Abstract
Reef-building coral species of the order Scleractinia play an important role in shallow tropical seas by providing an environmental base for the ecosystem. The molecular data of complete mitochondrial genome have become an important source for evaluating phylogenetic and evolutionary studies of Scleractinia. Here, the complete mitogenome of Homophyllia bowerbanki (Milne Edwards and Haime, 1857), collected from Nansha Islands of the South China Sea, was sequenced for the first time through a next-generation sequencing method. H. bowerbanki is the first species of its genus for which the mitogenome was sequenced. This mitogenome was 18,154 bp in size and included two transfer RNA genes (tRNAs), 13 protein-coding genes (PCGs), and two ribosomal RNA genes (rRNAs). It showed a similar gene structure and gene order to the other typical scleractinians. All 17 genes were encoded on the H strand and the total GC content was 33.86% in mitogenome. Phylogenetic analysis (maximum likelihood tree method) showed that H. bowerbanki belonged to the “Robust” clade and clustered together with other two species in the family Lobophylliidae based on 13 PCGs. The mitogenome can provide significant molecular information to clarify the evolutionary and phylogenetic relationships between stony corals and to facilitate their taxonomic classification; it can also support coral species monitoring and conservation efforts.
1. Introduction
Reef-building coral species of the order Scleractinia play a critical role in shallow tropical waters by supporting diverse and abundant marine life and providing an important environmental base for the ecosystem. The molecular data of complete mitochondrial genomes have become an important source for evaluating phylogenetic and evolutionary studies of Scleractinia as the cost of next-generation sequencing (NGS) technology decreases [1,2,3,4]. Multiple-gene analysis of nuclear and mitochondrial genes has already been used to infer phylogenetic relationships amongst scleractinians [5,6]. Nevertheless, only less than 130 complete mitogenomes of Scleractinia species can be obtained through the NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 17 February 2023)) to date, even though there are more than 1600 stony coral species [7].
H. bowerbanki (Milne Edwards and Haime, 1857) is a species of scleractinian coral with encrusting colonies. Its corallites are cerioid and usually with irregularly angular. Its septa are compact and columellae are small. It usually inhabits lower reef slopes protected from wave action. It shows different colours, such as pale grey, brown, or rust-coloured, and is often mottled [8]. H. bowerbanki was placed in the genus Acanthastrea by Veron (2000) and Budd et al. (2012) until Arrigoni et al. (2016) transferred it to Homophyllia. This genus belongs to the same family as Acanthastrea, Lobophylliidae based on a combination of morphological and three molecular data sets [8,9,10,11].
In this study, the complete mitochondrial genome of H. bowerbanki was sequenced and assembled for the first time through a next-generation sequencing method (NGS), in which its genome structure and nucleotide composition were characterized and analysed. Phylogenetic analyses of H. bowerbanki, based on 13 protein-coding genes (PCGs) of the mitogenome, will help determine its taxonomic classification and facilitate ongoing taxonomic revisions of scleractinian taxa [12]; analyses can also clarify the evolutionary and phylogenetic relationships amongst stony corals in favor of coral species monitoring and conservation efforts [13].
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
A specimen (Figure 1) of H. bowerbanki was collected from Nansha Islands of the South China Sea (9.9° N, 115.5° E) in 2020. Its fresh polyp was stored at −20 ℃ in 95% ethanol and then used to extract total genomic DNA (gDNA); its skeleton was kept in the Coral Sample Repository of Third Institute of Oceanography, Ministry of Natural Resources, with a unique code, 20200501-L1. The gDNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Shanghai, China). The integrity and concentration of the gDNA were measured using 1% agarose gel electrophoresis and NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).

Figure 1.
In situ live and skeleton photographs of H. bowerbanki.
