Abstract
Aerobic bacteria that degrade methylphosphonates and produce methane as a byproduct have emerged as key players in marine carbon and phosphorus cycles. Here, we present two new draft genome sequences of the genus Marivita that were assembled from metagenomes from hypersaline former industrial salterns and compare them to five other Marivita reference genomes. Phylogenetic analyses suggest that both of these metagenome-assembled genomes (MAGs) represent new species in the genus. Average nucleotide identities to the closest taxon were <85%. The MAGs were assembled with SPAdes, binned with MetaBAT, and curated with scaffold extension and reassembly. Both genomes contained the phnCDEGHIJLMP suite of genes encoding the full C-P lyase pathway of methylphosphonate degradation and were significantly more abundant in two former industrial salterns than in nearby reference and restored wetlands, which have lower salinity levels and lower methane emissions than the salterns. These organisms contain a variety of compatible solute biosynthesis and transporter genes to cope with high salinity levels but harbor only slightly acidic proteomes (mean isoelectric point of 6.48).
1. Introduction
Biogenic methane production in nature has historically been attributed to anaerobic decomposition of organic matter performed by archaea. While this is the dominant known source of naturally produced methane, recent work has elucidated additional pathways performed by members of the bacterial domain in aerobic environments. This body of work has its origins in observations of elevated methane emissions from aerobic marine environments, a phenomenon which became known as the “methane paradox” [1]. Subsequent work attempting to explain the methane paradox in both oceans and freshwater ecosystems over the past 15 years has elucidated two new pathways of aerobic methane production by bacteria—methylphosphonate degradation via the now well-described C-P lyase pathway [1,2,3,4,5], and methylamine degradation via a yet-undescribed biochemical pathway involving aspartate aminotransferase [6].
The more well-known and better-described C-P lyase pathway of methylphosphonate degradation involves the suite of genes phnCDE to first import methylphosphonate into the cytoplasm and then phnGHIJLM to catalyze a sequence of four reactions [4,5,7]. Methane is produced as a byproduct in the third reaction, in which PhnJ catalyzes the conversion of α-D-ribose-1-methylphosphonate 5-phosphate to 5-phospho-α-D-ribose 1,2-cyclic phosphate [4,5,8]. Importantly, methylphosphonates can be produced by abundant marine organisms, including Nitrosopumilus maritimus [9] and Pelagibacter ubique [10]. Methylphosphonates are known to be used by microbes, both in marine and soil environments, as a source of phosphorus when other forms of phosphorus such as inorganic phosphate are limited [11,12,13].
Many genera of archaea and bacteria contain the phnJ gene, from Euryarchaeota to Proteobacteria and Cyanobacteria [14,15]. At the time of writing, phnJ genes were annotated in 15,935 genomes on the Integrated Microbial Genomes and Microbiomes (IMG/M) database [16]; of these, 15,875 genomes are identified to genus level and span 18 archaeal genera and 730 bacterial genera. Marivita (Rhodobacteraceae) is one of the bacterial genera harboring these genes that has been described as abundant in marine environments and important for ocean biogeochemical cycles [17]. Marivita have been isolated from different saline habitats around the globe, including the Korean coast [17,18,19,20], the Chesapeake Bay estuary [21], the saline Tuosu Lake in China [22], and from the marine dinoflagellate Alexandrium catenella [23]. They have been described as having optimal growth at 2–4% sodium chloride (NaCl) but are capable of tolerating salinities up to 10% NaCl. The two new metagenome-assembled genomes (MAGs) presented here come from sediment samples with 147.6 ppt (14.76%) and 166.8 ppt (16.68%) salinity (Cl−), which expands the known salinity tolerance of the genus.
Here, we describe two new draft genomes of two novel Marivita species, given the names Marivita sp. SBSPR1 and Marivita sp. SBSPR2. Since these metagenome-assembled genomes were assembled from hypersaline former industrial solar salterns with elevated methane emissions, we focused on describing methylphosphonate cycling genes and halotolerance genes (i.e., genes for compatible solute synthesis) present in the two genomes. We also conducted a phylogenetic analysis and comparison of shared and unique orthologous gene groups of the two MAGs and the five Marivita sister taxa with currently and publicly available Marivita genomes.
2. Materials and Methods
Marivita sp. SBSPR1 (named after South Bay Salt Pond R1) was assembled from metagenomic DNA sequence from a 5–15 cm deep sediment sample R1_B_D2 (IMG/M ID 3300007712, National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) accession SRP098112) from an unrestored, hypersaline former industrial solar saltern as part of the study published by Zhou et al., 2021 [15]. Marivita sp. SBSPR2 (named after South Bay Salt Pond R2) was assembled from metagenomic DNA sequence from a 5–15 cm deep sediment sample R2_B_D2 (IMG/M ID 3300009061, NCBI SRA accession SRP118410), a 5–15 cm deep section of a sediment core from an adjacent unrestored hypersaline former industrial solar saltern. Industrial salt production occurred in these ponds from the 1850s to 2003. The two samples are characterized by high salinity (147.6 and 166.8 total ppt Cl− for R1 and R2 samples, respectively), moderate temperature (25.0 and 26.2 °C), hypoxic but not completely anaerobic conditions (1.35 and 3.08 mg L−1 DO), and near neutral pH (7.36 and 7.80) [15]. Elevated methane (CH4) emissions were measured from both cores in the field (R1 = 1347, R2 = 1607 µmol CH4 m−2 d−1) [15]. The metagenomic sequencing, processing, and binning has been described previously [15,24]. For these two draft genomes, we began with MAGs constructed with MetaBAT [25], with a completeness of 99.05% and 96.29% and contamination of 0.74% and 3.17%, for M. sp. SBSPR1 and M. sp. SBSPR2, respectively, as calculated by CheckM version 1.0.18 [26], which would be considered “high-quality” by accepted standards [27]. The original MAGs were initially classified to genus level as Marivita by both Bin Annotation Tool [28] and the GTDB-Tk classifier version 1.1.0 [29].
