Abstract
Pa0665 in Pseudomonas aeruginosa shares homologous sequences with that of the essential A-type iron–sulfur (Fe-S) cluster insertion protein ErpA in Escherichia coli. However, its essentiality in P. aeruginosa and its complementation with E. coli erpA has not been experimentally examined. To fulfill this task, we constructed plasmid-based ts-mutant Δpa0665/pTS-pa0665 using a three-step protocol. The mutant displayed growth defects at 42 °C, which were complemented by expressing ec.erpA. Microscopic observations indicated a petite cell phenotype for Δpa0665/pTS-pa0665 at 42 °C, correlated with the downregulation of the oprG gene. RNA sequencing revealed significant transcriptional changes in genes associated with the oxidative phosphorylation (OXPHOS) system, aligning with reduced ATP levels in Δpa0665/pTS-pa0665 under 42 °C. Additionally, the ts-mutant showed heightened sensitivity to H2O2 at 42 °C. Overall, our study demonstrates the essential role of pa0665 for OXPHOS function and is complemented by ec.erpA. We propose that the plasmid-based ts-allele is useful for genetic analysis of essential genes of interest in P. aeruginosa.
1. Introduction
Pseudomonas aeruginosa, a member of the Pseudomonadaceae family, is a Gram-negative, rod-shaped, motile bacterium that thrives in both aerobic and anaerobic conditions. It is widely found in nature, isolated from soil, plants, water, and animals, and is known for its versatility and opportunism [1]. The high mortality associated with Pseudomonas is mainly due to the emergence of drug-resistant strains, leading the WHO to prioritize it for research and new drug development [2].
Essential genes, which are crucial for microbial survival, are promising targets for drug development. Through the application of Tn-seq, Lee et al. identified 352 essential genes in P. aeruginosa [3], enriching the pool of potential targets for novel therapeutic interventions. Notably, 40 of these genes encode hypothetical proteins, whose functions and roles in cellular processes remain to be elucidated [3].
Hypothetical proteins, often identified across different lineages without functional validation, are a valuable resource for uncovering new biological insights and therapeutic targets [4,5]. The experimental analysis of these functionally unknown essential core genes will contribute to the understanding of new fundamental functions necessary for Pseudomonas growth [6]. This study is focused on pa0665, an essential gene coding for a hypothetical protein in P. aeruginosa PAO1. Comparative protein sequence analysis revealed that Pa0665 shares about 65.5% homologous sequences with ErpA, an A-type iron–sulfur (Fe-S) cluster insertion protein, deemed essential in Escherichia coli [7]; this is crucial for cellular respiration under aerobic conditions, as it contributes to the synthesis of isopentenyl diphosphate (IPP), a precursor for essential electron carriers like ubiquinone and menaquinone [8]. The absence of ErpA markedly diminishes complex I (NADH: ubiquinone oxidoreductase) content and NADH oxidase activity in the cytoplasmic membrane, significantly impacting the assembly of complex I [9], a testament to its essential role.
Despite the identified homology and the established essentiality of erpA in E. coli (ec.erpA), the functional importance of pa0665 in P. aeruginosa and its potential complementation with ec.erpA has not been empirically verified. Addressing this gap, we constructed the plasmid-based temperature-sensitive (ts) mutant of Δpa0665/pTS-pa0665 through our previously established three-step protocol [10,11]. We showed that Δpa0665/pTS-pa0665 has growth defects at restrictive temperature (42 °C), reversible by expressing ec.erpA. Microscopic observations revealed a smaller cell phenotype for Δpa0665/pTS-pa0665 at 42 °C, possibly linked to the downregulation of the oprG gene. RNA sequencing showed significant upregulation and downregulation of genes associated with cytochrome o ubiquinol oxidase and cytochrome c cbb3-type oxidase, indicating oxidative phosphorylation (OXPHOS) system disturbances in the pa0665 ts-mutant, consistent with reduced ATP in pa0665 ts-mutant cells. Taken together, our results highlight the essential role of pa0665 in the functioning of the OXPHOS system, which is complemented by ec.erpA. We propose that the plasmid-based ts-allele complementation presented here is a robust tool to unveil the molecular and biological function of unknown essential genes in P. aeruginosa.
2. Materials and Methods
2.1. DNA, Plasmids, and Bacterial Cultures
The oligonucleotides, plasmids, and bacterial strains used in this study are shown in Table 1. Strains were cultivated in LB (1 L: 10 g tryptone, 10 g NaCl, 5 g yeast extract, pH 7.0) liquid or solid medium supplemented with antibiotics (100 µg mL−1 ampicillin, 50 µg mL−1 gentamicin, and 100 µg mL−1 tetracycline) and chemicals (e.g., 10% sucrose or 0.2% arabinose) at 30 °C or 42 °C, as indicated.

Table 1.
Oligonucleotides, plasmids, and strains used in this study.