2.2. Mitogenome Sequencing, Annotation, and Analyses
The gDNA sample was sent to Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) for next-generation sequencing. The sequencing library was generated using NEBNext® UltraTM DNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA, Catalog #: E7370L) according to the standard protocols. Genomic DNA was sheared into fragments about 400 bp using a Focused-ultrasonicator M200 (Covaris, Woburn, MA, USA). Then, DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing, followed by further PCR amplification. Next, PCR products were purified by AMPure XP system (Beckman Coulter, Beverly, MA, USA). Library quality control was performed using Agilent 5400 system (Agilent, Santa Clara, CA, USA) and qPCR (1.5 nM). Then, the library was pooled based on the effective concentration and targeted data amount (12 Gb of raw data). Next, 5′-end of the library was phosphorylated and cyclized. Subsequently, loop amplification was performed to generate DNA nanoballs. These DNA nanoballs were finally loaded into flow cell with DNBSEQ-T7 for paired-end 150 bp sequencing. FastQC was used to assess the quality and quantity of raw data [14]. Fastp was used to remove reads containing poly-N regions, adapters, and low-quality reads; then, a series of filtered clean reads was obtained [15]. These clean reads were applied to reconstruct the mitochondrial genome via NOVOPlasty 4.3.1 [16] with a parameter of K-mer 33. A total of 66,914 of 79,646,476 raw reads (approximately 0.08%) were de novo assembled to produce the mitogenome with the guidance of seed sequence (COI gene from GenBank: MG792550); the average sequencing coverage was 676×.
The circularised contig of H. bowerbanki generated by NOVOPlasty 4.3.1 was then submitted to the MITOS Web Server (http://mitos.bioinf.uni-leipzig.de/index.py (accessed on 10 February 2023)) for preliminary annotation [17]. We also identified and annotated PCG and rRNA genes by alignments of homologous mitogenomes from other scleractinians, especially in the same family, through online BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 February 2023)). Meanwhile, we validated the tRNA genes through ARWEN [18]. Finally, the whole mitogenome structure of H. bowerbanki was mapped using the online CGView Server (https://proksee.ca/ (accessed on 10 February 2023)) [19]. We obtained base composition, nucleotide frequencies, and codon usage through MEGA11 [20]. The skewing of the nucleotide composition was measured in terms of AT skews ((A − T)/(A + T)) and GC skews ((G − C)/(G + C)) [21,22].
2.3. Phylogenetic Analyses
The phylogenetic positions of H. bowerbanki were inferred using thirteen tandem mitogenome PCGs together with another forty-one species of Scleractinia and two species of Corallimorpharia (outgroup), which we downloaded from https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 10 February 2023) (Table 1) [22]. We used MEGA11 to select the best-fitting model based on Akaike Information Criterion (AIC); subsequently, a maximum likelihood (ML) tree was constructed with 500 bootstrap replicates under GTR + G + I model.

Table 1.
Representative Scleractinians and the outgroup species of Corallimorpharia used in the maximum likelihood phylogenetic tree.
3. Results and Discussion
3.1. Characteristics and Composition of Mitogenome
The complete mitogenome of H. bowerbanki was identified as a circular molecule with a similar gene order and gene structure to other typical scleractinians [23,24], which included thirteen PCGs (ND5, ND1, Cyt b, ND2, ND6, ATP6, ND4, CO III, CO II, ND4L, ND3, ATP8, and COI), two tRNA genes (tRNAMet and tRNATrp), and two rRNA genes (12S and 16S). The mitochondrial genome size of H. bowerbanki was 18,154 bp. The base composition was 24.75% for A, 13.32% for C, 21.75% for G, and 40.17% for T, which showed a higher AT (66.14%) bias. All of the seventeen genes were encoded on H strand and the total GC content was 33.86% in mitogenome. The GC content was one of the important compositional features of different genome regions. Identifying the driving force that shaped the GC content and deciphering the biological meaning of variations in the GC content will help us understand genome evolution [25]. The mitogenome GC content of most scleractinians ranged from 30% to 40% [22,26,27,28]. The intergenic regions of different genes ranged from −1 to 1506 bp. Moreover, four overlaps were detected. ND6 overlapped ATP6 by 1 bp, ATP6 overlapped ND4 by 1 bp, COII overlapped ND4L by 19 bp, and ND5 overlapped tRNATrp by 2 bp (Figure 2 and Figure 3; Table 2 and Table 3).
