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Lee, J.; Kang, M.H.; Choi, D.M.; Marmagne, A.; Park, J.; Lee, H.; Gwak, E.; Lee, J.C.; Kim, J.I.; Masclaux-Daubresse, C.; et al. Phytochrome-interacting factors PIF4 and PIF5 directly regulate autophagy during leaf senescence in Arabidopsis. J. Exp. Bot. 2025, 76, 1068–1084. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef]
- Yang, M.K.; Zhu, X.J.; Chen, C.M.; Guo, X.; Xu, S.X.; Xu, Y.R.; Du, S.X.; Xiao, S.; Mueller-Roeber, B.; Huang, W.; et al. The plant circadian clock regulates autophagy rhythm through transcription factor LUX ARRHYTHMO. J. Integr. Plant Biol. 2022, 64, 2135–2149. [Google Scholar] [CrossRef]
- Li, X.; Liao, J.; Bai, H.; Bei, J.; Li, K.; Luo, M.; Shen, W.; Yang, C.; Gao, C. Arabidopsis flowering integrator SOC1 transcriptionally regulates autophagy in response to long-term carbon starvation. J. Exp. Bot. 2022, 73, 6589–6599. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 Coordinates Organellar Functions and Stress Responses by Reprogramming Retrograde Signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef]
- Yang, C.; Shen, W.; Yang, L.; Sun, Y.; Li, X.; Lai, M.; Wei, J.; Wang, C.; Xu, Y.; Li, F.; et al. HY5-HDA9 Module Transcriptionally Regulates Plant Autophagy in Response to Light-to-Dark Conversion and Nitrogen Starvation. Mol. Plant 2020, 13, 515–531. [Google Scholar] [CrossRef]
- Ullah, I.; Magdy, M.; Wang, L.; Liu, M.; Li, X. Genome-wide identification and evolutionary analysis of TGA transcription factors in soybean. Sci. Rep. 2019, 9, 11186. [Google Scholar] [CrossRef] [PubMed]
- Varadharajan, V.; Rajendran, R.; Muthuramalingam, P.; Runthala, A.; Madhesh, V.; Swaminathan, G.; Murugan, P.; Srinivasan, H.; Park, Y.; Shin, H.; et al. Multi-Omics Approaches Against Abiotic and Biotic Stress-A Review. Plants 2025, 14, 865. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Mittler, R.; Blumwald, E.; Zandalinas, S.I. Developing climate-resilient crops: Improving plant tolerance to stress combination. Plant J. 2022, 109, 373–389. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Samuel, M.A. A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal Behav. 2015, 10, e989749. [Google Scholar] [CrossRef]
- Peng, Y.; Guo, S.; Lei, B.; Yu, L.; Wang, Q. Autophagy deficiency confers freezing tolerance in Arabidopsis thaliana. BMC Plant Biol. 2025, 25, 994. [Google Scholar] [CrossRef]
- Zhang, Y.; Min, H.; Shi, C.; Xia, G.; Lai, Z. Transcriptome analysis of the role of autophagy in plant response to heat stress. PLoS ONE 2021, 16, e0247783. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, J.S.; Lee, M.H.; Seo, J.; Kim, J.; Yang, W.S.; Park, J.; Yoo, K.; Choi, J.; Seo, J.B.; et al. The N-degron pathway governs autophagy to promote thermotolerance in Arabidopsis. Nat. Commun. 2025, 16, 5889. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, J.; Yang, C.; Zhu, X.; Li, J.; Zheng, X.; Li, X.; Chen, S.; Feng, L.; Wang, P.; et al. A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat. Plants 2023, 9, 749–765. [Google Scholar] [CrossRef]