2.2. Plasmid Construction
We used the same deletion plasmid and rescue plasmid (or ts plasmid) constructed in the previous study [10,11]. To construct the pa0665 deletion and rescue plasmids, the deletion cassette and rescue cassette of pa0665 were cloned into the deletion plasmid and rescue plasmid, respectively, using the ClonExpress II one-step cloning kit (Vazyme, Nanjing, China). Overexpression plasmids were constructed by cloning the araC-PBAD promoter fragment and downstream gene fragment into the pBBR1MCS-5 plasmid [12] using the Vazyme cloning kit.
2.3. Plasmid-Based ts-Mutant Strain Construction
We used a three-step protocol that we developed previously [10,11] to construct the plasmid-based ts-lethal mutant strain Δpa0665/pTS-pa0665. Briefly, we first electroporated the deletion plasmid, which could not auto-replicate in P. aeruginosa, into the P. aeruginosa PAO1 strain, isolating integrants via a single crossover into the genome on a gentamicin-containing LB plate. Then, the rescue plasmid was transformed into the integrant on a tetracycline-containing plate. Subsequently, the counterselection of sacB generated the chromosomal Δpa0665 allele by looping out the integrated plasmid on a sucrose-containing plate. The resulting strains were PCR validated for the chromosomal Δpa0665 allele and assessed for ts-growth phenotype via spot-plating assay.
2.4. Spot-Plating Assay
The spot-plating assay [13] was employed to assess sensitivities to stress factors, including antibiotics, sucrose, hydrogen peroxide, and temperature. In brief, 10-fold serially diluted cultures were transferred onto LB plates supplemented with the relevant stress factors using a 48-pin replicator (V&P Scientific, Inc., San Diego, CA, USA) and incubated at 30 °C or 42 °C as required.
2.5. Fluorescence Microscopic Analysis
Cell morphology was investigated under the Olympus BX53 microscope (Olympus, Tokyo, Japan) using the phase contrast configuration. Nile red fluorescent dye was used to visualize the cytoplasmic membrane.
2.6. Fluorescence Activated Cell Sorting (FACS) Analysis
Cells were fixed with 70% ethanol for 30 min and washed with PBS 3 times. We utilized the green fluorescent dye PicoGreen (Solarbio, Beijing, China) to distinguish bacterial cells from other small particulate impurities in the liquid. Fixed cells were resuspended in PBS to a final concentration of OD600 = 0.6, and the green dye, PicoGreen, was added. The PicoGreen concentrated solution provided by the supplier was diluted 1:200 in dimethyl sulfoxide (DMSO) and added to cells at a ratio of 5 μL of diluted dye to 100 μL of cells. The cells were stained for 30 min at room temperature and diluted with 1 mL of PBS containing a 1:1000 dilution of PicoGreen. Stained cells were subjected to FACS analysis using Becton Dickinson FACS Calibur (BD Biosciences, San Diego, CA, USA) with a 488 nm laser. Data were processed with CellQuest software (version 5.1; BD Biosciences).
2.7. ATP Content Measurement
Cells were centrifuged at 10,000× g for 5 min at 4 °C. The pellets were treated with a lysis buffer from an ATP detection kit (Beyotime, Haimen, China) for 1 min at room temperature and then were centrifuged at 10,000× g for 5 min. The supernatant was transferred to a new 1.5 mL tube for an ATP test with the ATP detection kit purchased from Beyotime (China). The relative ATP content was determined using the formula: relative ATP content = ATP value/protein value. Protein concentration in the sample was quantified using a Bradford 1× Dye Reagent (Bio-Rad, Hercules, CA, USA), measured at a wavelength of 595 nm.
2.8. RNA Extraction and RNA-Seq Analysis
Total RNA was extracted in triplicate from four cell samples (wt and ts-mutant strains, incubated for 6 h at 30 °C and 42 °C after the second subculture) using a TaKaRa Bio Inc. RNA extraction kit and quality-checked with an Agilent 2100 Bioanalyzer. Afterward, samples underwent DNase I treatment (TaKaRa) and rRNA removal with an Epicentre Biotechnologies Ribo-Zero magnetic kit. The resulting rRNA-depleted RNA underwent RNA-seq on an Illumina HiSeq 2500 at the Shanghai Human Genome Centre (Shanghai, China) using paired-end (PE150) sequencing. For library construction, 100 ng of RNA was used with the NEB Next Ultra Directional RNA library prep kit. Data analysis was carried out on the RaNA-Seq [14] cloud platform, with raw data cleaned using Fastp software (version 0.2) [15] and mapped to the reference genome (Pseudomonas Genome Database version 22.1; www.pseudomonas.com accessed on 15 March 2024) using salmon [16]. Gene expression was normalized to TPM (transcripts per kilobase million), with differentially expressed genes (DEGs) identified using DEseq2 [17], based on a >4-fold change and p value < 0.01.
2.9. Statistics
Data are presented as mean ± standard error. The statistical significance of differences was assessed using an unpaired, two-tailed Student’s t-test. A p-value < 0.05 was deemed statistically significant.
2.10. Data Availability
The RNA-seq raw data sets were submitted to NCBI with the accession numbers PRJNA1085980 for wt and ts mutant at 30 °C and 42 °C.