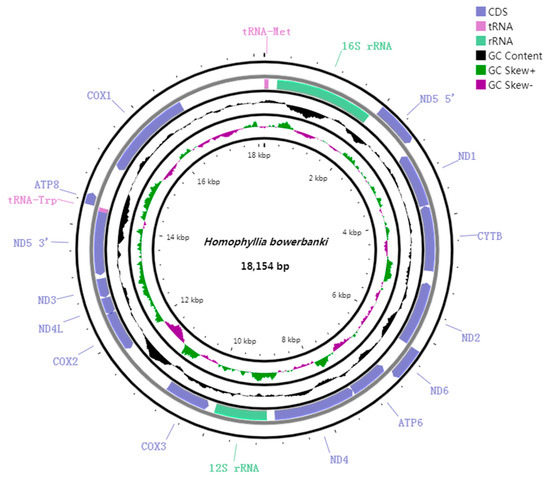
Figure 2.
Circular map of the mitogenome of H. bowerbanki. COX1, COX2, and COX3 refer to the cytochrome oxidase subunits, ND1–ND6 refer to NADH dehydrogenase components, and CYTB refers to cytochrome b. All of the seventeen genes are encoded on H strand.
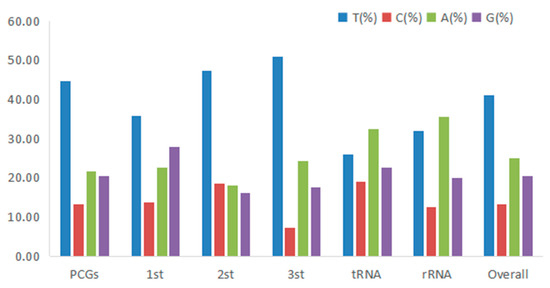
Figure 3.
Codon usage bias in different regions of the mitogenome of H. bowerbanki.

Table 2.
Organization of the mitogenome of H. bowerbanki.

Table 3.
Nucleotide composition in different regions of mitochondrial genome of H.bowerbanki.
3.2. Protein-Coding Genes
The total length of all 13 PCGs was 11,595 bp, with a base composition of 21.61% (A), 13.2% (C), 20.56% (G), and 44.63% (T), which showed that the PCGs preferred base AT. All the PCGs started with ATG, except for COIII, COI, and ND2. COIII started with GTG, COI and ND2 started with ATT. Five PCGs (ND1, Cyt b, ND4, COII, and ND5) terminated with TAG; the other eight PCGs (ND2, ND6, COIII, ATP6, ND3, ND4L, ATP8, and COI) stopped with TAA. Like other stony corals, the ND5 gene of H. bowerbanki also had an intron insertion with a length of 10,461 bp. ATP8 (198 bp) was the shortest gene and the longest gene was ND5 (1,815 bp) (Table 2 and Table 3). According to the analysis results of AT-skew and GC-skew, we can see both were negative (Table 3, Figure 4); meanwhile, all PCGs showed a stronger nucleotide asymmetry, with a higher AT-skew than GC-skew. Codon use frequency was higher amongst L, F, V, G, and S, accounting for 53.04% of a total of 3865 codons (Figure 5).
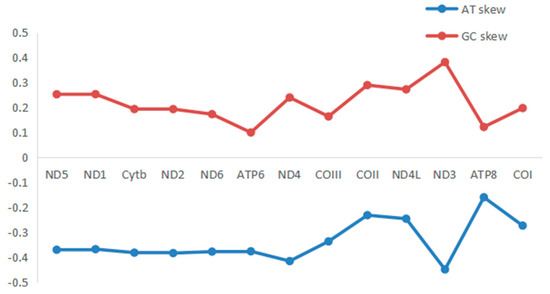
Figure 4.
AT−skew and GC−skew of PCGs in mitogenome of H. bowerbanki.
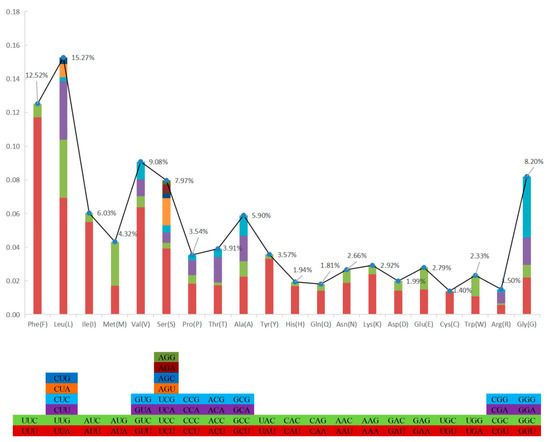
Figure 5.