We estimated the abundance of the two MAGs across the 24 samples collected by Zhou et al. (2021) using average read depth and then transformed this to counts per million of assembled reads [15]. The effect of site (4 separate wetlands) on MAG abundance was tested with a Kruskal–Wallis and Nemenyi post hoc tests, as assumptions for ANOVA and Tukey HSD were not met (Levene Test, p < 0.05, Shapiro–Wilk test, p < 0.05).
We used Geneious version 2021.1.1 [30] (Biomatters Ltd., Auckland, New Zealand) to remap paired-end reads to each scaffold and extend using reads partially overlapping the ends of the scaffolds as described elsewhere [24,31]. Performing two rounds of these steps improved the completeness of M. sp. SBSPR1 to 99.10% and M. sp. SBSPR2 to 97.34% and decreased the contamination of M. sp. SBSPR2 to 2.37%. Neither genome contained a full-length 16S rRNA gene, which can be difficult to recover during MAG assembly. The updated MAGs were again classified as Marivita by both Bin Annotation Tool and GTDB-Tk. The genomes were then uploaded to the Joint Genome Institute’s Integrated Microbial Genomes and Microbiomes database (IMG/M) [16] to undergo their comprehensive annotation procedure and are publicly available with IMG/M IDs 2930928012 and 2930931642.
To compare the two new genomes with their sister taxa in the Marivita genus, we used KBase [32] to download five reference genomes publicly available from the RefSeq [33] database. These reference genomes are M. hallyeonensis (NZ_FQXC01000020) [18,34], M. geojedonensis (NZ_PVTN01000001) [19,34], M. lacus (NZ_BMFC01000001) [22,35], M. cryptomonadis MP20-4 (NZ_JAFBXM010000001) [36], and M. cryptomonadis LZ-15-2 (NZ_SWKO01000011) [23]. For a broader phylogenetic perspective, the “Insert Set of Genomes Into SpeciesTree” tool (version 2.2.0) in Kbase [32] was used to place the seven Marivita genomes into a phylogenetic tree with 50 other genomes from RefSeq using a concatenated alignment of 49 single-copy clusters of orthologous groups (COGs). A more detailed phylogenetic tree of just the seven Marivita genomes was constructed by first identifying and aligning 120 universal single copy bacterial marker genes with GTDB-Tk and then building a consensus tree from the concatenated alignment with RAxML version 8.12 [37] with the PROTGAMMALG model of amino acid substitution, 1000 bootstraps, and Bacillus subtilis strain 168 (RefSeq GCF_000009045.1) as an outgroup. ProtTest3 version 3.4.2 [38] was used to select the best model of amino acid substitution (LG). Lastly, average nucleotide identity (ANI) was calculated with FastANI version 0.1.3 implemented in KBase. The KBase narrative for this project is publicly available under narrative ID 92727.
Prodigal [39] was used to predict protein-coding genes from the nucleotide sequences. We calculated mean isoelectric points for the seven proteomes with EMBOSS [40] using the “iep” tool on the Galaxy website platform (https://usegalaxy.org/ (accessed on 13 August 2021) [41] with a pH step of 0.5. Both proteinortho [42] and KEGG orthology (KO) [43] were used to analyze orthologous gene groups, and intersections among the genomes were plotted with ComplexUpset in R [44]. KO profiles were created for each genome with BLAST Koala [45]. Pfam [46] was used to classify genes from proteinortho. KOs involved in methylphosphonate cycling and salt tolerance were extracted from the KO tables. A list of KOs involved in methylphosphonate cycling was created based on the literature [4,5] and the BioCyc [47] and KEGG [43] databases. A list of KOs involved in salt tolerance was developed based on discussion of salt-tolerance-related genes and processes in the literature [48,49,50,51,52,53], and it includes KOs that are involved in compatible solute biosynthesis and transport (e.g., betaine and trehalose) and cation transporters. KO presence/absence was plotted as heatmaps with pheatmap in R [54].