3. Results
3.1. pa0665 Gene Is Essential for Growth on LB-Agar Plate
The construction of bacterial strains lacking essential genes typically led to the emergence of suppressors [18]. To circumvent this issue, we removed the gene pa0665 from the chromosome while keeping a complimentary copy in a temperature-sensitive (ts) suicide plasmid. Using a detailed three-step process [10,11], we successfully engineered a chromosomal deletion of pa0665 (Δpa0665), safeguarded by an identical, plasmid-based pa0665 gene under the control of its native promoter in plasmid pTS−pa0665 (Figure 1A). The Δpa0665/pTS-pa0665 ts-mutant strain was validated using PCR with primers F1/R1 and F2/R2 (Figure 1A), where F1/R1 sites are present, and F2/R2 sites are absent in the complementary copy of the rescue plasmid pTS-pa0665 (Figure 1B,C). Spot-plating assays showed that Δpa0665/pTS-pa0665 experienced reduced growth at 42 °C compared to the wild type, which displayed similar growth patterns at 30 °C (Figure 1D). These findings affirm pa0665’s essential role in growth in LB medium. Removing the high-copy ts-plasmid from the mutant’s cells would take several generations at higher temperatures; hence, we developed a sequential subculturing approach, reducing plasmid numbers in the first stage and allowing phenotype expression in the second subculture as described in our previous work [11]. The second subculture showed the growth defect of Δpa0665/pTS-pa0665 at 42 °C (Figure 1E).
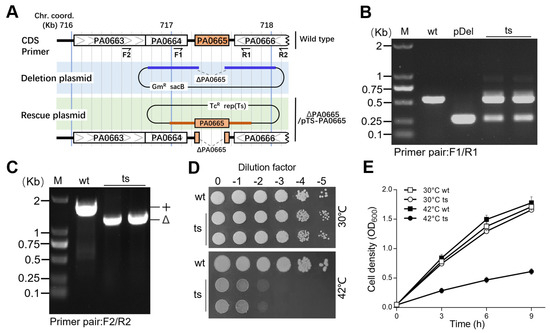
Figure 1.
Δpa0665/pTS-pa0665 exhibits growth defect on LB at the restrictive temperature. (A) The pa0665 deletion allele cassette (blue line) in the deletion plasmid and complementary sequences (brown line) in the rescue plasmid are shown in a physical map. Δpa0665/pTS-pa0665 (ts) carries a deletion allele, Δpa0665, on the chromosome, along with a complementary copy of pa0665 controlled by a native promoter on a temperature-sensitive plasmid. (B,C) PCR assays for pa0665 alleles use F1-R1 and F2-R2 primer pairs located inside and outside the pa0665 complementary sequences on the rescue cassette, revealing the chromosomal deletion allele in Δpa0665/pTS-pa0665 isolates. (D) Spot-plating assay displays Δpa0665/pTS-pa0665 growth, where 10-fold serial dilutions of wild-type and mutant cells were spotted on an LB plate and incubated overnight at 30 °C and 42 °C. (E) Growth curves of the second subcultures are shown, with time (h) on the x-axis and cell density (OD600) on the y-axis.
3.2. Putative Ortholog erpA in E. coli Functionally Complements the Defect of pa0665 in P. aeruginosa
Using BLASTp alignment (www.ncbi.nlm.nih.gov/blast accessed on 15 March 2024), we discovered that the protein sequences of Pa0665 from P. aeruginosa and ErpA from E. coli share 65.5% identity across 116 overlapping residues (Figure 2A). This level of identity, exceeding 40%, is sufficient to suggest a structural similarity between the two proteins [19]. To determine whether ec.erpA is a true ortholog of pa0665 in P. aeruginosa, a complementation experiment was performed. We engineered overexpression constructs for pa0665-OE and ec.erpA-OE, with transcription regulated by the arabinose-inducible PBAD promoter [20] in the multi-host pBBR1MCS-5 plasmid [12], resulting in pOE-pa0665 and pOE-ec.erpA constructs. Spot-plating assay indicated that under no inducer arabinose, multi-host plasmid pOE-ec.erpA at leakage expression level was sufficient to rescue the growth defect of Δpa0665/pTS-pa0665 at 42 °C, similar to the positive control plasmid pOE-pa0665 (Figure 2B, see arrowheads). Mild induction of ec.erpA with 0.02% arabinose rescued the growth defect of the Δpa0665/pTS-pa0665 pOE-ec.erpA strain at 42 °C (Figure 2B, see arrow), while strong induction of ec.erpA with 0.2% arabinose impeded the growth of Δpa0665/pTS-pa0665 pOE-erpA strain and the wild type (Figure 2B, see rectangles). These findings confirm that leaky expression and mild induction of ec.erpA can functionally compensate for the growth defect caused by pa0665 deficiency in P. aeruginosa.
