Abstract
Speckle-type POZ (pox virus and zinc finger protein) protein (SPOP) is a cullin 3-based E3 ubiquitin ligase adaptor protein that plays a crucial role in ubiquitin-mediated protein degradation. Recently, SPOP has attracted major research attention as it is frequently mutated in a range of cancers, highlighting pleiotropic tumorigenic effects and associations with treatment resistance. Structurally, SPOP contains a functionally critical N-terminal meprin and TRAF homology (MATH) domain for many SPOP substrates. SPOP has two other domains, including the internal Bric-a-brac-Tramtrack/Broad (BTB) domain, which is linked with SPOP dimerization and binding to cullin3, and a C-terminal nuclear localization sequence (NLS). The dysregulation of SPOP-mediated proteolysis is associated with the development and progression of different cancers since abnormalities in SPOP function dysregulate cellular signaling pathways by targeting oncoproteins or tumor suppressors in a tumor-specific manner. SPOP is also involved in genome stability through its role in the DNA damage response and DNA replication. More recently, studies have shown that the expression of SPOP can be modulated in various ways. In this review, we summarize the current understanding of SPOP’s functions in cancer and discuss how to design a rational therapeutic target.
1. Introduction
Protein degradation is an essential biological process that is involved in cellular homeostasis. Many diseases, including various cancers, result from the abnormal accumulation of proteins [1]. The ubiquitin proteasome system controls approximately 80% of the ubiquitination and degradation of intracellular proteins [2,3,4]. Ubiquitination is a conserved post-translational modification that is involved in regulating cell proliferation, differentiation, cell cycle progression, apoptosis, transcription, DNA damage and repair and drug resistance [2,3,5]. Ubiquitination is closely associated with carcinogenesis resulting from oncogenic alterations that disrupt the ubiquitination of proteins involved in key tumor suppressor pathways [6]. Proteasome-mediated degradation is a multi-step process that involves the labelling of target proteins with ubiquitin molecules (a single ubiquitin protein or multiple ubiquitin molecules), followed by the degradation of the target ubiquitinated substrates by the 26S proteasome complex [7,8].
Briefly, the first step in proteasomal degradation is the ATP-dependent activation of ubiquitin by the ubiquitin-activating E1 enzyme. The activated ubiquitin is transiently conjugated to a ubiquitin-conjugating E2 enzyme through a transthiolation reaction, after which a ubiquitin-protein E3 ligase transfers to the specific target substrate for ubiquitin ligation [9]. In mammalian cells, different numbers of ubiquitin ligases exist, including several E1 ligases, 30–40 E2 ligases and more than 600 putative E3 ubiquitin ligases [10,11]. Among these E3, the Cullin–RING ubiquitin ligases (CRLs) complex family is the most prominent group and consists of eight Cullin scaffold proteins (CRL1, 2, 3, 4A, 4B, 5, 7, 9) (CRL1, 2, 3, 4A, 4B, 5, 7, 9) [10,12,13]. Generally, the Cullin–RING ligase complex consists of the Cullin scaffold protein, a ring box protein, an adaptor, and the substrate receptor. In contrast to other CRLs, the Cullin3-RING consists of three primary components: the Cullin scaffold protein, a ring box protein called RBX1 and a Bric-a-brac-Tramtrack/Broad (BTB) protein that performs the dual functions of an adaptor and substrate receptor [13,14]. The speckle-type POZ protein (SPOP) is produced by the SPOP gene and was originally identified as a protein by Nagai et al. in 1997. SPOP is a BTB protein containing meprin and TRAF homology (MATH) and BTB domains that can bind Cullin 3 and other substrates [15].
An increasing amount of evidence indicates that SPOP may have dual functions in tumorigenesis. SPOP variants have been reported in many types of cancer, including prostate (PrCa) [16,17,18], endometrial cancer (EC) [19], ovarian [20,21], liver [22], thyroid [23,24], breast [25] and kidney cancers [26] amongst others. The majority of SPOP-related studies have been carried out in PrCa, where it has been reported that 6–15% of patients have point mutations in the substrate-binding MATH domain despite the discovery of a mutation in the BTB domain [27,28]. Mutations in the MATH domain have been recognized as an early event in the development and progression of PrCa, as SPOP mutations are associated with PrCa’s lack of rearrangements of TMPRSS2 ETS family genes [29,30,31].
Several genomic analyses have reported SPOP mutations in EC [32,33,34]. A previous study showed that SPOP mutations in EC fail to interact with and ubiquitinate estrogen receptor-α (ERα), thus facilitating tumorigenesis [35]. In contrast, a recent study suggested that SPOP mutations associated with EC increase the ability of three BETs (BRD2, BRD3 and BRD4) to bind and enhance polyubiquitination of these substrate proteins, leading to degradation and enhanced sensitivity to BET inhibitors [19]. These studies highlight that the pathophysiological function of SPOP mutations remains to be fully understood in EC.
SPOP mutants associated with ovarian, liver and thyroid cancers have been reported in several sequencing studies [20,22,23,24]. In breast cancer, studies have revealed that SPOP targets substrates such as c-Myc, steroid receptor coactivator 3 (SRC-3) and progesterone receptor (PR), thus functioning as a tumor suppressor [25,36,37]. However, SPOP can also directly interact with and mediate the ubiquitination and degradation of breast cancer metastasis suppressor 1 (BRMS1), which is a member of the metastasis suppressor family with known repressive effects on distant metastasis [38]. In contrast to other cancer types, studies have shown that SPOP is highly expressed in clear-cell renal cell carcinomas (RCCs). SPOP-mediated tumorigenesis in RCCs may be mediated by the degradation of cell proliferation suppressors and anti-apoptosis regulators such as Phosphatase and tensin homolog (PTEN), Daxx, Dual-Specificity Phosphatase 7 (DUSP7) and Gli2 [39]. These findings suggest that SPOP can act as a tumor suppressor in various solid cancers but also as a tumor promoter in RCCs. The molecular mechanism through which SPOP performs its functions is that its substrates mediate various cellular processes, including signaling pathways, transcriptional regulation, genome stability and so on. Additionally, SPOP can be regulated by MicroRNAs (miRNAs) and other molecules and can be post-transcriptionally modified by cationic processes such as phosphorylation and self-ubiquitination.
In this review, we introduce the structural features of SPOP and provide a foundation for understanding that SPOP interacts with and mediates efficient substrate ubiquitination in multiple cancers. We discuss the substrate-regulated processes that are involved and the underlying molecular mechanisms of oncogenesis. Finally, we discuss the regulatory mechanisms of SPOP gene expression.
2. The Modular Structure of SPOP
2.1. The SPOP MATH Domain
SPOP is a 374-residue protein that consists of four domains: an N-terminal MATH domain, a BTB/POZ domain, a BACK domain and a C-terminal NLS (Figure 1). The MATH domain (residues 31–164) has an antiparallel β-sandwich structure that selectively recruits substrates and is the predominant domain in which mutations occur [40]. Several critical residues such as Y87, F102, Y123, W131 and F133 are capable of binding the SPOP-binding consensus (SBC) motif from different substrates through the shallow central cleft [14,40]. The SBC motif comprises a five-residue Φ-π-S-S/T-S/T (Φ, denotes a nonpolar amino acid; π, denotes a polar amino acid) [40,41,42]. The interactions of the MATH domain and SBC motif rely on hydrophobic and polar interactions [40]. It has been reported that the binding affinity of one domain is independent of the other MATH domain within the dimer. Consequently, the dimeric configuration can increase the ability of binding to substrates in a substrate-specific manner, but only these substrates contain multiple suboptimal SBC motifs [6,43]. Additionally, it is important that polyubiquitination be followed by degradation when substrates have multiple SBC motifs, since substrates merely undergo monoubiquitination when they contain only one SBC motif [43]. Zhuang et al. reported that SBC phosphorylation in MacroH2A and Puc can block binding to SPOP, however; more studies are needed to verify these observations [40]. Exploratory analyses of SBC phosphorylation of other substrates at perhaps a range of sites and the responding biochemical functions are required.
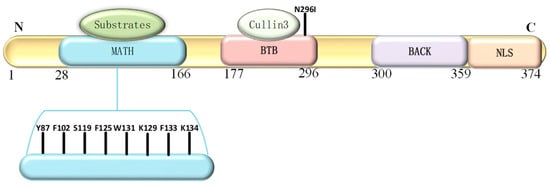
Figure 1.
Illustration of the structural features of Speckle-type POZ (pox virus and zinc finger protein) protein (SPOP). The modular structural arrangement of SPOP is shown, which includes the N-terminal meprin and TRAF homology (MATH) domain that selectively recognizes and recruits substrates. Somatic mutations are predominantly clustered in several key amino acids in the N-terminal MATH domain, including Y87, F102, S119, F125, K129, W131, F133 and K134. The Bric-a-brac-Tramtrack/Broad (BTB) domain is responsible for binding to cullin3 and SPOP dimerization, which also involves the BACK domain. The nuclear localization signal (NLS) is a C-terminal nuclear localization sequence.