Codon use frequency of PCGs in mitogenome of H. bowerbanki.
3.3. Transfer RNAs and Ribosomal RNAs
The 12S ribosomal RNA was 910 bp in size and located between ND4 and COIII; the 16S ribosomal RNA was 1,697 bp in size and located between tRNAMet and ND5. The base composition of two rRNAs was 35.52% A, 12.5% C, 20.02% G, and 31.95% T. The AT content of two rRNAs was 67.47%, and both AT-skew (0.053) and GC-skew (0.231) were positive. The two tRNA encoding genes (tRNAMet and tRNATrp) were 72 bp and 69 bp in size, respectively. The base composition of two tRNAs was 32.39% A, 19.01% C, 22.54% G, and 26.06% T. The AT content of two tRNAs was 58.45%, and both AT-skew (0.108) and GC-skew (0.085) were also positive. Both of the two tRNAs were folded into a typical cloverleaf structure, which included an amino acid accept arm, D loop, anticodon loop, and TψC loop (Table 3, Figure 6).
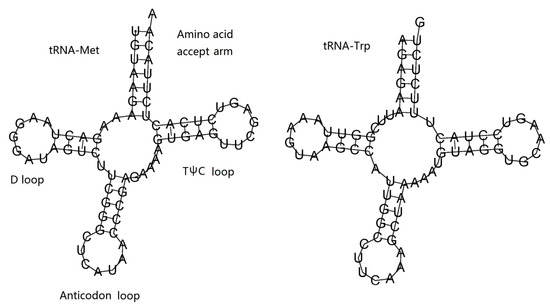
Figure 6.
Putative secondary structures of two tRNAs of H. bowerbanki.
3.4. Phylogenetic Analyses
In this study, the 13 PCGs in mitochondrial genomes of H. bowerbanki were tandem and established a phylogenetic relationship of Scleractinia. Numbers above the ML tree branches indicated bootstrap percentages (Figure 7). There were three distinct clades (“Complex,” “Robust,” and “Basal”) of Scleractinia in the tree; the ML topology tree showed that H. bowerbanki belonged to the “Robust” clade and clustered together with two other species in the family Lobophylliidae based on 13 PCGs. H. bowerbanki and S. maxima were once placed in the genus Acanthastrea, while a molecular method based on several tandem nuclear and mitochondrial DNA has helped to clarify their taxonomic classification [9,10,29,30]. As only fewer than a tenth of mitogenomes of scleractinians can be obtained through NCBI to date, more mitogenomes of other Scleractinia species should be sequenced. Furthermore, the continuous reduction of costs of NGS technologies can facilitate further studies on stony coral evolutionary and phylogenetic relationships and taxonomic classification.
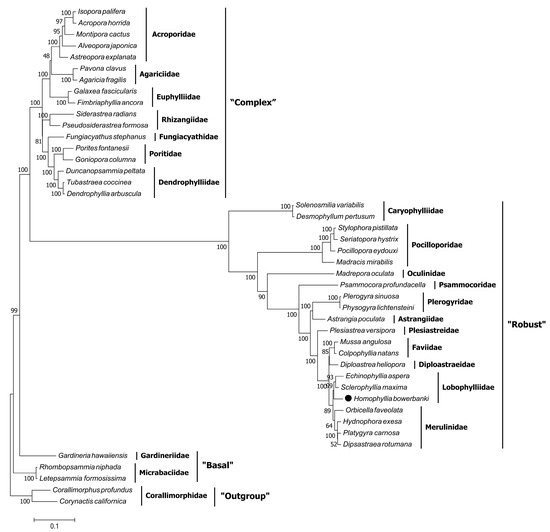
Figure 7.
Inferred maximum likelihood phylogenetic tree based on 13 tandem PCGs in mitochondrial genomes of H. bowerbanki. Numbers on branches are bootstrap percentages.