For a broader comparison of Marivita and phnJ abundances in the salterns to a wide variety of saline and hypersaline ecosystems, we downloaded available metagenomic datasets on IMG/M from sediment samples of marine/coastal habitats with at least two samples (n = 613) and at least marine salinity (e.g., lagoon, ocean, estuary, salt marsh), as well as inland saline habitats such as hypersaline lakes and hot springs with at least two samples. phnJ counts were acquired from assembled metagenomes using the “Statistical Analysis” tool on IMG/M set to analyze KO counts per genome. Counts were then normalized with DESeq2 [55] in R, and then the phnJ gene (K06163) was extracted from the full table. Marivita abundances were acquired by downloading the genus-level taxonomic tables with the “Statistical Analysis” tool and “Gene Count” measurement (not using coverage as it was not available for all metagenomes) on IMG/M and then calculating counts per million assembled metagenomic reads with the “Genome Size * Assembled” metadata column associated with each metagenome on IMG/M. Only metagenomes with at least 1000 genus-level classified reads prior to transformation were included (n = 590). Information about the metagenomes in this analysis can be found in Table S1. All R analyses were performed with version 4.0.2 [56].
3. Results
According to the phylogenetic tree constructed with 49 single copy COGs, the MAGs were placed within the Marivita genus (Figure S1), in agreement with MAG classification tools such as Bin Annotation Tool and GTDB-Tk. In the more detailed tree of just Marivita reference genomes and the two MAGs using the set of 120 universal bacterial single-copy protein coding genes in GTDB-Tk, both genomes were placed taxonomically with the Marivita genus, yet appear to be unique species in the genus (Figure 1). This result is supported by ANI analysis, with ANI values among the MAGs and reference genomes ranging from 78 to 84, which suggests new species in the Marivita genus (Figures S2 and S3) [57,58,59]. Both genomes were most closely related to M. lacus. M. sp. SBSPR1 has the largest genome of the seven Marivita genomes presented here, while M. sp. SBSPR2 has the smallest. The MAGs have two of the three highest GC contents among the seven genomes (Figure 1).
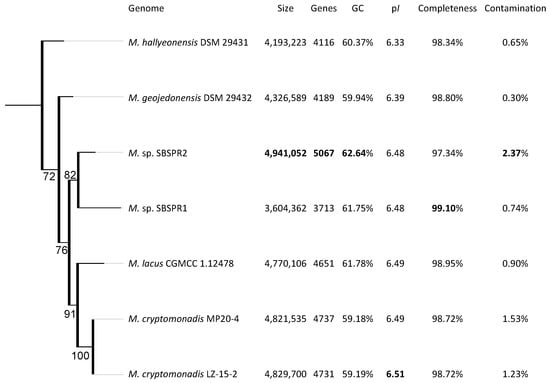
Figure 1.
RAxML phylogenetic tree of the genus Marivita. The tree was built with a concatenated alignment of 120 single-copy bacterial genes and the PROTGAMMALG model of amino acid substitution. Branch labels show the bootstrap support, calculated with 1000 bootstraps. The genome size, number of protein coding genes, percentage G + C content, percentage completeness and contamination estimates from CheckM, and RefSeq ID are also shown. Bolded values highlight the greatest values in each column. The tree is rooted with Bacillus subtilis strain 168 as an outgroup (not shown).
Across all seven genomes, proteinortho identified 5367 orthologous gene groups, while KEGG annotation identified 2312 KOs. Among the seven Marivita genomes included in this analysis, according to proteinortho, there were 2305 orthologous genes shared among all seven taxa; there were 279 genes missing from the Marivita sp. SBSPR1 genome, 64 genes missing from Marivita sp. SBSPR2, and 74 additional genes missing from both (Figure 2a). These missing genes encompass a wide variety of functions, including various transporters (ABC, periplasmic, binding-protein-dependent systems) and various enzymes (e.g., citrate synthase, Acyl-CoA dehydrogenase, dimethylsulfoniopropionate lyase) (Table S2). In terms of KOs, there were 91 KOs present in the other six genomes but not that of M. sp. SBSPR1, and likewise, 21 KOs were missing from M. sp. SBSPR2, as well as an additional 27 KOs missing from both genomes (Figure 2b). These included, for example, nitrate/nitrite transport system genes nrtABC, glucose/mannose transport system genes gtsABC, and N-methylhydantoinase hyuAB genes (Table S3). There were also 12 KOs present in both MAGs and missing from the other five reference genomes. Half of these were D-xylose (xylFGH) and putative multiple sugar transport system genes chvE and gguAB (Table S3). While genes unique to one genome were not calculated by proteinortho (by definition, as they are not shared), we identified 92 KOs unique to M. sp. SBSPR1 and 44 KOs unique to M. sp. SBSPR2 (Figure 2b, Table S2).
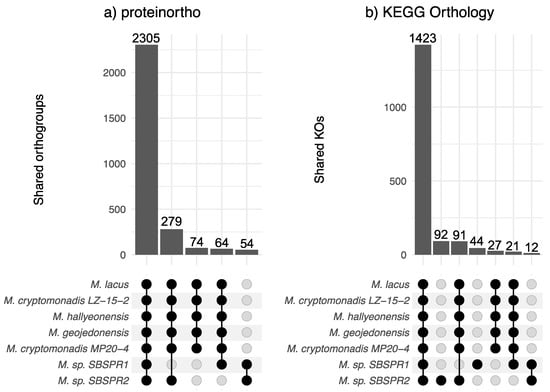
Figure 2.