2.2. The SPOP BTB Domain
The BTB domain is a versatile protein-protein interaction motif and a common structural element that was discovered in zinc finger transcription factors and Cul3 substrate adaptors. In the human genome, it is encoded by ~205 genes [8,44]. The BTB domain (residues 184–297) is responsible for SPOP dimerization and interactions. Specifically, an α3–β4 loop that consists of approximately ten amino acid residues in the BTB domain and is fundamental for the interactions [10,45,46,47].
Beyond the BTB domain, α-helices make up the three-box domain, which can enhance SPOP-Cul-3 interactions [48]. The BTB domain can dimerize to promote the dimerization of SPOP-cullin3 to generate an oligomeric CRL3 that is highly multivalent and has multiple catalytic centers for substrates [40,48,49]. There is a very low dissociation constant within BTB domain-mediated dimerization [43]. The dimerization capability is associated with α1-3 helices and is related to a strand-exchanged amino terminal region. Four essential hydrophobic BTB dimerization residues, L186, L190, L193 and I217, jointly contribute to the residues 184–297 to form SPOP dimers [14,40]. Dimerization-defective SPOP mutants exhibit impaired ubiquitination without a significant decrease in binding cullin3 affinity [40]. In the human proteome, only SPOP and its homolog, SPOP-like (SPOPL), have both BTB and MATH domains, despite BTB usually being linked to other interaction domains [44].
2.3. The SPOP BACK Domain
The BACK domain (residues 300–359) also mediates dimerization [49]. There are over 50 known human proteins that have BACK and BTB domain combinations [44]. Only the BACK domains within SPOP and SPOPL (which is a unique human SPOP analog) have an atypical truncation that allows dimerization of the BACK domain [50]. The dimerization interface consists of the BTB and BACK domains. The C-terminus acts independently to form higher-order SPOP oligomers capable of enhancing the ubiquitination of substrates [51]. These oligomers enable augmenting the E3 activity by increasing the substrate avidity and the availability of the E2 ubiquitin-conjugating enzyme [48,51].
2.4. The SPOP NLS Domain
SPOP contains a NLS at its C-terminus, in which amino acids 359–374 are essential for its location at nuclear speckles. The NLS is essential for the nuclear localization of SPOP and its interactions with nuclear-localizing substrates. SPOP lacking the NLS accumulates in the puncta of the cytosol [52].
3. SOP Expression in the Development, Progression and Treatment of Cancer
Multiple studies have suggested that SPOP has dual functions in tumorigenesis based on gene expression and clinical datasets. The analysis of datasets from The Cancer Genome Atlas (TCGA) and other sources has indicated that the majority of cancers have significantly decreased expression of SPOP compared to corresponding normal controls, including bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC) [Figure 2a].
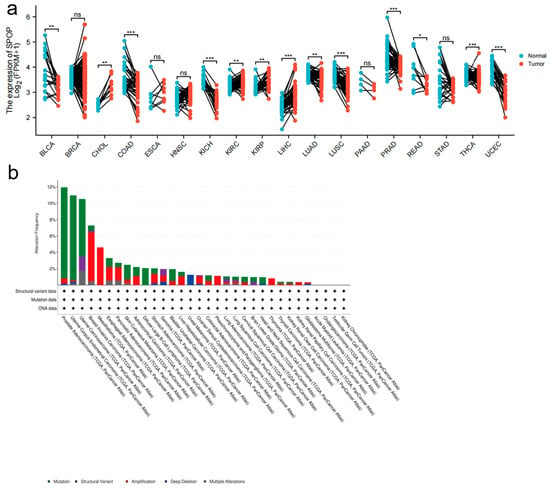
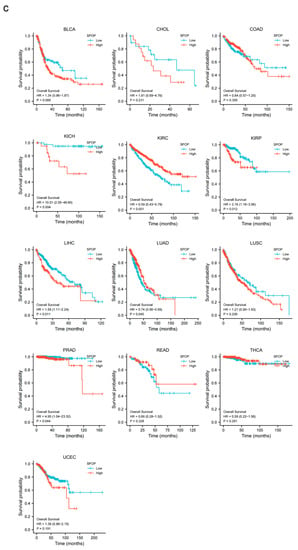
Figure 2.
SPOP mRNA expression in different cancers. (a) A graph showing the levels of SPOP mRNA expression levels in a variety of cancers along with corresponding normal controls; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CHOL, Cholangiocarcinoma; COAD, Colon Adenocarcinoma; ESCA, esophageal carcinoma; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; PAAD, Pancreatic adenocarcinoma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; STAD, Stomach adenocarcinoma; THCA, Thyroid carcinoma; UCEC, Uterine Corpus Endometrial Carcinoma; ns, not significant; * p < 0.05, ** p < 0.01, *** p < 0.001. (b) SPOP alterations occur across multiple cancer types (data from the cBioPortal database). (c) The correlation of SPOP with overall survival is further elevated (data from the The Cancer Genome Atlas (TCGA) portal or other published datasets of clinical cancer samples as indicated).
Cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), kidney renal clear cell carcinoma (KIRC) and kidney renal papillary cell carcinoma (KIRP) show a marked increase in the expression of SPOP, yet the ESCA variable was not statistically significant [Figure 2a].
Analysis of data in the cBioPortal database (www.cbioportal.org/) has shown that SPOP alterations occur in different tumor types. The alterations include mutation, structural variants, amplification, deep deletion and multiple alterations [Figure 2b]. SPOP mutations are most common in PrCa, in which the SPOP mutation rate is 12%, which is consistent with previous reports [27]. A previous clinical study suggested that SPOP mutations and downregulation were tightly associated with a poor prognosis in patients with PrCa [18]. From the data presented in Figure 2a, further analyses revealed that the upregulation of SPOP may predict favorable outcomes in KIRC and LUAD. In contrast, the low expression of SPOP is associated with improved outcomes in KICH, KIRP, LIHC and PRAD [Figure 2c]. However, patients with BLCA, CHOL, COAD, LUSC, READ, THCA and UCEC do not have significant associations between SPOP expression and overall survival (OS) [Figure 2c]. These findings require further validation before being used in routine patient management. In addition to the observations made in clinical cancer datasets, previous studies have also found that SPOP can have both oncogenic and tumor-suppressor functions [41].
In 2010, Kan et al. originally reported that SPOP is frequently mutated in PrCa [53]. Subsequently, whole-genome and exome sequencing analyses discovered that SPOP mutations occur as early events in the development of PrCa but do not occur in normal issues or prostate stroma [17,27,29,30,54,55]. In PrCa, SPOP mutations are located in the MATH domain, which typically contains Y87, F102, S199, F125, W131, K129, F133 and K143 mutations [Figure 1] that have been identified as biologically relevant events [27]. SPOP mutations can lead to genomic instability and impaired genome maintenance, resulting in the inactivation of BRCA1 and an impaired homology-directed repair (HDR) function [55]. Importantly, several have further indicated that SPOP acts as a tumor suppressor by promoting the ubiquitination and subsequent degradation of downstream substrates, including the Androgen receptor (AR) [56,57], steroid receptor coactivator 3 (SRC3) [58], TRIM24 [59], Gli3 [60,61,62,63], HIPK2 [64], 53BP1 [65], (Programmed death ligand 1) (PD-L1) [66], BMI1 [67], Macrohistone H2A1 (MacroH2A) [67,68], Pancreatic duodenal homeobox 1 (Pdx1) [69,70,71], SENP7 [72], ERG [73,74], BRD2/3/4 [19,75,76], DEK [77], DDIT3 [78], Nanog [79,80], Cdc20 [81], CYCLIN E1 [82], c-MYC [83], INF2 [84], EgIN2 [85], Activating Transcription Factor 2 (ATF2) [86], fatty acid synthase (FASN) [87], Caprin1 [52], 17βHSD4 [88], ITCH [89], GLP [90], CDCA5 [91], PDK1 [92], SQSTM1 [93] and PrLZ [94]. These substrates can regulate diverse cellular processes in PrCa [Figure 3a]. colorectal cancer, diffuse large B-cell lymphoma, lung cancer, liver cancer, choriocarcinoma, Ewing sarcoma and hepatoblastomthe downregulation of SPOP can promote the development and progression of PrCa, making it a potential therapeutic target.