4. Conclusions
H. bowerbanki is the first species of its genus for which the mitogenome was sequenced. This mitogenome was 18,154 bp in size and included two transfer RNA genes (tRNAs), 13 protein-coding genes (PCGs), and two ribosomal RNA genes (rRNAs). It showed a similar gene structure and gene order to other typical scleractinians. All 17 genes in the mitogenome were encoded on the H strand and the total GC content of the mitogenome was 33.86%, which showed a higher AT bias. Phylogenetic analysis showed that H. bowerbanki belonged to the “Robust” clade and clustered together with two other species in the family Lobophylliidae based on 13 PCGs. The mitogenome can provide significant molecular information to clarify the evolutionary and phylogenetic relationships among stony corals and to facilitate their taxonomic classification; it can also support coral species monitoring and conservation efforts.
Author Contributions
J.X. and W.N. designed this experiment and B.C., Z.X., F.S. and Z.J. took part in investigation, sample processing, and data analysis. The original draft of the manuscript was written by P.T. and W.W. J.X. and W.N. contributed to the revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific Research Foundation of Third Institute of Oceanography, Ministry of Natural Resources (grant number 2022024), the National Key Research and Development Program of China (2021YFC3100503), and the National Science Foundation of China (grant number 42106143, 42006098, 42006128).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mitochondrial genome of H. bowerbanki is available from GeneBank under the accession number OP821698. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA918871, SRR22996606, and SAMN32610146, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fukami, H.; Omori, M.; Hatta, M. Phylogenetic relationships in the coral family Acroporidae, reassessed by inference from mitochondrial genes. Zool. Sci. 2000, 17, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, C.R.C.; Davy, S.K.; Pilling, G.M.; Graham, N.A.J. The Biology of Coral Reefs; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Jex, A.R.; Hall, R.S.; Littlewood, D.T.; Gasser, R.B. An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res. 2009, 38, 522–533. [Google Scholar] [CrossRef]
- Niu, W.; Yu, S.; Tian, P.; Xiao, J. Complete mitochondrial genome of Echinophyllia aspera (Scleractinia, Lobophylliidae): Mitogenome characterization and phylogenetic positioning. ZooKeys 2018, 793, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, M.V.; Fukami, H.; Benzoni, F.; Huang, D. The new systematics of Scleractinia: Integrating Molecular and Morphological Evidence. In The Cnidaria, Past, Present and Future; Springer, Cham: Switzerland, 2016; pp. 41–59. [Google Scholar] [CrossRef]
- Arrigoni, R.; Berumen, M.L.; Mariappan, K.G.; Beck, P.S.; Hulver, A.M.; Montano, S.; Pichon, M.; Strona, G.; Terraneo, T.I.; Benzoni, F. Towards a rigorous species delimitation framework for Scleractinian corals based on RAD sequencing: The case study of leptastrea from the Indo-Pacific. Coral Reefs 2020, 39, 1001–1025. [Google Scholar] [CrossRef]
- Hoeksema, B.W.; Cairns, S. Word List of Scleractinia. Available online: https://www.marinespecies.org/scleractinia/ (accessed on 14 February 2023).
- Veron, J.E.N.; Stafford-Smith, M.G.; Turak, E.; DeVantier, L.M. Corals of the World. Available online: http://www.coralsoftheworld.org/species_factsheets/?version=0.01 (accessed on 14 February 2023).