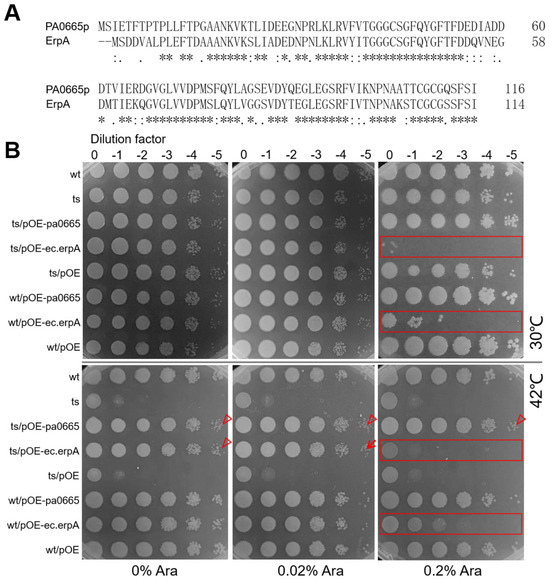
Figure 2.
Putative ortholog erpA from E. coli rescues the growth defect of Δpa0665/pTS-pa0665 at 42 °C. (A) Protein sequence alignment between Pa0665 and ErpA. Asterisk (*) indicates positions where residues are identical across all sequences; Colon (:) indicates conservation between groups of strongly similar properties; Dot (.) indicates conservation between groups of weakly similar properties. (B) Spot-plating assay. No induction (0 arabinose) or mild induction (0.02% arabinose) of erpA-OE rescues the growth defect of Δpa0665/pTS-pa0665 at 42 °C (see down row arrowhead and arrow). No mild and strong induction (0.2% arabinose) of pa0665-OE rescues the growth defect of Δpa0665/pTS-pa0665 at 42 °C (see up row arrowheads). Strong induction of erpA-OE hampers the growth of Δpa0665/pTS-pa0665 and wild-type cells (see rectangles); wt/pOE and ts/pOE served as the plasmid control.
3.3. The Δpa0665/pTS-pa0665 Mutant Exhibits Petite Cell Morphology under Restrictive Temperature
To investigate the impact of pa0665 depletion on cell morphology, we examined the terminal phenotype of Δpa0665/pTS-pa0665 at 42 °C. Both the mutant and wild-type strains underwent a temperature shift from 30 °C to 42 °C. Samples from the second subculture at 0 h, 3 h, 6 h, and 9 h at 42 °C were fixed and stained with Nile red for fluorescence microscopy. At 0 h, Δpa0665/pTS-pa0665 displayed a wild-type-like rod-shaped morphology (Figure 3A, top row). At 3 h and 6 h, the mutant cells exhibited a petite phenotype (Figure 3A, middle two rows, see arrowheads). By 9 h, ghost cells or lysed cells began to appear alongside petite cells (Figure 3A, bottom row, see arrowheads). After growth for 9 h in the second subculture, in addition to the petite cells, ghost cells or lysed cells started to appear (Figure 3A, bottom row, see arrowheads). Fluorescence-activated cell sorting (FACS) analysis corroborated the presence of the petite phenotype in Δpa0665/pTS-pa0665 cells under 42 °C (Figure 3B). These findings suggest that the depletion of pa0665 in P. aeruginosa leads to a significant reduction in cell size.
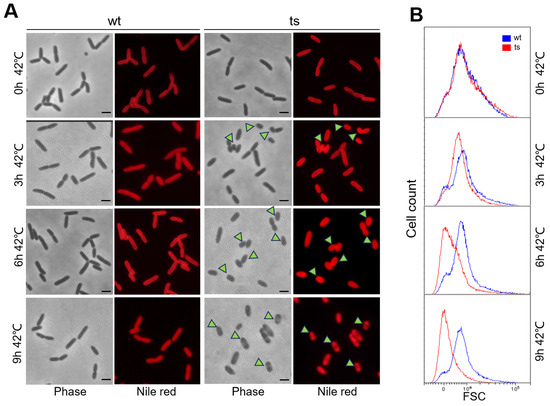
Figure 3.
Δpa0665/pTS-pa0665 exhibits petite cell morphology under restrictive temperature. (A) The cells were examined prior to the temperature shift from 30 °C to 42 °C during the first subculture or at 0 h, 3 h, 6 h, and 9 h after the second subculture began at 42 °C. Petite cell morphology is indicated by arrowheads, with a 1 μm scale bar shown. (B) FACS analysis of cell size, with the x-axis representing cell size and the y-axis indicating cell count.
3.4. ATP Content Was Decreased in Δpa0665/pTS-pa0665 Mutant under Restrictive Temperature
Fe-S cluster insertion proteins were required for an oxidative phosphorylation (OXPHOS) system that generates energy molecule ATP for cell growth. To evaluate whether the pa0665 deficiency impacts cellular ATP levels, we measured the ATP content in the Δpa0665/pTS-pa0665 mutant at 42 °C. The results showed that at 3 h, 6 h, and 9 h of incubation at 42 °C after a second subculture, the Δpa0665/pTS-pa0665 mutant exhibited a significant reduction in ATP content, which was not observed at 30 °C, indicating a temperature-sensitive phenotype (Figure 4A). In addition, we evaluated whether supplementing ATP could rescue the growth defect of Δpa0665/pTS-pa0665 at 42 °C. Serial dilutions of cultures were spot-plated on LB-agar supplemented with 0, 1 mM, or 2 mM ATP. Contrary to our expectations, exogenous ATP with 1 mM or 2 mM did not restore the growth of the Δpa0665/pTS-pa0665 mutant at 42 °C (Figure 4B), suggesting that the growth defect at the restrictive temperature is not simply due to ATP depletion but likely involves more complex metabolic disturbances.