- Li, B.; Niu, F.; Zeng, Y.; Tse, M.K.; Deng, C.; Hong, L.; Gao, S.; Lo, S.W.; Cao, W.; Huang, S.; et al. Ufmylation reconciles salt stress-induced unfolded protein responses via ER-phagy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2208351120. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Fan, S.; Yang, C.; Hu, W.; Liu, F. Proteomics revealed novel functions and drought tolerance of Arabidopsis thaliana protein kinase ATG1. BMC Biol. 2025, 23, 48. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.J.; Huang, L.; Tan, W.J.; Zhai, N.; Yuan, L.B.; Zhou, Y.; Yu, L.J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 2007, 3, 257–258. [Google Scholar] [CrossRef]
- Katz, E.; Chamovitz, D.A. Wounding of Arabidopsis leaves induces indole-3-carbinol-dependent autophagy in roots of Arabidopsis thaliana. Plant J. 2017, 91, 779–787. [Google Scholar] [CrossRef]
- Calero-Munoz, N.; Exposito-Rodriguez, M.; Collado-Arenal, A.M.; Rodriguez-Serrano, M.; Laureano-Marin, A.M.; Santamaria, M.E.; Gotor, C.; Diaz, I.; Mullineaux, P.M.; Romero-Puertas, M.C.; et al. Cadmium induces reactive oxygen species-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. 2019, 42, 2696–2714. [Google Scholar] [CrossRef]
- Robert, G.; Yagyu, M.; Lascano, H.R.; Masclaux-Daubresse, C.; Yoshimoto, K. A proposed role for endomembrane trafficking processes in regulating tonoplast content and vacuole dynamics under ammonium stress conditions in Arabidopsis root cells. Plant Signal Behav. 2021, 16, 1924977. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, D.; Tanoi, K.; Yoshimoto, K. Optimal Distribution of Iron to Sink Organs via Autophagy Is Important for Tolerance to Excess Zinc in Arabidopsis. Plant Cell Physiol. 2021, 62, 515–527. [Google Scholar] [CrossRef]
- Chen, J.; Chen, S.; Xu, C.; Yang, H.; Achom, M.; Wang, X. A key virulence effector from cyst nematodes targets host autophagy to promote nematode parasitism. New Phytol. 2023, 237, 1374–1390. [Google Scholar] [CrossRef]
- Wu, G.; Jia, Z.; Ding, K.; Zheng, H.; Lu, Y.; Lin, L.; Peng, J.; Rao, S.; Wang, A.; Chen, J.; et al. Turnip mosaic virus co-opts the vacuolar sorting receptor VSR4 to promote viral genome replication in plants by targeting viral replication vesicles to the endosome. PLoS Pathog. 2022, 18, e1010257. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef]