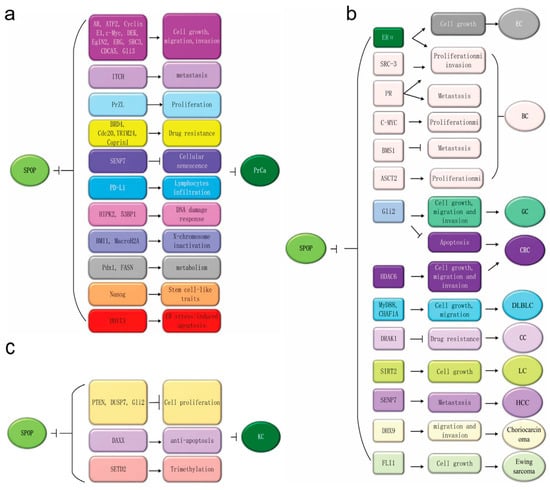
Figure 3.
The potential oncogenic functions of Speckle-type POZ (pox virus and zinc finger protein) protein (SPOP) in kidney cancer and tumor-suppressive roles in other cancer types. (a) SPOP exerts potential antitumor activity by regulating proliferation, migration, invasion, metastasis, apoptosis, drug resistance, cell senescence and lymphocyte infiltration via modulating various substrates in PrCa. (b) SPOP has tumor-suppressive effects across multiple cancer types, including endometrial, gastric, colorectal, cervical and lung cancers, hepatocellular carcinoma, choriocarcinoma, Ewing sarcoma and diffuse large B-cell lymphoma. It has dual roles in breast cancer that regulate the degradation of its various substrates. (c) SPOP exerts its potential oncogenic activity by enhancing the ubiquitination and degradation of Phosphatase and tensin homolog (PTEN), Dual-Specificity Phosphatase 7 (DUSP7), Gli2 and SETD2 to promote renal carcinogenesis.
The majority of cancer studies involving SPOP have been in PrCa; however, multiple studies have reported SPOP mutations in other cancer types including EC, breast cancer (BC), gastric cancer (GC), colorectal cancer, diffuse large B-cell lymphoma, lung cancer, liver cancer, choriocarcinoma, Ewing sarcoma and hepatoblastoma. The SPOP substrates in these cancers include: ERα [35,95], and BRD 2/3/4 in endometrial cancer [19], ERα [96], PR [37], SRC3 [25], c-MYC [36], BRMS1 [38], ASCT2 in breast cancer [97], Gli2 in gastric cancer [98], Gli2 [99], HDAC6 in colorectal cancer (CRC) [100], and MyD88 and CHAF1A in diffuse large B-cell lymphoma (DLBLC) [101,102,103,104]. Fas-associated death domain protein (FADD) [105], SIRT2 in lung cancer (LC) [106], SENP7 in hepatocellular carcinoma (HCC) [107], DHX9 in choriocarcinoma [108], FLI1 in Ewing sarcoma [109], and SLC7A1 [110], DRAK1 and PIPKIIβ in cervical cancer (CC) [111,112]. Figure 3b summarizes the different SPOP substrates and their functions in cellular processes reported in different cancers. The in-depth investigations are necessary to validate the definitive role of SPOP in these cancer types before designing a clear rationale for therapeutic strategies.
SPOP could potentially have tumor-suppressive roles in several cancers. Emerging evidence demonstrates an oncogenic function of SPOP in kidney cancer (KC). Studies have reported the overexpression and accumulation of SPOP in renal cell carcinoma (RCC) in primary and metastatic tissues that is significantly associated with EMT and poor prognosis [26,113,114,115]. In contrast, SPOP silencing in the A498 and ACHN RCC cells induces cell apoptosis, inhibits cell viability, and decreases colony formation and migration ability whilst also increasing sensitivity to Sorafenib [116]. In addition, Li et al. reported that the SPOP protein can accumulate in the cytoplasm under hypoxia to promote tumorigenesis via the degradation of tumor suppressors such as PTEN, DUSP7, ERK phosphatases and Gli2 as well as the proapoptotic protein Daxx in clear cell renal cell carcinoma (ccRCC) [39]. SPOP can also be responsible for the polyubiquitination of the tumor suppressor SETD2 and can negatively regulate H3K36me3 levels in RCC [117,118]. In summary, SPOP possesses an oncogenic role in RCC via overexpression, cytoplasmic accumulation and subsequent ubiquitination and degradation of multiple tumor suppressive substrates [Figure 3c]. Given these findings, SPOP inhibitors may be a potential new therapeutic in the treatment of RCC.
SPOP has oncogenic or tumor suppressive functions that are cancer type specific. The development of targeted therapies enables treatment for different human cancers. Previous structural analyses have indicated that SPOP binds to the SBC motif of specific substrates via its N-terminal MATH domain [40]. On this basis, Guo et al. identified 109 small molecule inhibitors by computational screening that can disrupt SPOP-substrate interactions [119]. Among the identified molecules, compound 6a was considered an initial hit for binding SPOP that competed with the puc_SBC1 peptide. Additional synthetic optimization of 6a resulted in a compound that had strong inhibitory activity in disrupting SPOP’s binding to PTEN and DUSP7. This resulted in the inactivation of downstream signaling pathways such as the PTEN-phosphoinositide 3-kinase/AKT pathway and the dephosphorylation of extracellular signal-regulated kinase (ERK) [119]. The small-molecule inhibitor 6b was also cytotoxic in A498 ccRCC cells and stabilizes SPOP substrates [119]. Recently, Rincon et al. identified novel therapeutic natural compounds that can specifically target stable MCF-7 SPOP knockdown cells by disrupting GLI3-dependent SHH signaling [120].
4. SPOP-Regulated Processes
SPOP serves as a regulatory hub that mainly works on two necessary processes, specifically intracellular signaling pathways and the regulation of genomic function (Table 1) [14].

Table 1.
Summary of SPOP substrates and their cellular functions. Androgen receptor—AR, progesterone receptor—PR, steroid receptor coactivator 3—SRC3, Macrohistone H2A1—MacroH2A, Phosphatase and tensin homolog—PTEN, Dual-Specificity Phosphatase 7—DUSP7, Fas-associated death domain protein—FADD, Programmed death ligand 1—PD-L1, Pancreatic duodenal homeobox 1—Pdx1, fatty acid synthase—FASN, breast cancer metastasis suppressor 1—BRMS1.
4.1. SPOP-Related Intracellular Signaling Pathways
Increasing evidence has shown that SPOP substrates are implicated in the biological processes of cells, including signaling pathways that are essential to maintain normal cellular functions. For example, the Hedgehog (Hh) signaling pathway is a largely conserved signal transduction pathway during metazoan evolution that intimately regulates cell growth and differentiation [63,121]. Cai et al. showed that SPOP plays a critical role in skeletal development and remodeling and that SPOP mutations can increase the level of the repressor Gli3R, which has been further explored as a substrate of SPOP but not Gli2 [63]. As a result, Ihh signaling is compromised, suggesting an important positive regulator in SPOP in Hh signaling [63]. These researchers also demonstrated that SPOP mutations lead to a significant increase in the level of the activator Gli3, which promotes Shh signaling, suggesting a negative role for SPOP in cord patterning [122].
4.2. SPOP’s Role in Genome Stability
Impaired DNA damage repair (DDR) can result in genome instability. Initially, it was reported that SPOP participates in DDR in a predominantly ATM-dependent manner [123]. SPOP can be recruited to DNA double-strand break (DSB) foci, where it partially colocalizes with γ-H2AX foci in response to ionizing irradiation (IR) or the topoisomerase 1 inhibitor, camptothecin [123]. SPOP depletion can lead to impaired DDR, resulting in hypersensitivity to IR [123]. However, it is unclear how SPOP terminates DDR upon IR exposure. Furthermore, Boysen et al. demonstrated the role of SPOP in DDR, with SPOP mutations causing homology-directed repair (HDR) of DSB defects by BRCA1 inactivation and RAD51 foci decrease [55]. The researchers found that the loss function of SPOP results in an increased sensitivity to PARP inhibition after DSB induction [55]. Additionally, SPOP can induce non-degradable ubiquitination of HIPK2 and 53BP1, which are both involved in DDR, and degradable ubiquitination of STED2, which regulates the trimethylation of histone H3K36 [64,65,117,118] that is associated with HDR [117,118]. These results suggest that SPOP plays a role in genome stability via multiple pathways affecting DDR.
Gene rearrangements caused by DNA replication stress can also result in genome instability. Previous studies have reported that DSBs induced by the collaboration of TOP2A, 2B and AR signaling lead to gene rearrangements that contribute to genome instability [124,125]. SPOP can remove TOP2A from the TOP2A-DNA cleavage complex in normally growing cells [42]. However, SPOP mutations fail to remove TOP2A from DNA, thus causing genome instability.
5. The Regulation of SPOP
The regulation of SPOP expression at different levels includes DNA methylation that affects transcription, miRNAs that affect translation, and phosphorylation and self-ubiquitination that affect posttranscriptional modifications.