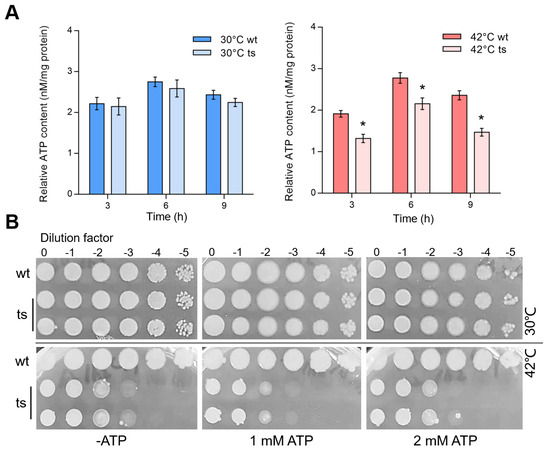
Figure 4.
Reduced ATP levels in Δpa0665/pTS-pa0665 mutant at 42°C and exogenous ATP fails to rescue lethal phenotype. (A) Intracellular ATP Level after the temperature shift to 42 °C compared to that at 30 °C in Δpa0665/pTS-pa0665 (ts) and wild-type (wt) cells. Cells for ATP assays were collected at 3 h, 6 h, and 9 h after the second subculture was started at 42 °C. *, p < 0.05; n = 3. (B) Spot-plating assay shows that supplementing with 1 mM ATP or 2 mM ATP concentrations does not compensate Δpa0665/pTS-pa0665 growth defect at 42 °C.
3.5. The Δpa0665/pTS-pa0665 Mutant Is Hypersensitive to Oxidative Stress Mediated by H2O2
Iron–sulfur (Fe-S) cluster proteins are critical for a variety of cellular processes, including electron transfer, enzyme activation, and the regulation of gene expression [21,22]. These proteins play a significant role in antioxidant activities by facilitating redox reactions and protecting cells from oxidative damage [23]. To evaluate the oxidative stress tolerance of the Δpa0665/pTS-pa0665 mutant at 42 °C, we conducted a growth curve experiment over a period of 5 h to assess bacterial sensitivity to hydrogen peroxide (H2O2). This duration was strategically chosen considering the known degradation of H2O2 over time. Since H2O2’s most pronounced effects occur shortly after its application, we standardized the initial OD600 of all strain samples, ensuring an equal oxidative challenge during H2O2’s effective period. We exposed both the wild-type and Δpa0665/pTS-pa0665 strains to 0.03% H2O2 in LB medium for 5 h at 30 °C and 42 °C following a second subculture. Upon exposure to 0.03% H2O2, both strains demonstrated a minor growth decline (Figure 5). Notably, at 42 °C, the Δpa0665/pTS-pa0665 mutant displayed a pronounced vulnerability to H2O2, indicating a higher sensitivity at 42 °C in comparison to 30 °C (Figure 5). These findings suggest that the presence of pa0665 is critical for protection against oxidative stress caused by H2O2.
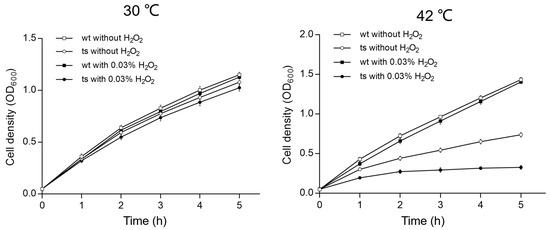
Figure 5.
Sensitivity of the Δpa0665/pTS-pa0665 mutant to H2O2 compared to wild type. Growth curves of Δpa0665/pTS-pa0665 (ts) and wild-type (wt) at 30 °C and 42 °C after second subcultures. The x and y axes show the time (h) treated with or without H2O2 and cell density (OD600), respectively.
3.6. Transcriptomic Analysis Reveals Impaired Oxidative Phosphorylation in pa0665-Deficient P. aeruginosa
To elucidate the impact of pa0665 deletion on the gene expression profile of P. aeruginosa and identify genes potentially associated with oxidative phosphorylation, we performed transcriptome sequencing (RNA-seq) analysis under both 30 °C and 42 °C for the wild type and the Δpa0665/pTS-pa0665 mutant in triplicate (Figure 6A). This comprehensive analysis revealed 721 genes with significant transcriptional changes between the two temperatures in the Δpa0665/pTS-pa0665 (ts) mutant, diverging markedly from the wild type (Rts/Rwt change > 4-fold, FDR-adjusted p-value < 0.01, n = 3) (Figure 6B). Notably, the top differentially expressed genes include those encoding components of the cytochrome o ubiquinol oxidase and cytochrome c cbb3-type oxidase (Figure 6C,D), underscoring a substantial disturbance in the oxidative phosphorylation (OXPHOS) system of the mutant. These findings suggest that pa0665 plays a critical role in maintaining OXPHOS functionality.