Shared orthologous gene groups in the genus Marivita. Intersections were calculated with (a) proteinortho and (b) KEGG orthology profiles. Displayed are intersections among all seven genomes, six other genomes but not Marivita sp. SBSPR1, six other genomes but not Marivita sp. SBSPR2, genes unique to both M. sp. SBSPR1 and SBSPR2, or genes present in only M. sp. SBSPR1 or M. sp. SBSPR2 (only applicable for shared KOs).
With respect to methylphosphonate cycling genes, none of the genomes examined here contained the mpnS gene for methylphosphonate biosynthesis, but all contained the three methylphosphonate transporter genes phnCDE (Figure 3). Five of the seven genomes, including both MAGs, contained the full suite of genes phnGHILMJ for the C-P lyase methylphosphonate degradation pathway. It is also notable that the two unrestored salterns sampled here had among the highest phnJ gene counts of any saline or hypersaline sediment metagenomes on IMG/M (Figure 4a). Following production of methane during the formation of 5-phospho-a-ribosyl-1,2-cyclic phosphate catalyzed by phnJ, the degradation can proceed to produce either a-D-ribose 1,5-biphosphate or D-ribofuranose 5-phosphate. All seven genomes contained phnP to produce the former, while they all lacked phnPP to produce the latter. Alternatively, there is another described pathway of methylphosphonate degradation that does not involve C-P lyases [60]. However, none of the seven genomes contained phnY or phnZ1, the two genes involved in this non C-P lyase pathway.
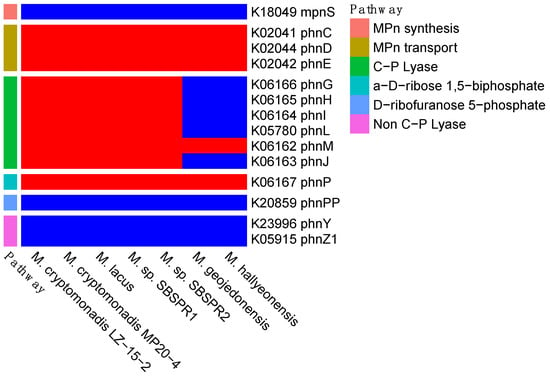
Figure 3.
Presence (red) or absence (blue) methylphosphonate synthesis, transport, and degradation genes, with their abbreviated names and KEGG K number assignments. Shown here are the core C-P lyase pathway genes followed by two different post-methane pathways (phnP or phnPP), as well as the non-C-P lyase pathway.
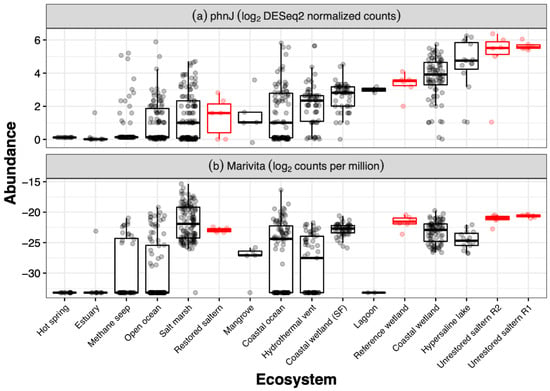
Figure 4.
Abundance of (a) phnJ gene (K06163) and (b) Marivita from metagenomic sediment samples from various saline to hypersaline environments. Abundances are gene counts that were extracted from publicly available metagenomes on IMG/M using the “Statistical Analysis” tool and selecting feature type “KO” for (a) and “Genus” for (b) (see methods). Highlighted in red are the restored saltern, reference wetland, and the two unrestored salterns (R2, R1) described in Zhou et al. (2021). Note that open ocean sediments are separated from coastal ocean sediments. Furthermore, coastal wetlands from the San Francisco Bay and Delta (“Coastal wetland (SF)”) are separated from other coastal wetland samples due to their proximity to our sampling sites.
The two MAGs were more abundant in two unrestored former salterns compared to a reference wetland and a restored saltern (Figure 5a). Furthermore, generally Marivita counts per million were greater in the two hypersaline former salterns sampled here than in other saline to hypersaline sediments such as lagoons, mangroves, open ocean sediments, or hypersaline lakes (Figure 4b), although some specific salt marshes and marine samples had greater counts. Greater abundances of the MAGs were associated with the greater methane emissions in the salterns, but these relationships were not linear (Figure 5b). MAG abundances followed a bell-shaped curve in relation to salinity, close to zero at near ocean salinity, peaking between 100 and 150 ppt Cl−, and declining in the most hypersaline saltern sediments with 200+ ppt Cl− (Figure 5c).
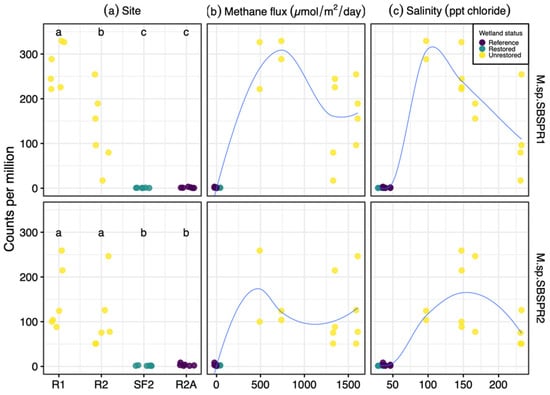
Figure 5.