Hypermethylation is a common modification of DNA that silences the expression of tumor suppressor genes in most cancer types [126]. DNA methylation can regulate tumor suppressor SPOP gene transcription by affecting the binding affinity between RXRA and the SPOP promoter in CRC and RXRA as a transcription factor [99]. A similar study in LC suggests that the transcription factor C/EBPα can bind the SPOP promoter [127]. In non-small cell lung cancer (NSCLC), the combination of this transcriptional regulatory element and DNA methylation regulates the expression and function of SPOP [127].
miRNAs are small single-stranded non-coding RNAs that have been shown to negatively modulate SPOP expression by targeting the 3′-untranslated region (3′-UTR) of its mRNA. Huang et al. in 2014 showed the post-transcriptional regulation of SPOP expression by miR-145 [128]. In 2018 and 2019, several miRNAs were identified that can regulate SPOP expression in RCC, oral squamous cell carcinoma (OSCC), CRC, NSCLC and GC. In RCC, carcinogenesis is controlled through the E2F1-miR-520/372/373-SPOP axis. The miR-520/372/373 family is downregulated, and SPOP was upregulated, resulting in a significant decrease of PTEN and DUSP7 levels with subsequent increased proliferation, invasion/migration and metastasis [129]. Recently, a study suggested that miRNA-17-5p can target SPOP and that upregulation of miRNA-17-5p can downregulate SPOP to promote proliferation and inhibit anti-tumor immunity in CRC through the overexpression of PD-L1 [130]. Intriguingly, in OSCC, miR373 was upregulated in samples as well as cell lines and negatively regulates the expression of the SPOP protein but not mRNA, promoting proliferation, invasion and migration of corresponding cells [131]. Another study showed that miR-372/373 enhances CRC cell stemness by targeting factors important for cell differentiation, including SPOP [132]. Additionally, miR-543 and miR-520b are upregulated in GC and LC, respectively [133,134]. SPOP has been identified as a direct target of these miRNAs that act to promote proliferation and metastasis by decreasing SPOP expression [133,134].
Post-transcriptional modifications of the SPOP protein consist of phosphorylation and self-ubiquitination. Zhang et al. reported that cyclin D-CDK4 can mediate the phosphorylation of SPOP to promote SPOP degradation by regulating PD-L1 protein abundance [66]. More recently, everal have further indicated that SPOP acts as a tumor suppressorhas been shown to stabilize its interaction with 53BP1 [65]. Subsequently, SPOP induces polyubiquitination of 53BP1 and dissociation of 53BP1 from DSBs to activate HDR and consequently induce genome stability [65]. Interestingly, another study found that snails can facilitate SPOP self-ubiquitination and degradation in a cullin3-dependent manner [135].
SPOP function can also be regulated by the liquid-liquid phase separation (LLPS) of proteins as a novel regulatory mechanism [136,137]. Increasing evidence has indicated that various nuclear bodies, such as nuclear speckles, DNA damage loci and other substrate-containing bodies, contain the SPOP protein [15,49]. LLPS is critical for SPOP-substrate ubiquitination and the subsequent degradation in cells [136]. In contrast, tumor-associated SPOP mutations may disrupt LLPS and thereby inhibit the ubiquitin-dependent proteolysis of substrates.
Mukhopadhyay et al. showed that the GTPase Activating Protein (SH3 Domain) Binding Protein 1 (G3BP1) can interact with SPOP and inhibit SPOP ubiquitin ligase in PrCa [138]. Ji et al. have also found that heparan sulfate 3-O-sulfotransferase 1 (HS3ST1) inhibits SPOP expression and also inhibits activation of the NF-κB pathway activation mediated by FADD degradation in NSCLC [139].
6. Summary and Perspectives
In this review, we summarize the molecular functions and dual roles of SPOP in promoting or inhibiting tumorigenesis in a cancer type-specific manner by targeting multiple proteins. These roles suggest that SPOP and its substrates are potentially viable targets for the development of cancer therapeutics. The complex oncogenic and tumor suppressive roles of SPOP indicate that the design of therapeutic strategies is cancer type specific. For example, SPOP inhibitors could be used to treat kidney cancer as SPOP is an oncogene in RCC. As a putative tumor suppressor in other cancer types, SPOP promoters may also be a desirable approach. An alternative approach is to target the factors upstream of SPOP; for instance, the upregulation of miR-520/372/373 could cause the downregulation of SPOP in CRC cells, inhibiting proliferation, invasion and metastasis by increasing the levels of PTEN and DUSP7 [129]. Conversely, the downregulation of miR-17-5p, miR-543 and miR-520b can inhibit proliferation and metastasis via the upregulation of SPOP [130,133,134].
SPOP binds to the cullin 3–RING box 1 through the BTB domain to increase E3 ubiquitin ligase activity by promoting the dimerization of SPOP [40,48]. It would be an attractive option to design small-molecule inhibitors that target cullin 3–RING box 1 to affect its affinity for SPOP and thereby decrease SPOP dimerization.
The majority of investigations have focused on the role of SPOP in PrCa. Further studies are needed in other cancer types, including the systematic exploration of SPOP substrates. Future studies will undoubtedly provide further insight into the functional mechanisms of SPOP in human cancers and uncover a rationale for the design of therapeutic interventions.
Author Contributions
All authors have substantially contributed to the work reported. Individual author contributions were as follows: conceptualization: X.Y.; validation: X.Y. and Q.Z.; writing—original draft preparation, X.Y.; writing—review and editing, X.Y. and Q.Z.; supervision, Q.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, Grant/Award Numbers: 82072481 and 81874223; 1.3.5 project for disciplines of excellence (West China Hospital, Sichuan University), Grant/Award Number: ZYJC21042.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Christianson, J.C.; Ye, Y. Cleaning up in the endoplasmic reticulum: Ubiquitin in charge. Nat. Struct. Mol. Biol. 2014, 21, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, P.; Inuzuka, H.; Wei, W. Roles of F-box proteins in cancer. Nat. Cancer 2014, 14, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Chen, R.-H. Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications. Front. Oncol. 2016, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagan, J.; Pagano, M. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 2014, 13, 889–903. [Google Scholar] [CrossRef]
- DeSalle, L.M.; Pagano, M. Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett. 2001, 490, 179–189. [Google Scholar] [CrossRef]
- Clark, A.; Burleson, M. SPOP and cancer: A systematic review. Am. J. Cancer Res. 2020, 10, 704–726. [Google Scholar]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat. Rev. Mol. Cell Biol. 2008, 9, 679–689. [Google Scholar] [CrossRef]
- Mani, A.; Gelmann, E.P. The Ubiquitin-Proteasome Pathway and Its Role in Cancer. J. Clin. Oncol. 2005, 23, 4776–4789. [Google Scholar] [CrossRef]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin–proteasome system as drug targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