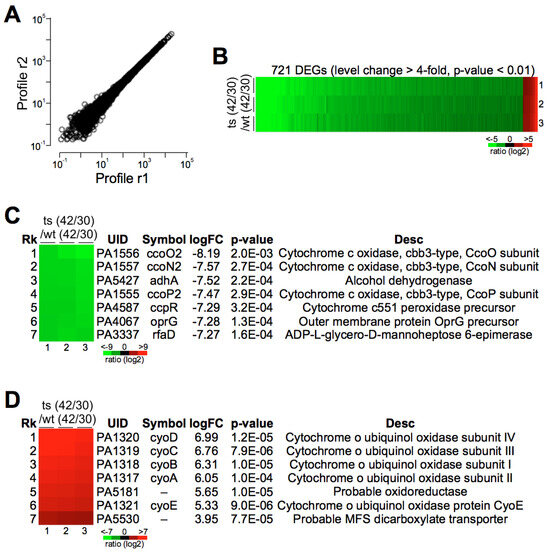
Figure 6.
Transcriptional changes in Δpa0665/pTS−pa0665 mutant compared to wild-type at 42°C and 30 °C. (A) Reproducibility of samples. The scatter plot represents the correlation of gene expression data. The x and y axes, respectively, represent the gene expression levels that were randomly taken from two technical replicates, illustrating a high degree of linear correlation indicative of good reproducibility. (B) Heatmap of differentially expressed genes (DEGs). Displaying 721 genes identified with differential expression (fold change > 4, p-value < 0.01) between the Δpa0665/pTS-pa0665 and wild-type strains. The color gradient in the heatmap corresponds to the log2 fold change in expression levels, ranging from −5 (red) to +5 (green). (C,D) Top 7 most upregulated or downregulated genes in the Δpa0665/pTS-pa0665 compared to the wild-type.
3.7. Impairment of pa4067/oprG Possibly Linked to the Altered Morphology of Δpa0665/pTS-pa0665 Mutant at 42 °C
Transcriptomic analyses revealed that pa3337/rfaD and pa4067/oprG were among the top seven significantly downregulated genes (Figure 6C), both critical for the structural integrity of the bacterial outer membrane. oprG plays a key role in forming outer membrane channels in Gram-negative bacteria, crucial for molecular transport and affecting bacterial virility and antibiotic resistance [24], while rfaD is vital for lipopolysaccharide (LPS) biosynthesis, a key outer membrane component in Gram-negative bacteria [25,26]. To investigate the impact of disrupting pa3337/rfaD and pa4067/oprG on cellular morphology, we constructed knockout plasmids containing sequences approximately 500 bp identical to the N-terminal coding sequences of pa3337/rfaD and pa4067/oprG. These plasmids were then inserted into the corresponding genes of the wild-type P. aeruginosa strain through homologous recombination, creating knockout mutants for pa3337/rfaD and pa4067/oprG. Fluorescence microscopy revealed a decrease in cell size for the pa4067/oprG mutants compared to wild-type cells (Figure 7A). In contrast, cell size in pa3337/rfaD mutants did not differ significantly from that of wild-type cells, a conclusion supported by flow cytometry analysis (Figure 7B,C). These findings suggest that mutations in pa4067/oprG lead to decreased cell size, indicating a potential connection between diminished oprG expression and the smaller cell size observed in Δpa0665/pTS-pa0665 mutants at 42 °C.
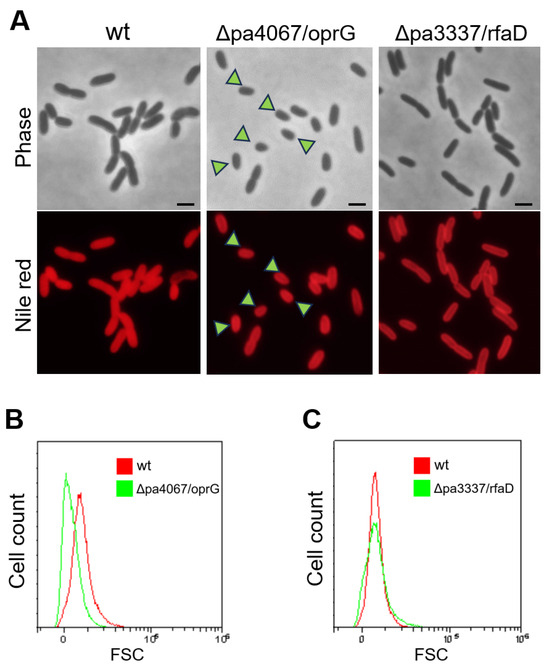
Figure 7.
Cellular morphology analysis of pa4067/oprG and pa3337/rfaD mutants. (A) Fluorescence microscopy analysis of Δpa4067/oprG and Δpa3337/rfaD mutant strains. Arrowheads indicate the petite cell morphology. A scale bar of 1 μm is shown. (B,C) FACS analysis of cell size. X and Y axes indicate cell size and cell count, respectively.