Abundance of Marivita sp. SBSPR1 and SBSPR2, expressed as counts per million assembled reads, across 24 metagenomic samples from Zhou et al. (2021) [15], organized by (a) site, (b) methane flux, and (c) salinity. Lines from loess functions are shown in (b,c). R1, R2, SF2, and R2A refer to site names as in Zhou et al. (2021). Different letters in (a) denote statistically significant pairwise differences (Nemenyi posthoc test, p < 0.05).
The two MAGs and the other five reference genomes contained genes for the synthesis and transport of a variety of compatible solutes including betaine, ectoine, glutamine, glutamate, hydroxyectoine, proline, and trehalose, as well as potassium uptake uniporters (Figure 6). There were some differences in the presence/absence of these genes between the two MAGs and between the MAGs and the other reference genomes. For example, M. sp. SBSPR1 lacked genes for trehalose synthesis and transport but was the only genome containing the sdmt gene for betaine biosynthesis from sarcosine. On the other hand, M. sp. SBSPR2 lacked trehalose synthesis genes but was the only genome containing trehalose transporter genes. M. sp. SBSPR2 was also the only genome containing Vnx1, a sodium and hydrogen antiporter for sodium extrusion. M. sp. SBSPR2 and M. lacus were the only two genomes with the ectD gene for hydroxyectoine synthesis. The proteomes were only slightly acidic, with mean isoelectric points ranging from pH 6.33 to 6.51 (Figure 1), which is greater than the numbers reported for specialized halophilic archaea using the “salt in” strategy [61]. However, the genomes do show asymmetrical bimodal isoelectric point profiles typical of halophilic organisms with a large peak around 5 and a smaller peak around 10 (Figure S4).
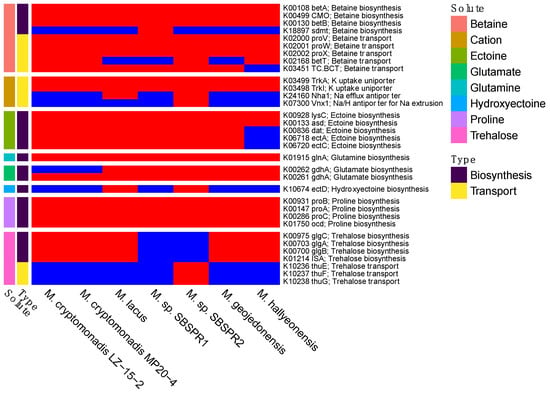
Figure 6.
Presence (red) or absence (blue) of compatible solute or salt biosynthesis and transport genes, with their abbreviated names and KEGG K number assignments. Compounds include betaine, cations (K+, Na+, H+), ectoine, glutamine, glutamate, hydroxyectoine, proline, and trehalose.
4. Discussion
Marivita taxa have been isolated from diverse marine environments and as part of the Roseobacter clade are important players in marine biogeochemical cycles [17,62,63]. Indeed, in a broad survey of other saline metagenomes on IMG/M, Marivita were present in most sample types except for lagoon sediments and inland saline hot spring sediments, the latter of which is not surprising due to the extreme temperatures. Previous laboratory growth experiments on Marivita cultures demonstrated a salt tolerance up to at least 10% in two taxa [22] but optimal growth at 2–4% salinity and maximum tolerances between 5 and 9% for the majority of the described Marivita species [18,19,20,21,22,23]. It is thus notable that we found two Marivita MAGs in samples with up to 23% salinity, which expands the known salt tolerance of the genus.
All seven Marivita genomes contained genes for the synthesis and transport of multiple compatible solutes, including betaine, ectoine, glutamine, glutamate, and proline, and some genomes additionally contained genes for trehalose and hydroxyectoine. These highly soluble, low-molecular-weight compounds are used by bacteria to maintain osmotic equilibrium with the environment without increasing intracellular salt concentrations [64]. The Marivita proteomes were slightly acidic on average, similar to other aerobic marine bacteria such as Aliivibrio fischeri (6.52) and Alteromonas macleodii (6.46) [61], with asymmetric bimodal isoelectric profiles (Figure S4). Their average isoelectric points (pIs) of 6.48 are considerably higher than extremely halophilic organisms such as the archaeon Halobacterium NRC-1 (5.03) and the bacterium Salinibacter ruber (5.92), both of which require high salt concentrations and utilize the “salt in” strategy of increasing cytoplasm salt concentrations with potassium [61]. Furthermore, 6.48 is also higher than the pI of a Methanosarcinaceae sp. genome isolated from the same Pond R1, which had a mean pI of 5.90 [24], similar to Salinibacter ruber. As with pI, GC contents of the Marivita MAGs (~62%) were also similar to the other Marivita genomes and are in the range of other reported halophilic organisms [65]. Thus, neither of these MAGs possess salinity adaptation mechanisms clearly distinct from those found in other Marivita species. A broad analysis of gene content did not suggest any additional adaptation mechanisms, although numerous differences in transporter genes suggest these may help these organisms survive in this unique biogeochemical environment.
All of these data together suggest that despite being found in 15–23% salinities, these two Marivita MAGs are capable of growing in a wide range of salinities and are not specialized to only extremely hypersaline environments, but this remains to be confirmed by laboratory growth experiments. We plan to conduct follow up studies on the results presented in this paper with efforts to culture these organisms and run growth experiments with methylphosphonate substrates, different levels of salinity, and other standard variables.