- Mani, R.-S. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov. Today 2014, 19, 1498–1502. [Google Scholar] [CrossRef]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, Y.; Pan, C.; Yan, L.; Wang, Z.-W.; Zhu, X. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol. Cancer 2020, 19, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Wu, H.-L.; Xia, Q.-D.; Zhou, P.; Wang, S.-G.; Yu, X.; Hu, J. Novel insights into the SPOP E3 ubiquitin ligase: From the regulation of molecular mechanisms to tumorigenesis. Biomed. Pharmacother. 2022, 149, 112882. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Kojima, T.; Muro, Y.; Hachiya, T.; Nishizawa, Y.; Wakabayashi, T.; Hagiwara, M. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 1997, 418, 23–26. [Google Scholar] [CrossRef]
- Brenner, J.C.; Chinnaiyan, A.M. Disruptive Events in the Life of Prostate Cancer. Cancer Cell 2011, 19, 301–303. [Google Scholar] [CrossRef]
- Berger, M.F.; Lawrence, M.S.; Demichelis, F.; Drier, Y.; Cibulskis, K.; Sivachenko, A.Y.; Sboner, A.; Esgueva, R.; Pflueger, D.; Sougnez, C.; et al. The genomic complexity of primary human prostate cancer. Nature 2011, 470, 214–220. [Google Scholar] [CrossRef]
- García-Flores, M.; Casanova-Salas, I.; Rubio-Briones, J.; Calatrava, A.; Domínguez-Escrig, J.; Rubio, L.; Ramírez-Backhaus, M.; Fernández-Serra, A.; García-Casado, Z.; López-Guerrero, J.A. Clinico-pathological significance of the molecular alterations of the SPOP gene in prostate cancer. Eur. J. Cancer 2014, 50, 2994–3002. [Google Scholar] [CrossRef]
- Janouskova, H.; El Tekle, G.; Bellini, E.; Udeshi, N.D.; Rinaldi, A.; Ulbricht, A.; Bernasocchi, T.; Civenni, G.; Losa, M.; Svinkina, T.; et al. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat. Med. 2017, 23, 1046–1054. [Google Scholar] [CrossRef]
- Arildsen, N.S.; Jönsson, J.-M.; Bartuma, K.; Ebbesson, A.; Westbom-Fremer, S.; Måsbäck, A.; Malander, S.; Nilbert, M.; Hedenfalk, I.A. Involvement of Chromatin Remodeling Genes and the Rho GTPases RhoB and CDC42 in Ovarian Clear Cell Carcinoma. Front. Oncol. 2017, 7, 109. [Google Scholar] [CrossRef]
- Hu, X.; Yang, Z.; Zeng, M.; Liu, Y.; Yang, X.; Li, Y.; Li, X.; Yu, Q. Speckle-type POZ (pox virus and zinc finger protein) protein gene deletion in ovarian cancer: Fluorescence in situ hybridization analysis of a tissue microarray. Oncol. Lett. 2016, 12, 658–662. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jia, D.; Dong, R.; Jing, Y.; Xu, D.; Wang, Q.; Chen, L.; Li, Q.; Huang, Y.; Zhang, Y.; Zhang, Z.; et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology 2014, 60, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-K.; Lee, S.; Kim, S.-J.; Jee, H.-G.; Kim, B.-A.; Cho, H.; Song, Y.S.; Cho, S.W.; Won, J.-K.; Shin, J.-Y.; et al. Comprehensive Analysis of the Transcriptional and Mutational Landscape of Follicular and Papillary Thyroid Cancers. PLoS Genet. 2016, 12, e1006239. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhou, X.; Huang, F.; Wang, W.; Qi, Y.; Xu, H.; Shu, Y.; Shen, L.; Fei, X.; Xie, J.; et al. The genetic landscape of benign thyroid nodules revealed by whole exome and transcriptome sequencing. Nat. Commun. 2017, 8, 15533. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ao, J.; Fu, J.; Lee, D.-F.; Xu, J.; Lonard, D.; O’Malley, B.W. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 2011, 30, 4350–4364. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ghanim, M.; Xue, L.; Brown, C.D.; Iossifov, I.; Angeletti, C.; Hua, S.; Nègre, N.; Ludwig, M.; Stricker, T.; et al. Analysis of Drosophila Segmentation Network Identifies a JNK Pathway Factor Overexpressed in Kidney Cancer. Science 2009, 323, 1218–1222. [Google Scholar] [CrossRef]
- Barbieri, C.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.-P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef]
- Zuhlke, K.A.; Johnson, A.M.; Tomlins, S.A.; Palanisamy, N.; Carpten, J.D.; Lange, E.M.; Isaacs, W.B.; Cooney, K.A. Identification of a novel germline SPOP mutation in a family with hereditary prostate cancer. Prostate 2014, 74, 983–990. [Google Scholar] [CrossRef]
- Romanel, A.; Garritano, S.; Stringa, B.; Blattner, M.; Dalfovo, D.; Chakravarty, D.; Soong, D.; Cotter, K.; Petris, G.; Dhingra, P.; et al. Inherited determinants of early recurrent somatic mutations in prostate cancer. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Jung, S.-H.; Shin, S.; Kim, M.S.; Baek, I.-P.; Lee, J.Y.; Lee, S.H.; Kim, T.-M.; Lee, S.H.; Chung, Y.-J. Genetic Progression of High Grade Prostatic Intraepithelial Neoplasia to Prostate Cancer. Eur. Urol. 2016, 69, 823–830. [Google Scholar] [CrossRef]
- Wei, X.; Fried, J.; Li, Y.; Hu, L.; Gao, M.; Zhang, S.; Xu, B. Functional roles of Speckle-Type Poz (SPOP) Protein in Genomic stability. J. Cancer 2018, 9, 3257–3262. [Google Scholar] [CrossRef] [PubMed]
- Le Gallo, M.; Program, N.I.S.C.C.S.; O’Hara, A.J.; Rudd, M.L.; Urick, M.E.; Hansen, N.F.; O’Neil, N.J.; Price, J.C.; Zhang, S.; England, B.M.; et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012, 44, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Le Gallo, M.; Rudd, M.L.; Urick, M.E.; Hansen, N.F.; Zhang, S.; Lozy, F.; Sgroi, D.C.; Bel, A.V.; Matias-Guiu, X.; Broaddus, R.R.; et al. Somatic mutation profiles of clear cell endometrial tumors revealed by whole exome and targeted gene sequencing. Cancer 2017, 123, 3261–3268. [Google Scholar] [CrossRef]
- DeLair, D.F.; Burke, K.A.; Selenica, P.; Lim, R.S.; Scott, S.N.; Middha, S.; Mohanty, A.S.; Cheng, D.T.; Berger, M.F.; Soslow, R.A.; et al. The genetic landscape of endometrial clear cell carcinomas. J. Pathol. 2017, 243, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Gao, K.; Jin, X.; Ma, J.; Peng, J.; Wumaier, R.; Tang, Y.; Zhang, Y.; An, J.; Yan, Q.; et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-α protein turnover. Cell Death Dis. 2015, 6, e1687. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Tang, H.; Ling, L.; Li, N.; Jia, X.; Zhang, Z.; Wang, X.; Shi, L.; Yin, J.; Qiu, N.; et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene 2018, 37, 6166–6179. [Google Scholar] [CrossRef]
- Gao, K.; Jin, X.; Tang, Y.; Ma, J.; Peng, J.; Yu, L.; Zhang, P.; Wang, C. Tumor suppressor SPOP mediates the proteasomal degradation of progesterone receptors (PRs) in breast cancer cells. Am. J. Cancer Res. 2015, 5, 3210–3220. [Google Scholar]
- Kim, B.; Nam, H.J.; Pyo, K.E.; Jang, M.J.; Kim, I.S.; Kim, D.; Boo, K.; Lee, S.H.; Yoon, J.-B.; Baek, S.H.; et al. Breast cancer metastasis suppressor 1 (BRMS1) is destabilized by the Cul3–SPOP E3 ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 2011, 415, 720–726. [Google Scholar] [CrossRef]
- Li, G.; Ci, W.; Karmakar, S.; Chen, K.; Dhar, R.; Fan, Z.; Guo, Z.; Zhang, J.; Ke, Y.; Wang, L.; et al. SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer. Cancer Cell 2014, 25, 455–468. [Google Scholar] [CrossRef]
- Zhuang, M.; Calabrese, M.F.; Liu, J.; Waddell, M.B.; Nourse, A.; Hammel, M.; Miller, D.J.; Walden, H.; Duda, D.M.; Seyedin, S.N.; et al. Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases. Mol. Cell 2009, 36, 39–50. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Y.; Ye, M.; Dai, X.; Zhu, X.; Wei, W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat. Rev. Urol. 2020, 17, 339–350. [Google Scholar] [CrossRef]
- Maekawa, M.; Higashiyama, S. The Roles of SPOP in DNA Damage Response and DNA Replication. Int. J. Mol. Sci. 2020, 21, 7293. [Google Scholar] [CrossRef] [PubMed]