4. Discussion
This study contributed to the understanding of pa0665, a hypothetical protein in P. aeruginosa, by building on foundational work that identified essential genes in the PAO1 strain of P. aeruginosa, including pa0665 [3]. Our investigation provides insights into the functional significance of this gene, which was predicted to be essential but had not been experimentally validated. We constructed the plasmid-based temperature-sensitive (ts) lethal mutant strain, Δpa0665/pTS-pa0665, using a three-step protocol that we previously developed [10,11]. This approach enabled us to suggest the importance of pa0665 for the growth and survival of P. aeruginosa (Figure 1) and to conduct a functional complementation analysis with ec.erpA, a key A-type iron–sulfur (Fe-S) cluster insertion protein [8], and to validate their orthologship (Figure 2). The functional complementation by ec.erpA indicates a potential evolutionary conservation between these proteins and hints at common vulnerabilities in bacteria that might be of interest for further research in antimicrobial strategies.
Iron–sulfur (Fe–S) proteins are crucial for prokaryotic and eukaryotic cell metabolism [21,27,28]. In E. coli, erpA is involved in various metabolic pathways, including respiratory metabolism [8,9,29]. Our transcriptomic analysis of the pa0665 temperature-sensitive mutant revealed changes in gene expression related to the oxidative phosphorylation system, including alterations in the expression of genes involved in the electron transport chain (Figure 6), mainly cytochrome o ubiquinol oxidase and cytochrome c cbb3-type oxidase, that participate in the electron transport chain’s final steps [30,31]. These alterations align with the observed growth defects and reduced ATP levels (Figure 4) of Δpa0665/pTS-pa0665 under 42 °C, suggesting a possible role of pa0665 in oxidative phosphorylation functionality and cellular energy metabolism.
In response to oxidative stress, like exposure to hydrogen peroxide, E. coli activates defense mechanisms, including the expression of antioxidant enzymes. For example, the isc operon, which specifies Fe-S cluster formation and repair activities, is known to be induced by hydrogen peroxide, independent of the common oxidative stress regulators OxyR and SoxRS [32]. This suggests a direct link between Fe-S cluster biogenesis and the cellular response to oxidative stress. In this work, we found that the Δpa0665/pTS-pa0665 mutant displayed heightened sensitivity to oxidative stress under restrictive temperature, indicating a potential role for pa0665 in protecting against oxidative damage. It seems that disruption of pa0665 might impact Fe-S cluster assembly or repair, affecting enzymes involved in detoxifying reactive oxygen species.
Additionally, our results indicated a reduction in cell size associated with pa0665 depletion (Figure 3), correlated with the downregulation of oprG in our transcriptomic analysis (Figure 6), a gene encoding a major outer membrane protein [24,33]. This observation suggests a role for pa0665 in maintaining not only metabolic processes but also cellular structure and integrity.
In summary, while our study contributed to the understanding of the role of pa0665 in P. aeruginosa, it also highlighted areas for future investigation. These included its function in energy metabolism, oxidative stress response, and cellular integrity. The insights provided could contribute to the broader field of research on hypothetical proteins and essential genes and may inform future developments in antimicrobial therapies. The methodology used in this work, particularly the plasmid-based ts-allele approach, might offer a useful framework for genetic analysis in understanding the roles of essential genes in bacterial biology.
5. Conclusions
This study provided a foundational understanding of the pa0665 gene in P. aeruginosa, elucidating its significant role in cell morphology and oxidative phosphorylation. Our findings revealed that the Δpa0665/pTS-pa0665 mutant exhibits a distinct petite cell phenotype and altered ATP production under restrictive temperatures (42 °C). The lethal phenotype caused by the deletion of pa0665 can be reversed through the expression of the homologous ec.erpA gene, which underscores the potential functional similarities between these organisms. However, the specific mechanisms by which pa0665 influences cellular processes in P. aeruginosa require further investigation. This study contributed to the broader understanding of essential genes in bacteria and offers a basis for future research into the complex biology of P. aeruginosa, with potential implications for targeted antibiotic development.
Author Contributions
J.Z. and H.Z. carried out the biological and biochemical studies. Z.Y. conceived of the study, participated in its design and coordination, and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ24C010003.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
All authors declare no conflicts of interest.