- Jeon, H.S.; Jang, E.; Kim, J.; Kim, S.H.; Lee, M.H.; Nam, M.H.; Tobimatsu, Y.; Park, O.K. Pathogen-induced autophagy regulates monolignol transport and lignin formation in plant immunity. Autophagy 2023, 19, 597–615. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.D.; Vierstra, R.D.; Nurnberger, T.; Gust, A.A. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal Behav. 2011, 6, 1040–1042. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Pandey, S.; Mishra, S.K.; Giri, V.P.; Agarwal, L.; Dwivedi, S.; Pandey, A.K.; Nautiyal, C.S.; Mishra, A. Omics-Based Mechanistic Insight Into the Role of Bioengineered Nanoparticles for Biotic Stress Amelioration by Modulating Plant Metabolic Pathways. Front. Bioeng. Biotechnol. 2020, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Samperna, S.; Masi, M.; Vurro, M.; Evidente, A.; Marra, M. Cyclopaldic Acid, the Main Phytotoxic Metabolite of Diplodia cupressi, Induces Programmed Cell Death and Autophagy in Arabidopsis thaliana. Toxins 2022, 14, 474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal Behav. 2011, 6, 1408–1410. [Google Scholar] [CrossRef]
- Wang, Y.; Nishimura, M.T.; Zhao, T.; Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011, 68, 74–87. [Google Scholar] [CrossRef]
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287. [Google Scholar] [CrossRef]
- Wang, F.X.; Luo, Y.M.; Ye, Z.Q.; Cao, X.; Liang, J.N.; Wang, Q.; Wu, Y.; Wu, J.H.; Wang, H.Y.; Zhang, M.; et al. iTRAQ-based proteomics analysis of autophagy-mediated immune responses against the vascular fungal pathogen Verticillium dahliae in Arabidopsis. Autophagy 2018, 14, 598–618. [Google Scholar] [CrossRef]
- Yang, M.; Wang, L.; Chen, C.; Guo, X.; Lin, C.; Huang, W.; Chen, L. Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress. Sci. Rep. 2021, 11, 22933. [Google Scholar] [CrossRef]
- Zhao, W.; Song, J.; Wang, M.; Chen, X.; Du, B.; An, Y.; Zhang, L.; Wang, D.; Guo, C. Alfalfa MsATG13 Confers Cold Stress Tolerance to Plants by Promoting Autophagy. Int. J. Mol. Sci. 2023, 24, 12033. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877. [Google Scholar] [CrossRef]
- Wu, X.; Chen, S.; Zhang, Z.; Zhou, W.; Sun, T.; Ning, K.; Xu, M.; Ke, X.; Xu, P. A viral small interfering RNA-host plant mRNA pathway modulates virus-induced drought tolerance by enhancing autophagy. Plant Cell 2024, 36, 3219–3236. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhang, Y.; Wu, H.; Bai, J. Drought stress-induced autophagy gene expression is correlated with carbohydrate concentrations in Caragana korshinskii. Protoplasma 2020, 257, 1211–1220. [Google Scholar] [CrossRef]
- Mo, F.; Li, H.; Li, Y.; Chen, X.; Wang, M.; Li, Z.; Deng, N.; Yang, Y.; Huang, X.; Zhang, R.; et al. Physiological, biochemical, and transcriptional regulation in a leguminous forage Trifolium pratense L. responding to silver ions. Plant Physiol. Biochem. 2021, 162, 531–546. [Google Scholar] [CrossRef]
- Hashimi, S.M.; Huang, M.J.; Amini, M.Q.; Wang, W.X.; Liu, T.Y.; Chen, Y.; Liao, L.N.; Lan, H.J.; Liu, J.Z. Silencing GmATG7 Leads to Accelerated Senescence and Enhanced Disease Resistance in Soybean. Int. J. Mol. Sci. 2023, 24, 16508. [Google Scholar] [CrossRef]