- Pierce, W.K.; Grace, C.R.; Lee, J.; Nourse, A.; Marzahn, M.R.; Watson, E.R.; High, A.A.; Peng, J.; Schulman, B.A.; Mittag, T. Multiple Weak Linear Motifs Enhance Recruitment and Processivity in SPOP-Mediated Substrate Ubiquitination. J. Mol. Biol. 2016, 428, 1256–1271. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.S.; Nandra, S.K.; Privé, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.-H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [CrossRef]
- Furukawa, M.; He, Y.J.; Borchers, C.; Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nature 2003, 5, 1001–1007. [Google Scholar] [CrossRef]
- Pintard, L.; Willis, J.H.; Willems, A.; Johnson, J.-L.F.; Srayko, M.; Kurz, T.; Glaser, S.; Mains, P.E.; Tyers, M.; Bowerman, B.; et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 2003, 425, 311–316. [Google Scholar] [CrossRef]
- Errington, W.J.; Khan, M.Q.; Bueler, S.A.; Rubinstein, J.; Chakrabartty, A.; Privé, G. Adaptor Protein Self-Assembly Drives the Control of a Cullin-RING Ubiquitin Ligase. Structure 2012, 20, 1141–1153. [Google Scholar] [CrossRef]
- Marzahn, M.R.; Marada, S.; Lee, J.; Nourse, A.; Kenrick, S.; Zhao, H.; Ben-Nissan, G.; Kolaitis, R.; Peters, J.L.; Pounds, S.; et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016, 35, 1254–1275. [Google Scholar] [CrossRef]
- Cuneo, M.J.; Mittag, T.; Cuneo, M.J.; Mittag, T. The ubiquitin ligase adaptor SPOP in cancer. FEBS J. 2019, 286, 3946–3958. [Google Scholar] [CrossRef]
- Van Geersdaele, L.K.; Stead, M.; Harrison, C.M.; Carr, S.B.; Close, H.J.; Rosbrook, G.O.; Connell, S.D.; Wright, S.C. Structural basis of high-order oligomerization of the cullin-3 adaptor SPOP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Zhu, Y.; Ma, J.; Chang, K.; Ding, D.; Bai, Y.; Gao, K.; Zhang, P.; Mo, R.; Feng, K.; et al. Prostate Cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol. Cancer 2019, 18, 170. [Google Scholar] [CrossRef] [PubMed]
- Kan, Z.; Jaiswal, B.S.; Stinson, J.; Janakiraman, V.; Bhatt, D.; Stern, H.M.; Yue, P.; Haverty, P.M.; Bourgon, R.; Zheng, J.; et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010, 466, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Hjorth-Jensen, K.; Maya-Mendoza, A.; Dalgaard, N.; Sigurðsson, J.O.; Bartek, J.; Iglesias-Gato, D.; Olsen, J.; Flores-Morales, A. SPOP promotes transcriptional expression of DNA repair and replication factors to prevent replication stress and genomic instability. Nucleic Acids Res. 2018, 46, 9484–9495. [Google Scholar] [CrossRef] [PubMed]
- Boysen, G.; Barbieri, C.E.; Prandi, D.; Blattner, M.; Chae, S.-S.; Dahija, A.; Nataraj, S.; Huang, D.; Marotz, C.; Xu, L.; et al. SPOP mutation leads to genomic instability in prostate cancer. eLife 2015, 4, e09207. [Google Scholar] [CrossRef]
- Geng, C.; Rajapakshe, K.; Shah, S.S.; Shou, J.; Eedunuri, V.K.; Foley, C.; Fiskus, W.; Rajendran, M.; Chew, S.A.; Zimmermann, M.; et al. Androgen Receptor Is the Key Transcriptional Mediator of the Tumor Suppressor SPOP in Prostate Cancer. Cancer Res. 2014, 74, 5631–5643. [Google Scholar] [CrossRef]
- An, J.; Wang, C.; Deng, Y.; Yu, L.; Huang, H. Destruction of Full-Length Androgen Receptor by Wild-Type SPOP, but Not Prostate-Cancer-Associated Mutants. Cell Rep. 2014, 6, 657–669. [Google Scholar] [CrossRef]
- Geng, C.; He, B.; Xu, L.; Barbieri, C.E.; Eedunuri, V.K.; Chew, S.A.; Zimmermann, M.; Bond, R.; Shou, J.; Li, C.; et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl. Acad. Sci. USA 2013, 110, 6997–7002. [Google Scholar] [CrossRef]
- Groner, A.C.; Cato, L.; de Tribolet-Hardy, J.; Bernasocchi, T.; Janouskova, H.; Melchers, D.; Houtman, R.; Cato, A.C.; Tschopp, P.; Gu, L.; et al. TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer Cell 2016, 29, 846–858. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, L.; Wang, B.; Ou, C.-Y.; Chien, C.-T.; Jiang, J. A Hedgehog-Induced BTB Protein Modulates Hedgehog Signaling by Degrading Ci/Gli Transcription Factor. Dev. Cell 2006, 10, 719–729. [Google Scholar] [CrossRef]
- Burleson, M.; Deng, J.J.; Qin, T.; Duong, T.M.; Yan, Y.; Gu, X.; Das, D.; Easley, A.; Liss, M.A.; Yew, P.R.; et al. GLI3 Is Stabilized by SPOP Mutations and Promotes Castration Resistance via Functional Cooperation with Androgen Receptor in Prostate Cancer. Mol. Cancer Res. 2022, 20, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, Q.; Chen, Y.; Yue, T.; Li, S.; Wang, B.; Jiang, J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2009, 106, 21191–21196. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 14751–14756. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Qing, S.; Li, Q.; Zhuang, H.; Shen, L.; Li, J.; Qi, H.; Lin, T.; Lin, Z.; Wang, J.; et al. Prostate cancer-associated SPOP mutations lead to genomic instability through disruption of the SPOP–HIPK2 axis. Nucleic Acids Res. 2021, 49, 6788–6803. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ma, J.; Botuyan, M.V.; Cui, G.; Yan, Y.; Ding, D.; Zhou, Y.; Krueger, E.W.; Pei, J.; Wu, X.; et al. ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication. Sci. Adv. 2021, 7, eabd9208. [Google Scholar] [CrossRef]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D–CDK4 kinase destabilizes PD-L1 via cullin 3–SPOP to control cancer immune surveillance. Nature 2018, 553, 91–95. [Google Scholar] [CrossRef]
- Hernández-Muñoz, I.; Lund, A.H.; van der Stoop, P.; Boutsma, E.; Muijrers, I.; Verhoeven, E.; Nusinow, D.A.; Panning, B.; Marahrens, Y.; van Lohuizen, M. Stable X Chromosome Inactivation Involves the PRC1 Polycomb Complex and Requires Histone MACROH2A1 and the CULLIN3/SPOP Ubiquitin E3 Ligase. Proc. Natl. Acad. Sci. USA 2005, 102, 7635–7640. [Google Scholar] [CrossRef]
- Takahashi, I.; Kameoka, Y.; Hashimoto, K. MacroH2A1.2 binds the nuclear protein Spop. Biochim. et Biophys. Acta 2002, 1591, 63–68. [Google Scholar] [CrossRef][Green Version]
- Liu, A.; Desai, B.M.; Stoffers, D.A. Identification of PCIF1, a POZ Domain Protein That Inhibits PDX-1 (MODY4) Transcrip-tional Activity. Mol. Cell Biol. 2004, 24, 4372–4383. [Google Scholar] [CrossRef]
- Claiborn, K.C.; Sachdeva, M.M.; Cannon, C.E.; Groff, D.N.; Singer, J.D.; Stoffers, D.A. Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. J. Clin. Investig. 2010, 120, 3713–3721. [Google Scholar] [CrossRef]
- Liu, A.; Oliver-Krasinski, J.; Stoffers, D.A. Two Conserved Domains in PCIF1 Mediate Interaction with Pancreatic Transcrip-tion Factor PDX-1. FEBS Lett. 2006, 580, 6701–6706. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Ren, S.; Bitler, B.G.; Aird, K.M.; Tu, Z.; Skordalakes, E.; Zhu, Y.; Yan, J.; Sun, Y.; Zhang, R. SPOP E3 Ubiquitin Ligase Adaptor Promotes Cellular Senescence by Degrading the SENP7 deSUMOylase. Cell Rep. 2015, 13, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Dai, X.; Lunardi, A.; Li, Z.; Inuzuka, H.; Liu, P.; Varmeh, S.; Zhang, J.; Cheng, L.; Sun, Y.; et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol. Cell 2015, 59, 917–930. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ren, S.; Murphy, S.J.; Dalangood, S.; Chang, C.; Pang, X.; Cui, Y.; Wang, L.; Pan, Y.; Zhang, X.; et al. Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol. Cell 2015, 59, 904–916. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, D.; Zhao, Y.; Ren, S.; Gao, K.; Ye, Z.; Wang, S.; Pan, C.-W.; Zhu, Y.; Yan, Y.; et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT–mTORC1 activation. Nat. Med. 2017, 23, 1055–1062. [Google Scholar] [CrossRef]
- Dai, X.; Gan, W.; Li, X.; Wang, S.; Zhang, W.; Huang, L.; Liu, S.; Zhong, Q.; Guo, J.; Zhang, J.; et al. Prostate cancer–associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 2017, 23, 1063–1071. [Google Scholar] [CrossRef]