Given that the hypersaline unrestored salterns studied here had elevated methane emissions [15], we were particularly interested in the capability of Marivita to produce methane from methylphosphonate degradation. Indeed, both MAGs as well as genomes of three other taxa (M. lacus, M. cryptomonadis MP20-4, and M. cryptomonadis LZ-15-2) contained genes to import methylphosphonate into the cytosol (phnCDE) and degrade it to a-D-ribose 1,5-biphosphate via the C-P lyase pathway (phnGHILMJP). From the genomic data alone, M. hallyeonensis and M. geojedonensis do not appear to be capable of degrading methylphosphonate.
The phnJ gene involved in the methane production step of methylphosphonate degradation was notably high in unrestored salterns compared to all of the other ecosystem types, which can be attributed to the higher abundances of phnJ-containing taxa such as the two Marivita genomes presented here as well as other taxa such as Roseobacter [15], likely as a result of phosphate-limited conditions. phnJ abundances have been found to be negatively correlated with inorganic phosphate concentrations in both ocean water [11] and soils [12]. In our data, this trend is also true; phnJ abundance increases with increasing phosphorus limitation as measured by both decreasing inorganic phosphate levels and increasing inorganic N:P ratios (Figure S5). The unrestored salterns are phosphorus-limited relative to the reference wetland where both total N:total P ratios and inorganic N:inorganic P ratios are much lower than in the unrestored ponds [15] (Figure S5). In such phosphate-limited conditions, methylphosphonates can provide an alternative source of phosphorus for Marivita, as has been shown for several other taxa such as the cyanobacteria Nodularia spumigena [14] and Trichodesmium [66], the Alphaproteobacteria Pelagibacterales [13], Agrobacterium, Rhizobium [5], and the Gammaproteobacteria Pantoea [5].
Neither of the two Marivita MAGs nor other Marivita reference genomes contained mpnS to synthesize methylphosphonate. It has been previously shown that other abundant marine taxa such as the ammonia-oxidizing Thaumarchaeota taxa Nitrosopumilus maritimus and Pelagibacter ubique can synthesize methylphosphonate, so these or other marine organisms could be a source of methylphosphonates in this system [9,10]. While Nitrosopumilus was not abundant in the unrestored salterns [15], it was abundant in the reference wetland and in San Francisco Bay area wetlands in general, and thus there could be both historic stores of methylphosphonates from before the ponds became hypersaline, or inputs from surrounding areas. In fact, the mpnS gene (K18049) was completely absent in saltern R1 metagenomes and nearly absent (total counts of 0–6) in saltern R2 metagenomes [15], so it is unclear if there is currently any methylphosphonate production in the unrestored ponds themselves or if the only current inputs are influxes from the surrounding environments.
5. Conclusions
Here, we have presented the draft genome sequences of two new Marivita MAGs, designated Marivita sp. SBSPR1 and Marivita sp. SBSPR2, and compared them to their sister taxa in the Marivita genus. Both of these genomes likely represent new species in the Marivita genus; while clearly placed in Marivita in a reference genome tree, ANI values were less than 85%. Marivita sp. SBSPR1 and Marivita sp. SBSPR2 both contain genetic architecture for compatible solute transport and biosynthesis, likely enabling them to grow in the hypersaline samples from which the metagenomes were sourced. Marivita are ubiquitous marine taxa, which may play important biogeochemical roles including methylphosphonate degradation, not only in the open ocean but also in human-altered coastal wetlands.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13010148/s1, Figure S1: The seven genomes analyzed in this study (highlighted) placed within a reference tree set to display 50 other genomes. The tree was built using the “Insert Genomes into Species Tree” tool in KBase, which uses 49 single-copy COGs to build the tree. Figure S2: Average nucleotide identity (ANI) between the new Marivita sp. SBSPR1 and SBSPR2 genomes and five other Marivita reference genomes. ANI was calculated with the FastANI tool in KBase. Figure S3: MASH clustering of average nucleotide identities, which were calculated with gANI in dRep. Figure S4: Isoelectric point profiles for the seven genomes analyzed in this study. All genomes show an acidic bias with an asymmetrical bimodal distribution. Histograms were created with 30 bins. Figure S5: phnJ gene abundance (normalized by DESeq2) as a function of inorganic N (NH4+ + NO3−) to inorganic P (phosphate extracted with Olsen method) ratio and inorganic P (phosphate extracted with Olsen method). Table S1: Information for the metagenomes downloaded from IMG/M for the comparative analysis. Table S2: Pfam results for 74 genes missing from Marivita sp. SBSPR1 and Marivita sp. SBSPR2 according to proteinortho, Table S3: Kos missing from or unique to Marivita sp. SBSPR1 and Marivita sp. SBSPR2.