- Wang, B.; Sumit, R.; Sahu, B.B.; Ngaki, M.N.; Srivastava, S.K.; Yang, Y.; Swaminathan, S.; Bhattacharyya, M.K. Arabidopsis Novel Glycine-Rich Plasma Membrane PSS1 Protein Enhances Disease Resistance in Transgenic Soybean Plants. Plant Physiol. 2018, 176, 865–878. [Google Scholar] [CrossRef]
- Gomez, S.K.; Javot, H.; Deewatthanawong, P.; Torres-Jerez, I.; Tang, Y.; Blancaflor, E.B.; Udvardi, M.K.; Harrison, M.J. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Xiao, D.; Liu, D.; Chai, W.; Gong, Q.; Wang, N.N. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS ONE 2012, 7, e37217. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Ishibashi, Y.; Nakagawa, A.C.; Tomita, Y.; Iwaya-Inoue, M.; Arima, S.; Zheng, S.H. Nitrogen redistribution and its relationship with the expression of GmATG8c during seed filling in soybean. J. Plant Physiol. 2016, 192, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Wleklik, K.; Stefaniak, S.; Nuc, K.; Pietrowska-Borek, M.; Borek, S. Identification and Potential Participation of Lipases in Autophagic Body Degradation in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds. Int. J. Mol. Sci. 2023, 25, 90. [Google Scholar] [CrossRef]
- Quezada-Rodriguez, E.H.; Gomez-Velasco, H.; Arthikala, M.K.; Lara, M.; Hernandez-Lopez, A.; Nanjareddy, K. Exploration of Autophagy Families in Legumes and Dissection of the ATG18 Family with a Special Focus on Phaseolus vulgaris. Plants 2021, 10, 2619. [Google Scholar] [CrossRef]
- Hernandez-Lopez, A.; Diaz, M.; Rodriguez-Lopez, J.; Guillen, G.; Sanchez, F.; Diaz-Camino, C. Uncovering Bax inhibitor-1 dual role in the legume-rhizobia symbiosis in common bean roots. J. Exp. Bot. 2019, 70, 1049–1061. [Google Scholar] [CrossRef]
- Nanjareddy, K.; Blanco, L.; Arthikala, M.K.; Alvarado-Affantranger, X.; Quinto, C.; Sanchez, F.; Lara, M. A Legume TOR Protein Kinase Regulates Rhizobium Symbiosis and Is Essential for Infection and Nodule Development. Plant Physiol. 2016, 172, 2002–2020. [Google Scholar] [CrossRef]
- Semenova, M.G.; Petina, A.N.; Fedorova, E.E. Autophagy and Symbiosis: Membranes, ER, and Speculations. Int. J. Mol. Sci. 2024, 25, 2918. [Google Scholar] [CrossRef]
- Xue, Q.; Shen, C.; Liu, Q.; Liu, P.; Guo, D.; Zheng, L.; Liu, J.; Liu, C.; Ye, Q.; Wang, T.; et al. The PtdIns3P phosphatase MtMP promotes symbiotic nitrogen fixation via mitophagy in Medicago truncatula. iScience 2023, 26, 107752. [Google Scholar] [CrossRef]