- Theurillat, J.-P.P.; Udeshi, N.D.; Errington, W.J.; Svinkina, T.; Baca, S.C.; Pop, M.; Wild, P.J.; Blattner, M.; Groner, A.C.; Rubin, M.A.; et al. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 2014, 346, 85–89. [Google Scholar] [CrossRef]
- Zhang, P.; Gao, K.; Tang, Y.; Jin, X.; An, J.; Yu, H.; Wang, H.; Zhang, Y.; Wang, D.; Huang, H.; et al. Destruction of DDIT3/CHOP Protein by Wild-Type SPOP but Not Prostate Cancer-Associated Mutants. Hum. Mutat. 2014, 35, 1142–1151. [Google Scholar] [CrossRef]
- Wang, X.; Jin, J.; Wan, F.; Zhao, L.; Chu, H.; Chen, C.; Liao, G.; Liu, J.; Yu, Y.; Teng, H.; et al. AMPK Promotes SPOP-Mediated NANOG Degradation to Regulate Prostate Cancer Cell Stemness. Dev. Cell 2018, 48, 345–360.e7. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Zhu, Y.; Dai, X.; Dang, F.; Ren, J.; Ren, S.; Shulga, Y.V.; Beca, F.; Gan, W.; et al. SPOP Promotes Nanog Destruction to Suppress Stem Cell Traits and Prostate Cancer Progression. Dev. Cell 2018, 48, 329–344.e5. [Google Scholar] [CrossRef]
- Wu, F.; Dai, X.; Gan, W.; Wan, L.; Li, M.; Mitsiades, N.; Wei, W.; Ding, Q.; Zhang, J. Prostate cancer-associated mutation in SPOP impairs its ability to target Cdc20 for poly-ubiquitination and degradation. Cancer Lett. 2017, 385, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.-G.; Zhu, Y.; Long, Q.-Y.; Li, X.-J.; Lin, X.; Tang, S.-B.; Yin, L.; Xiao, Y.; Wang, X.; Li, L.; et al. SPOP suppresses prostate cancer through regulation of CYCLIN E1 stability. Cell Death Differ. 2018, 26, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Kaochar, S.; Li, M.; Rajapakshe, K.; Fiskus, W.; Dong, J.; Foley, C.; Dong, B.; Zhang, L.; Kwon, O.-J.; et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene 2017, 36, 4767–4777. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, J.; Gao, K.; Zhang, P.; Yao, L.; Tang, Y.; Tang, L.; Ma, J.; Xiao, J.; Zhang, E.; et al. Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. PLoS Genet. 2017, 13, e1006748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, S.; Dai, X.; Gan, W.; Nie, X.; Wei, W.; Hu, G.; Guo, J. Tumor suppressor SPOP ubiquitinates and degrades EglN2 to compromise growth of prostate cancer cells. Cancer Lett. 2017, 390, 11–20. [Google Scholar] [CrossRef]
- Ma, J.; Chang, K.; Peng, J.; Shi, Q.; Gan, H.; Gao, K.; Feng, K.; Xu, F.; Zhang, H.; Dai, B.; et al. SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J. Exp. Clin. Cancer Res. 2018, 37, 145. [Google Scholar] [CrossRef]
- Gang, X.; Xuan, L.; Zhao, X.; Lv, Y.; Li, F.; Wang, Y.; Wang, G. Speckle-type POZ protein suppresses lipid accumulation and prostate cancer growth by stabilizing fatty acid synthase. Prostate 2019, 79, 864–871. [Google Scholar] [CrossRef]
- Shi, L.; Yan, Y.; He, Y.; Yan, B.; Pan, Y.; Orme, J.J.; Zhang, J.; Xu, W.; Pang, J.; Huang, H. Mutated SPOP E3 Ligase Promotes 17βHSD4 Protein Degradation to Drive Androgenesis and Prostate Cancer Progression. Cancer Res. 2021, 81, 3593–3606. [Google Scholar] [CrossRef]
- Ma, J.; Cai, M.; Mo, Y.; Fried, J.S.; Tan, X.; Ma, Y.; Chen, J.; Han, S.; Xu, B. The SPOP-ITCH Signaling Axis Protects Against Prostate Cancer Metastasis. Front. Oncol. 2021, 11, 658230. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, K.; Xie, H.; Wang, D.; Zhang, P.; Wei, T.; Yan, Y.; Pan, Y.; Ye, W.; Chen, H.; et al. SPOP mutation induces DNA methylation via stabilizing GLP/G9a. Nat. Commun. 2021, 12, 5716. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, J.; Zhu, Y.; Sun, X.; He, C.; Cai, M.; Ma, J.; Wang, Y.; Han, S. SPOP promotes CDCA5 degradation to regulate prostate cancer progression via the AKT pathway. Neoplasia 2021, 23, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Zheng, N.; Bu, L.; Zhang, X.; Zhang, X.; Wu, Y.; Su, Y.; Wang, L.; Ren, S.; Dai, X.; et al. SPOP-mediated ubiquitination and degradation of PDK1 suppresses AKT kinase activity and oncogenic functions. Mol. Cancer 2021, 20, 100. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Jin, X.; Zhang, P.; Li, Q.; Lv, Z.; Ding, Y.; He, H.; Wang, Y.; He, Y.; Zhao, X.; et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ. 2022, 29, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Hou, T.; Dan, W.; Zhu, Y.; Liu, B.; Wei, Y.; Wang, Z.; Gao, Y.; Zeng, J.; Li, L. ERK1/2 inhibits Cullin 3/SPOP-mediated PrLZ ubiquitination and degradation to modulate prostate cancer progression. Cell Death Differ. 2022, 29, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Byun, B.; Jung, Y. Repression of Transcriptional Activity of Estrogen Receptor α by a Cullin3/SPOP Ubiquitin E3 Ligase Complex. Mol. Cells 2008, 25, 289–293. [Google Scholar] [PubMed]
- Zhang, N.; Sun, P.; Xu, Y.; Li, H.; Liu, H.; Wang, L.; Cao, Y.; Zhou, K.; Wang, T. The GPER1/SPOP axis mediates ubiquitination-dependent degradation of ERα to inhibit the growth of breast cancer induced by oestrogen. Cancer Lett. 2021, 498, 54–69. [Google Scholar] [CrossRef]
- Zhou, Q.; Lin, W.; Wang, C.; Sun, F.; Ju, S.; Chen, Q.; Wang, Y.; Chen, Y.; Li, H.; Wang, L.; et al. Neddylation inhibition induces glutamine uptake and metabolism by targeting CRL3SPOP E3 ligase in cancer cells. Nat. Commun. 2022, 13, 3034. [Google Scholar] [CrossRef]
- Zeng, C.; Wang, Y.; Lu, Q.; Chen, J.; Zhang, J.; Liu, T.; Lv, N.; Luo, S. SPOP suppresses tumorigenesis by regulating Hedgehog/Gli2 signaling pathway in gastric cancer. J. Exp. Clin. Cancer Res. 2014, 33, 75. [Google Scholar] [CrossRef]
- Zhi, X.; Tao, J.; Zhang, L.; Tao, R.; Ma, L.; Qin, J. Silencing speckle-type POZ protein by promoter hypermethylation decreases cell apoptosis through upregulating Hedgehog signaling pathway in colorectal cancer. Cell Death Dis. 2016, 7, e2569. [Google Scholar] [CrossRef][Green Version]
- Tan, Y.; Ci, Y.; Dai, X.; Wu, F.; Guo, J.; Liu, D.; North, B.J.; Huo, J.; Zhang, J. Cullin 3SPOP ubiquitin E3 ligase promotes the poly-ubiquitination and degradation of HDAC6. Oncotarget 2017, 8, 47890–47901. [Google Scholar] [CrossRef]
- Jin, X.; Shi, Q.; Li, Q.; Zhou, L.; Wang, J.; Jiang, L.; Zhao, X.; Feng, K.; Lin, T.; Lin, Z.; et al. CRL3–SPOP ubiquitin ligase complex suppresses the growth of diffuse large B-cell lymphoma by negatively regulating the MyD88/NF-κB signaling. Leukemia 2020, 34, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-H.; Wang, Y.; Wang, F.; Dong, Y.-M.; Jiang, W.-L.; Wang, Y.-P.; Zhong, X.; Ma, L.-X. SPOP negatively regulates Toll-like receptor-induced inflammation by disrupting MyD88 self-association. Cell. Mol. Immunol. 2020, 18, 1708–1717. [Google Scholar] [CrossRef]
- Guillamot, M.; Ouazia, D.; Dolgalev, I.; Yeung, S.T.; Kourtis, N.; Dai, Y.; Corrigan, K.; Zea-Redondo, L.; Saraf, A.; Florens, L.; et al. The E3 ubiquitin ligase SPOP controls resolution of systemic inflammation by triggering MYD88 degradation. Nat. Immunol. 2019, 20, 1196–1207. [Google Scholar] [CrossRef]
- Yan, W.; Shi, X.; Wang, H.; Liao, A. Aberrant SPOP-CHAF1A ubiquitination axis triggers tumor autophagy that endows a therapeutical vulnerability in diffuse large B cell lymphoma. J. Transl. Med. 2022, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, B.; Gao, C.-X.; Xie, H.-K.; Han, C.-N.; Zhou, C.-C. SPOP promotes FADD degradation and inhibits NF-κB activity in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 504, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Bao, Y.-C.; Ji, X.-X.; Chen, B.; Deng, Q.-F.; Zhou, S.-W. SPOP promotes SIRT2 degradation and suppresses non-small cell lung cancer cell growth. Biochem. Biophys. Res. Commun. 2017, 483, 880–884. [Google Scholar] [CrossRef]