Author Contributions
Conceptualization, C.P.B.d.M. and S.G.T.; Data curation, C.P.B.d.M.; Formal analysis, C.P.B.d.M., J.Z. and S.T.; Funding acquisition, S.G.T.; Methodology, C.P.B.d.M., J.Z., S.T. and S.G.T.; Project administration, S.G.T.; Resources, S.G.T.; Supervision, S.G.T.; Visualization, C.P.B.d.M.; Writing—original draft, C.P.B.d.M.; Writing—review and editing, C.P.B.d.M. and S.G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the DOE Early Career Research Program, grant no. KP/CH57/1, and includes work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The two new genomes are publicly available at NCBI (JAIPUS000000000 and JAIPUT000000000) and IMG/M (2930928012 and 2930931642), with detailed metadata provided in GOLD (Ga0500780 and Ga0500781) for M. sp. SBSPR1 and M. sp. SBSPR2, respectively. The metagenomes from which they were derived are also available on IMG/M (3300026165 and 3300026251; GOLD IDs Gp0125921 and Gp0125927) and NCBI (SAMN06264691 and SAMN07631159) for M. sp. SBSPR1 and M. sp. SBSPR2, respectively. Some aspects of the analysis are publicly available on Kbase narrative 92727.
Acknowledgments
We thank the production and sequence analysis teams at JGI for sequencing and bioinformatic support and Frederik Schulz for phylogenomic advice. We thank John Bourgeois, Donna Ball, and John Krause for help accessing the field sites. We thank Markus Göker for permission to use the M. hallyeonensis and M. geojedonensis genomes and acknowledge the United States Department of Energy and German Research Council for funding the sequencing and prior work done on those genomes. We thank Alma Parada and Anne Dekas for permission to use metagenomes from their project (IMG/M Study Name “Marine sediment microbial communities off the coast of San Francisco, CA, United States”) in the comparative analysis. We thank Brandi Reese and Rachel Weisend for permission to use metagenomes from their project (IMG/M Study Name “Sediment microbial communities from coastal wetland in Texas, United States”), funded by the Texas General Land Office, in the comparative analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karl, D.M.; Beversdorf, L.; Björkman, K.M.; Church, M.J.; Martinez, A.; Delong, E.F. Aerobic Production of Methane in the Sea. Nat. Geosci. 2008, 1, 473–478. [Google Scholar] [CrossRef]
- Del Valle, D.; Karl, D. Aerobic Production of Methane from Dissolved Water-Column Methylphosphonate and Sinking Particles in the North Pacific Subtropical Gyre. Aquat. Microb. Ecol. 2014, 73, 93–105. [Google Scholar] [CrossRef]
- Repeta, D.J.; Ferrón, S.; Sosa, O.A.; Johnson, C.G.; Repeta, L.D.; Acker, M.; DeLong, E.F.; Karl, D.M. Marine Methane Paradox Explained by Bacterial Degradation of Dissolved Organic Matter. Nat. Geosci. 2016, 9, 884–887. [Google Scholar] [CrossRef]
- Martínez, A.; Ventouras, L.-A.; Wilson, S.T.; Karl, D.M.; Delong, E.F. Metatranscriptomic and Functional Metagenomic Analysis of Methylphosphonate Utilization by Marine Bacteria. Front. Microbiol. 2013, 4, 340. [Google Scholar] [CrossRef]
- Yao, M.; Henny, C.; Maresca, J.A. Freshwater Bacteria Release Methane as a By-Product of Phosphorus Acquisition. Appl. Environ. Microbiol. 2016, 82, 6994–7003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Alowaifeer, A.; Kerner, P.; Balasubramanian, N.; Patterson, A.; Christian, W.; Tarver, A.; Dore, J.E.; Hatzenpichler, R.; Bothner, B.; et al. Aerobic Bacterial Methane Synthesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2019229118. [Google Scholar] [CrossRef] [PubMed]
- Kamat, S.S.; Williams, H.J.; Raushel, F.M. Intermediates in the Transformation of Phosphonates to Phosphate by Bacteria. Nature 2011, 480, 570–573. [Google Scholar] [CrossRef]
- Kamat, S.S.; Williams, H.J.; Dangott, L.J.; Chakrabarti, M.; Raushel, F.M. The Catalytic Mechanism for Aerobic Formation of Methane by Bacteria. Nature 2013, 497, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, W.W.; Griffin, B.M.; Cicchillo, R.M.; Gao, J.; Janga, S.C.; Cooke, H.A.; Circello, B.T.; Evans, B.S.; Martens-Habbena, W.; Stahl, D.A.; et al. Synthesis of Methylphosphonic Acid by Marine Microbes: A Source for Methane in the Aerobic Ocean. Science 2012, 337, 1104–1107. [Google Scholar] [CrossRef]
- Born, D.A.; Ulrich, E.C.; Ju, K.-S.; Peck, S.C.; van der Donk, W.A.; Drennan, C.L. Structural Basis for Methylphosphonate Biosynthesis. Science 2017, 358, 1336–1339. [Google Scholar] [CrossRef] [PubMed]
- Sosa, O.A.; Repeta, D.J.; DeLong, E.F.; Ashkezari, M.D.; Karl, D.M. Phosphate-Limited Ocean Regions Select for Bacterial Populations Enriched in the Carbon-Phosphorus Lyase Pathway for Phosphonate Degradation. Environ. Microbiol. 2019, 21, 2402–2414. [Google Scholar] [CrossRef]