- Gallardo, K.; Besson, A.; Klein, A.; Le Signor, C.; Aubert, G.; Henriet, C.; Terezol, M.; Pateyron, S.; Sanchez, M.; Trouverie, J.; et al. Transcriptional Reprogramming of Pea Leaves at Early Reproductive Stages. Front. Plant Sci. 2019, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Bhattacharyya, S.; Kumar, R.; Kumar, A.; Ibanez, F.; Wang, J.; Guo, B.; Sudini, H.K.; Gopalakrishnan, S.; DasGupta, M.; et al. Molecular Basis of Root Nodule Symbiosis between Bradyrhizobium and ‘Crack-Entry’ Legume Groundnut (Arachis hypogaea L.). Plants 2020, 9, 276. [Google Scholar] [CrossRef]
- Zhen, X.; Zheng, N.; Yu, J.; Bi, C.; Xu, F. Autophagy mediates grain yield and nitrogen stress resistance by modulating nitrogen remobilization in rice. PLoS ONE 2021, 16, e0244996. [Google Scholar] [CrossRef]
- Zhen, X.; Li, X.; Yu, J.; Xu, F. OsATG8c-Mediated Increased Autophagy Regulates the Yield and Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2019, 20, 4956. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zheng, X.; Xie, D.; Zhou, H.; Shao, S.; Zhou, J. Autophagic pathway contributes to low-nitrogen tolerance by optimizing nitrogen uptake and utilization in tomato. Hortic. Res. 2022, 9, uhac068. [Google Scholar] [CrossRef]
- Huang, W.; Ma, D.N.; Liu, H.L.; Luo, J.; Wang, P.; Wang, M.L.; Guo, F.; Wang, Y.; Zhao, H.; Ni, D.J. Genome-Wide Identification of CsATGs in Tea Plant and the Involvement of CsATG8e in Nitrogen Utilization. Int. J. Mol. Sci. 2020, 21, 7043. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jia, X.; Huo, L.; Che, R.; Gong, X.; Wang, P.; Ma, F. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018, 41, 469–480. [Google Scholar] [CrossRef]
- Thanthrige, N.; Bhowmik, S.D.; Williams, B. ‘Friend versus foe’-does autophagy help regulate symbiotic plant-microbe interactions and can it be manipulated to improve legume cultivation? FEBS Lett. 2025, 599, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Liu, Y.; Fan, Y.; Qiu, D.; Li, Z.; Gong, F.; Yin, D. Research Progress on High-Protein Peanut (Arachis hypogaea L.) Varieties in China. Plants 2025, 14, 2917. [Google Scholar] [CrossRef]
- Huang, R.; Li, H.; Gao, C.; Yu, W.; Zhang, S. Advances in omics research on peanut response to biotic stresses. Front. Plant Sci. 2023, 14, 1101994. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Dong, J.; Wang, T. Autophagy in Plant Abiotic Stress Management. Int. J. Mol. Sci. 2021, 22, 4075. [Google Scholar] [CrossRef]
- Gross, A.S.; Raffeiner, M.; Zeng, Y.; Ustun, S.; Dagdas, Y. Autophagy in Plant Health and Disease. Annu. Rev. Plant Biol. 2025, 76, 197–227. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Pei, D.; Zhang, W.; Sun, H.; Wei, X.; Yue, J.; Wang, H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014, 33, 1697–1710. [Google Scholar] [CrossRef]
- Chung, T.; Suttangkakul, A.; Vierstra, R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009, 149, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Song, I.; Hong, S.; Huh, S.U. Identification and Expression Analysis of the Solanum tuberosum StATG8 Family Associated with the WRKY Transcription Factor. Plants 2022, 11, 2858. [Google Scholar] [CrossRef] [PubMed]