- Ji, P.; Liang, S.; Li, P.; Xie, C.; Li, J.; Zhang, K.; Zheng, X.; Feng, M.; Li, Q.; Jiao, H.; et al. Speckle-type POZ protein suppresses hepatocellular carcinoma cell migration and invasion via ubiquitin-dependent proteolysis of SUMO1/sentrin specific peptidase 7. Biochem. Biophys. Res. Commun. 2018, 502, 30–42. [Google Scholar] [CrossRef]
- Yuan, D.; Chen, Y.; Yang, Z.; Li, G.; Wu, M.; Jiang, J.; Li, D.; Yu, Q. SPOP Attenuates Migration and Invasion of Choriocar-cinoma Cells by Promoting DHX9 Degradation. Am. J. Cancer Res. 2020, 10, 2428–2445. [Google Scholar]
- Su, S.; Chen, J.; Jiang, Y.; Wang, Y.; Vital, T.; Zhang, J.; Laggner, C.; Nguyen, K.T.; Zhu, Z.; Prevatte, A.W.; et al. SPOP and OTUD7A Control EWS–FLI1 Protein Stability to Govern Ewing Sarcoma Growth. Adv. Sci. 2021, 8, 2004846. [Google Scholar] [CrossRef]
- He, W.; Zhang, J.; Liu, B.; Liu, X.; Liu, G.; Xie, L.; He, J.; Wei, M.; Li, K.; Ma, J.; et al. S119N Mutation of the E3 Ubiquitin Ligase SPOP Suppresses SLC7A1 Degradation to Regulate Hepatoblastoma Progression. Mol. Ther. Oncolytics 2020, 19, 149–162. [Google Scholar] [CrossRef]
- Pang, K.; Lee, J.; Kim, J.; Park, J.; Park, Y.; Hong, E.; An, H.; Ooshima, A.; Son, M.; Park, K.-S.; et al. Degradation of DRAK1 by CUL3/SPOP E3 Ubiquitin ligase promotes tumor growth of paclitaxel-resistant cervical cancer cells. Cell Death Dis. 2022, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Bunce, M.W.; Boronenkov, I.V.; Anderson, R.A. Coordinated Activation of the Nuclear Ubiquitin Ligase Cul3-SPOP by the Generation of Phosphatidylinositol 5-Phosphate. J. Biol. Chem. 2008, 283, 8678–8686. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Bhattacharyya, S.; Ojha, R.; Mandal, A.K.; Singh, S.K. Speckle-type POZ protein as a diagnostic biomarker in renal cell carcinoma. J. Cancer Res. Ther. 2018, 14, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhou, J.; Deng, Z.; Gao, Y.; Cheng, Y. SPOP promotes tumor progression via activation of β-catenin/TCF4 complex in clear cell renal cell carcinoma. Int. J. Oncol. 2016, 49, 1001–1008. [Google Scholar] [CrossRef]
- Harb, O.A.; Elfeky, M.A.; El Shafaay, B.S.; Taha, H.F.; Osman, G.; Harera, I.S.; Gertallah, L.M.; Abdelmonem, D.M.; Embaby, A. SPOP, ZEB-1 and E-cadherin expression in clear cell renal cell carcinoma (cc-RCC): Clinicopathological and prognostic significance. Pathophysiology 2018, 25, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Liu, X.; Sun, X. RNA interference-mediated silencing of speckle-type POZ protein promotes apoptosis of renal cell cancer cells. OncoTargets Ther. 2016, 9, 2393–2402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Lei, P.-J.; Ju, L.-G.; Wang, X.; Huang, K.; Yang, B.; Shao, C.; Zhu, Y.; Changwei, S.; Fu, X.-D.; et al. SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 2016, 45, 92–105. [Google Scholar] [CrossRef]
- Pfister, S.X.; Ahrabi, S.; Zalmas, L.-P.; Sarkar, S.; Aymard, F.; Bachrati, C.Z.; Helleday, T.; Legube, G.; La Thangue, N.B.; Porter, A.C.; et al. SETD2-Dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell Rep. 2014, 7, 2006–2018. [Google Scholar] [CrossRef]
- Guo, Z.-Q.; Zheng, T.; Chen, B.; Luo, C.; Ouyang, S.; Gong, S.; Li, J.; Mao, L.-L.; Lian, F.; Yang, Y.; et al. Small-Molecule Targeting of E3 Ligase Adaptor SPOP in Kidney Cancer. Cancer Cell 2016, 30, 474–484. [Google Scholar] [CrossRef]
- Araujo Rincon, M.; Burleson, M. Natural Compounds as a Promising Therapeutic Agent for SPOP Downregulated Breast Cancer. FASEB J. 2022, 36 (Suppl. 1). [Google Scholar] [CrossRef]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef]
- Cai, H.; Liu, A. Spop regulates Gli3 activity and Shh signaling in dorsoventral patterning of the mouse spinal cord. Dev. Biol. 2017, 432, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, H.; Sun, M.; Yang, J.; Zhang, W.; Han, S.; Xu, B. Speckle-type POZ protein, SPOP, is involved in the DNA damage response. Carcinogenesis 2014, 35, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Haffner, M.C.; Aryee, M.J.; Toubaji, A.; Esopi, D.M.; Albadine, R.; Gurel, B.; Isaacs, W.B.; Bova, G.S.; Liu, W.; Xu, J.; et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010, 42, 668–675. [Google Scholar] [CrossRef]
- Schaefer-Klein, J.L.; Murphy, S.J.; Johnson, S.H.; Vasmatzis, G.; Kovtun, I.V. Topoisomerase 2 α Cooperates with Androgen Receptor to Contribute to Prostate Cancer Progression. PLoS ONE 2015, 10, e0142327. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The Epigenomics of Cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Chen, X.; Chen, J.; Guan, Y.; Liu, Y.; Chen, J.; Lv, X. Speckle-type POZ protein functions as a tumor suppressor in non-small cell lung cancer due to DNA methylation. Cancer Cell Int. 2018, 18, 213. [Google Scholar] [CrossRef]
- Huang, C.-J.; Chen, H.-Y.; Lin, W.-Y.; Choo, K.B. Differential expression of speckled POZ protein, SPOP: Putative regulation by miR-145. J. Biosci. 2014, 39, 401–413. [Google Scholar] [CrossRef]
- Ding, M.; Lu, X.; Wang, C.; Zhao, Q.; Ge, J.; Xia, Q.; Wang, J.; Zen, K.; Zhang, C.-Y.; Zhang, C. The E2F1–miR-520/372/373–SPOP Axis Modulates Progression of Renal Carcinoma. Cancer Res. 2018, 78, 6771–6784. [Google Scholar] [CrossRef]
- Sun, W.; Cui, J.; Ge, Y.; Wang, J.; Yu, Y.; Han, B.; Liu, B. Tumor stem cell-derived exosomal microRNA-17-5p inhibits anti-tumor immunity in colorectal cancer via targeting SPOP and overexpressing PD-L1. Cell Death Discov. 2022, 8, 223. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Jin, Y.; Song, J.-L.; Deng, F. MiR-373 promotes proliferation and metastasis of oral squamous cell carcinoma by targeting SPOP. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5270–5276. [Google Scholar]
- Wang, L.; Yu, P.; Li, B.; Guo, Y.; Liang, Z.; Zheng, L.; Yang, J.; Xu, H.; Liu, S.; Zheng, L.; et al. miR-372 and miR-373 enhance the stemness of colorectal cancer cells by repressing differentiation signaling pathways. Mol. Oncol. 2018, 12, 1949–1964. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, F.; Wang, X.; He, Z.; Zhu, X. miRNA-543 promotes cell migration and invasion by targeting SPOP in gastric cancer. OncoTargets Ther. 2018, 11, 5075–5082. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, J.; Zhang, X.; Tong, Y.; Gan, X. MiR-520b promotes the progression of non-small cell lung cancer through activating Hedgehog pathway. J. Cell. Mol. Med. 2018, 23, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Huan, M.; Yang, W.; Gao, Y.; Wang, K.; Xu, S.; Zhang, M.; Ma, J.; Wang, X.; Chen, Y.; et al. Snail promotes prostate cancer migration by facilitating SPOP ubiquitination and degradation. Biochem. Biophys. Res. Commun. 2020, 529, 799–804. [Google Scholar] [CrossRef]
- Bouchard, J.J.; Otero, J.H.; Scott, D.C.; Szulc, E.; Martin, E.W.; Sabri, N.; Granata, D.; Marzahn, M.R.; Lindorff-Larsen, K.; Salvatella, X.; et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 2018, 72, 19–36.e8. [Google Scholar] [CrossRef]
- Chong, P.A.; Forman-Kay, J.D. Liquid–liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 2016, 41, 180–186. [Google Scholar] [CrossRef]
- Mukhopadhyay, C.; Zhou, P. G3BP1 modulates SPOP to promote prostate tumorigenesis. Mol. Cell. Oncol. 2022, 9, 2030171. [Google Scholar] [CrossRef]
- Ji, X.; Cheng, K.; Gao, C.; Xie, H.; Zhu, R.; Luo, J. HS3ST1 Promotes Non-Small-Cell Lung Cancer Progression by Targeting the SPOP/FADD/NF-κB Pathway. BioMed Res. Int. 2022, 2022, 5509346. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).