References
- Vasil, M.L. Pseudomonas aeruginosa: Biology, mechanisms of virulence, epidemiology. J. Pediatr. 1986, 108, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Gallagher, L.A.; Thongdee, M.; Staudinger, B.J.; Lippman, S.; Singh, P.K.; Manoil, C. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 5189–5194. [Google Scholar] [CrossRef] [PubMed]
- Ijaq, J.; Chandrasekharan, M.; Poddar, R.; Bethi, N.; Sundararajan, V.S. Annotation and curation of uncharacterized proteins-challenges. Front. Genet. 2015, 6, 115944. [Google Scholar] [CrossRef] [PubMed]
- Naveed, M.; Chaudhry, Z.; Ali, Z.; Amjad, M. Annotation and curation of hypothetical proteins: Prioritizing targets for experimental study. Adv. Life Sci. 2018, 5, 73–87. [Google Scholar]
- Galperin, M.Y.; Koonin, E.V. ‘Conserved hypothetical’ proteins: Prioritization of targets for experimental study. Nucleic Acids Res. 2004, 32, 5452–5463. [Google Scholar] [CrossRef]
- Goodall, E.C.; Robinson, A.; Johnston, I.G.; Jabbari, S.; Turner, K.A.; Cunningham, A.F.; Lund, P.A.; Cole, J.A.; Henderson, I.R. The essential genome of Escherichia coli K-12. mBio 2018, 9, e02096-17. [Google Scholar] [CrossRef] [PubMed]
- Loiseau, L.; Gerez, C.; Bekker, M.; Ollagnier-de Choudens, S.; Py, B.; Sanakis, Y.; Teixeira de Mattos, J.; Fontecave, M.; Barras, F. ErpA, an iron–sulfur (Fe–S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 13626–13631. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, S.; Höfflin, S.; Friedrich, T. ErpA is important but not essential for the Fe/S cluster biogenesis of Escherichia coli NADH: Ubiquinone oxidoreductase (complex I). Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148286. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Z.; Zhu, J.; Ma, Y.; Wang, J.; Liu, J. Analysis of the Plasmid-Based ts Allele of PA0006 Reveals Its Function in Regulation of Cell Morphology and Biosynthesis of Core Lipopolysaccharide in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2022, 88, e00480-22. [Google Scholar] [CrossRef]
- Tian, L.; Yang, Z.; Wang, J.; Liu, J. Analysis of the Plasmid-Based ts-Mutant ΔfabA/pTS-fabA Reveals Its Lethality under Aerobic Growth Conditions That Is Suppressed by Mild Overexpression of desA at a Restrictive Temperature in Pseudomonas aeruginosa. Microbiol. Spectr. 2023, 11, e01338-23. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop II, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Hung, C.-W.; Martínez-Márquez, J.Y.; Javed, F.T.; Duncan, M.C. A simple and inexpensive quantitative technique for determining chemical sensitivity in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 11919. [Google Scholar] [CrossRef]
- Prieto, C.; Barrios, D. RaNA-Seq: Interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics 2020, 36, 1955–1956. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Charbon, G.; Riber, L.; Cohen, M.; Skovgaard, O.; Fujimitsu, K.; Katayama, T.; Løbner-Olesen, A. Suppressors of DnaAATP imposed overinitiation in Escherichia coli. Mol. Microbiol. 2011, 79, 914–928. [Google Scholar] [CrossRef]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Guzman, L.-M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Brzóska, K.; Meczyńska, S.; Kruszewski, M. Iron-sulfur cluster proteins: Electron transfer and beyond. Acta Biochim. Pol. 2006, 53, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Lill, R. The role of mitochondria in cellular iron–sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 2013, 5, a011312. [Google Scholar] [CrossRef]
- Saninjuk, K.; Romsang, A.; Duang-Nkern, J.; Wongsaroj, L.; Leesukon, P.; Dubbs, J.M.; Vattanaviboon, P.; Mongkolsuk, S. Monothiol Glutaredoxin Is Essential for Oxidative Stress Protection and Virulence in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2023, 89, e01714-22. [Google Scholar] [CrossRef]
- Touw, D.S.; Patel, D.R.; van den Berg, B. The crystal structure of OprG from Pseudomonas aeruginosa, a potential channel for transport of hydrophobic molecules across the outer membrane. PLoS ONE 2010, 5, e15016. [Google Scholar] [CrossRef]
- Coleman Jr, W.G. The rfaD gene codes for ADP-L-glycero-D-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J. Biol. Chem. 1983, 258, 1985–1990. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.; Guan, N.; Song, Y.; Li, Y.; Xie, X. A lipopolysaccharide synthesis gene rfaD from Mesorhizobium huakuii is involved in nodule development and symbiotic nitrogen fixation. Microorganisms 2022, 11, 59. [Google Scholar] [CrossRef]
- Barras, F.; Loiseau, L.; Py, B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 2005, 50, 41–101. [Google Scholar] [PubMed]
- Roche, B.; Aussel, L.; Ezraty, B.; Mandin, P.; Py, B.; Barras, F. Reprint of: Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1827, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Pinske, C.; Sawers, R.G. A-type carrier protein ErpA is essential for formation of an active formate-nitrate respiratory pathway in Escherichia coli K-12. J. Bacteriol. 2012, 194, 346–353. [Google Scholar] [CrossRef]
- Chepuri, V.; Lemieux, L.; Au, D.; Gennis, R.B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J. Biol. Chem. 1990, 265, 11185–11192. [Google Scholar] [CrossRef]
- Kučera, I.; Sedláček, V. Involvement of the cbb 3-type terminal oxidase in growth competition of Bacteria, biofilm formation, and in switching between denitrification and aerobic respiration. Microorganisms 2020, 8, 1230. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Wang, X.; Templeton, L.J.; Smulski, D.R.; LaRossa, R.A.; Storz, G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001, 183, 4562–4570. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Tamber, S.; Bains, M.; Maier, E.; Gellatly, S.; Lo, A.; Benz, R.; Hancock, R.E. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol. Lett. 2009, 296, 241–247. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).