- Honig, A.; Avin-Wittenberg, T.; Galili, G. Selective autophagy in the aid of plant germination and response to nutrient starvation. Autophagy 2012, 8, 838–839. [Google Scholar] [CrossRef]
- Miklaszewska, M.; Zienkiewicz, K.; Klugier-Borowska, E.; Rygielski, M.; Feussner, I.; Zienkiewicz, A. CALEOSIN 1 interaction with AUTOPHAGY-RELATED PROTEIN 8 facilitates lipid droplet microautophagy in seedlings. Plant Physiol. 2023, 193, 2361–2380. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Weckwerth, W.; Varshney, R.K. Proteomics and Metabolomics: Two Emerging Areas for Legume Improvement. Front. Plant Sci. 2015, 6, 1116. [Google Scholar] [CrossRef] [PubMed]

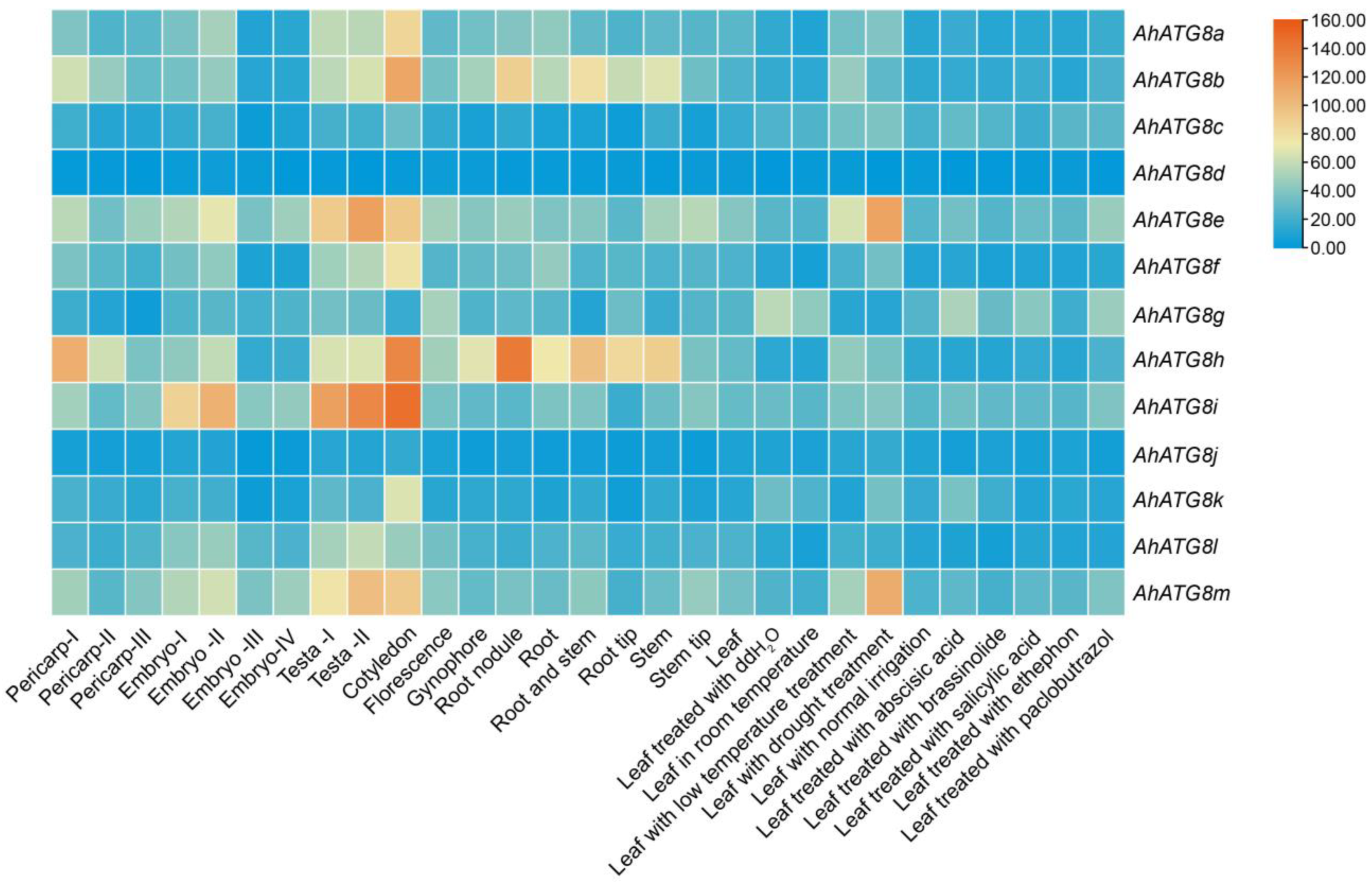
| Member | Locus | Chromosomal Location | Gene Length/bp | Protein Length/bp |
|---|---|---|---|---|
| AhATG8a | arahy.Tifrunner.gnm1.ann1.JG4KI1.1 | Chr01:39903752-39907161 | 3410 | 156 |
| AhATG8b | arahy.Tifrunner.gnm1.ann1.EXRI64.1 | Chr01:103592801-103594956 | 2156 | 119 |
| AhATG8c | arahy.Tifrunner.gnm1.ann1.IDQ1YW.1 | Chr04:4930217-4936705 | 6489 | 230 |
| AhATG8d | arahy.Tifrunner.gnm1.ann1.6U9WS0.1 | Chr06:103978667-103981122 | 2456 | 125 |
| AhATG8e | arahy.Tifrunner.gnm1.ann1.63V2PN.1 | Chr09:18979110-18985083 | 5974 | 118 |
| AhATG8f | arahy.Tifrunner.gnm1.ann1.0A7WSD.1 | Chr11:48330080-48333436 | 3357 | 122 |
| AhATG8g | arahy.Tifrunner.gnm1.ann1.SY21CD.1 | Chr11:132703325-132704747 | 1423 | 123 |
| AhATG8h | arahy.Tifrunner.gnm1.ann1.8SMT51.1 | Chr11:137923012-137926868 | 3857 | 177 |
| AhATG8i | arahy.Tifrunner.gnm1.ann1.X3Q019.1 | Chr13:17577237-17580265 | 3029 | 120 |
| AhATG8j | arahy.Tifrunner.gnm1.ann1.CQM6UK.1 | Chr14:6182864-6185138 | 2275 | 119 |
| AhATG8k | arahy.Tifrunner.gnm1.ann1.E4CLSM.1 | Chr15:15136078-15138924 | 2847 | 114 |
| AhATG8l | arahy.Tifrunner.gnm1.ann1.ZFW017.1 | Chr18:7807375-7809476 | 2102 | 120 |
| AhATG8m | arahy.Tifrunner.gnm1.ann1.HJA00B.1 | Chr19:24321913-24327978 | 6066 | 136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Cui, X.; Gao, M.; Wang, Z. From Model to Crop: Roles of Macroautophagy in Arabidopsis and Legumes. Genes 2025, 16, 1343. https://doi.org/10.3390/genes16111343
Feng L, Cui X, Gao M, Wang Z. From Model to Crop: Roles of Macroautophagy in Arabidopsis and Legumes. Genes. 2025; 16(11):1343. https://doi.org/10.3390/genes16111343
Chicago/Turabian StyleFeng, Lanlan, Xiaowei Cui, Meng Gao, and Zhenyu Wang. 2025. "From Model to Crop: Roles of Macroautophagy in Arabidopsis and Legumes" Genes 16, no. 11: 1343. https://doi.org/10.3390/genes16111343
APA StyleFeng, L., Cui, X., Gao, M., & Wang, Z. (2025). From Model to Crop: Roles of Macroautophagy in Arabidopsis and Legumes. Genes, 16(11), 1343. https://doi.org/10.3390/genes16111343