- Arrigoni, R.; Benzoni, F.; Huang, D.; Fukami, H.; Chen, C.A.; Berumen, M.L.; Hoogenboom, M.; Thomson, D.P.; Hoeksema, B.W.; Budd, A.F.; et al. When forms meet genes: Revision of the scleractinian genera Micromussa and Homophyllia (Lobophylliidae) with a description of two new species and one new genus. Contrib. Zool. 2016, 85, 387–422. [Google Scholar] [CrossRef]
- Huang, D.; Arrigoni, R.; Benzoni, F.; Fukami, H.; Knowlton, N.; Smith, N.D.; Stolarski, J.; Chou, L.M.; Budd, A.F. Taxonomic classification of the reef coral family Lobophylliidae (Cnidaria: Anthozoa: Scleractinia). Zool. J. Linn. Soc. 2016, 178, 436–481. [Google Scholar] [CrossRef]
- Budd, A.F.; Fukami, H.; Smith, N.D.; Knowlton, N. Taxonomic classification of the reef coral family Mussidae (Cnidaria: Anthozoa: Scleractinia). Zool. J. Linn. Soc. 2012, 166, 465–529. [Google Scholar] [CrossRef]
- Juszkiewicz, D.J.; White, N.E.; Stolarski, J.; Benzoni, F.; Arrigoni, R.; Hoeksema, B.W.; Wilson, N.G.; Bunce, M.; Richards, Z.T. Phylogeography of recent Plesiastrea (Scleractinia: Plesiastreidae) based on an integrated taxonomic approach. Mol. Phylogenetics Evol. 2022, 172, 107469. [Google Scholar] [CrossRef]
- Colin, L.; Yesson, C.; Head, C. Complete mitochondrial genomes of three reef forming Acropora corals (Acroporidae, Scleractinia) from Chagos Archipelago, Indian Ocean. Biodivers. Data J. 2021, 9, e72762. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 February 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Novoplasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. Arwen: A program to detect trna genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2007, 24, 172–175. [Google Scholar] [CrossRef]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2004, 21, 537–539. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11, Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Tian, P.; Jia, Z.; Cao, B.; Wang, W.; Xiao, J.; Niu, W. Complete mitochondrial genome sequences of Physogyra lichtensteini (Milne Edwards &Amp; Haime, 1851) and Plerogyra sinuosa (Dana, 1846) (Scleractinia, Plerogyridae): Characterisation and phylogenetic analysis. ZooKeys 2022, 1114, 21–34. [Google Scholar] [CrossRef]
- Lin, M.F.; Kitahara, M.V.; Tachikawa, H.; Fukami, H.; Miller, D.J.; Chen, C.A. Novel organization of the mitochondrial genome in the deep-sea coral, Madrepora oculata (Hexacorallia, Scleractinia, Oculinidae) and its taxonomic implications. Mol. Phylogenetics Evol. 2012, 65, 323–328. [Google Scholar] [CrossRef]
- Lin, M.F.; Kitahara, M.V.; Luo, H.; Tracey, D.; Geller, J.; Fukami, H.; Miller, D.J.; Chen, C.A. Mitochondrial genome rearrangements in the Scleractinia/ Corallimorpharia complex: Implications for coral phylogeny. Genome Biol. Evol. 2014, 6, 1086–1095. [Google Scholar] [CrossRef]
- Singh, R.; Ming, R.; Yu, Q. Comparative analysis of GC content variations in plant genomes. Trop. Plant Biol. 2016, 9, 136–149. [Google Scholar] [CrossRef]
- Niu, W.; Xiao, J.; Tian, P.; Yu, S.; Guo, F.; Wang, J.; Huang, D. Characterization of the complete mitochondrial genome sequences of three Merulinidae corals and novel insights into the phylogenetics. PeerJ 2020, 8, e8455. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Xiao, J.; Tian, P.; Guo, F. The complete mitochondrial genome of Plesiastrea versipora (Scleractinia, Plesiastreidae) sheds light on its phylogeny and taxonomy of the family Plesiastreidae. Saudi J. Biol. Sci. 2020, 27, 1830–1834. [Google Scholar] [CrossRef]
- Wang, W.; Cao, B.; Xu, Z.; Jia, Z.; Yu, S.; Tian, P.; Niu, W.; Xiao, J. The complete mitochondrial genome of Montipora vietnamensis (Scleractinia, Acroporidae). Biodivers. Data J. 2022, 10, e91531. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, J. Indo-Pacific Corals; Joe Rowlett: Chicago, IL, USA, 2020. [Google Scholar]
- Arrigoni, R.; Berumen, M.L.; Terraneo, T.I.; Caragnano, A.; Bouwmeester, J.; Benzoni, F. Forgotten in the taxonomic literature: Resurrection of the scleractinian coral genus Sclerophyllia (scleractinia, Lobophylliidae) from the Arabian Peninsula and its phylogenetic relationships. Syst. Biodivers. 2014, 13, 140–163. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).