- Oliverio, A.M.; Bissett, A.; McGuire, K.; Saltonstall, K.; Turner, B.L.; Fierer, N. The Role of Phosphorus Limitation in Shaping Soil Bacterial Communities and Their Metabolic Capabilities. mBio 2020, 11, e01718-20. [Google Scholar] [CrossRef]
- Carini, P.; White, A.E.; Campbell, E.O.; Giovannoni, S.J. Methane Production by Phosphate-Starved SAR11 Chemoheterotrophic Marine Bacteria. Nat. Commun. 2014, 5, 4346. [Google Scholar] [CrossRef] [PubMed]
- Teikari, J.E.; Fewer, D.P.; Shrestha, R.; Hou, S.; Leikoski, N.; Mäkelä, M.; Simojoki, A.; Hess, W.R.; Sivonen, K. Strains of the Toxic and Bloom-Forming Nodularia Spumigena (Cyanobacteria) Can Degrade Methylphosphonate and Release Methane. ISME J. 2018, 12, 1619–1630. [Google Scholar] [CrossRef]
- Zhou, J.; Theroux, S.M.; Bueno de Mesquita, C.P.; Hartman, W.H.; Tringe, S.G. Microbial Drivers of Methane Emissions from Unrestored Industrial Salt Ponds. ISME J. 2021, 16, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An Integrated Data Management and Comparative Analysis System for Microbial Genomes and Microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Bae, G.D.; Yih, W.; Cho, B.C.Y. Marivita Cryptomonadis Gen. Nov., Sp. Nov. and Marivita Litorea Sp. Nov., of the Family Rhodobacteraceae, Isolated from Marine Habitats. Int. J. Syst. Evol. Microbiol. 2009, 59, 1568–1575. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoon, J.-H.; Kang, S.-J.; Lee, S.-Y.; Jung, Y.-T.; Lee, J.-S.; Oh, T.-K. Marivita Hallyeonensis Sp. Nov., Isolated from Seawater, Reclassification of Gaetbulicola Byunsanensis as Marivita Byunsanensis Comb. Nov. and Emended Description of the Genus Marivita. Int. J. Syst. Evol. Microbiol. 2012, 62, 839–843. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kang, S.-J.; Lee, J.-S. Marivita Geojedonensis Sp. Nov., Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2013, 63, 423–427. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kang, S.-J.; Jung, Y.-T.; Oh, T.-K. Gaetbulicola Byunsanensis Gen. Nov., Sp. Nov., Isolated from Tidal Flat Sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 196–199. [Google Scholar] [CrossRef]
- Budinoff, C.R.; Dunlap, J.R.; Hadden, M.; Buchan, A. Marivita Roseacus Sp. Nov., of the Family Rhodobacteraceae, Isolated from a Temperate Estuary and an Emended Description of the Genus Marivita. J. Gen. Appl. Microbiol. 2011, 57, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.-P.; Liu, Y.; Hou, T.-T.; Liu, H.-C.; Zhou, Y.-G.; Wang, F.; Liu, Z.-P. Marivita Lacus Sp. Nov., Isolated from a Saline Lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, X.-A.; Jiang, Z.-W.; Yang, X.; Zhang, X.-L.; Yang, Q. Combined Characterization of a New Member of Marivita Cryptomonadis Strain LZ-15-2 Isolated from Cultivable Phycosphere Microbiota of Highly Toxic HAB Dinoflagellate Alexandrium Catenella LZT09. Braz. J. Microbiol. 2021, 52, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Bueno de Mesquita, C.P.; Zhou, J.; Theroux, S.M.; Tringe, S.G. Methanogenesis and Salt Tolerance Genes of a Novel Halophilic Methanosarcinaceae Metagenome-Assembled Genome from a Former Solar Saltern. Genes 2021, 12, 1609. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum Information about a Single Amplified Genome (MISAG) and a Metagenome-Assembled Genome (MIMAG) of Bacteria and Archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Von Meijenfeldt, F.A.B.; Arkhipova, K.; Cambuy, D.D.; Coutinho, F.H.; Dutilh, B.E. Robust Taxonomic Classification of Uncharted Microbial Sequences and Bins with CAT and BAT. Genome Biol. 2019, 20, 217. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chen, L.-X.; Anantharaman, K.; Shaiber, A.; Eren, A.M.; Banfield, J.F. Accurate and Complete Genomes from Metagenomes. Genome Res. 2020, 30, 315–333. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ma, J. The Global Catalogue of Microorganisms (GCM) 10K Type Strain Sequencing Project: Providing Services to Taxonomists for Standard Genome Sequencing and Annotation. Int. J. Syst. Evol. Microbiol. 2019, 69, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Feng, X.; Chen, Y.; Li, L.; Wang, X.; Hu, Z.; Wang, H.; Luo, H. Metapopulation Structure of Diatom-Associated Marine Bacteria. bioRxiv 2021. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Blankenberg, D.; Taylor, J.; Schenck, I.; He, J.; Zhang, Y.; Ghent, M.; Veeraraghavan, N.; Albert, I.; Miller, W.; Makova, K.D.; et al. A Framework for Collaborative Analysis of ENCODE Data: Making Large-Scale Analyses Biologist-Friendly. Genome Res. 2007, 17, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)Orthologs in Large-Scale Analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Krassowski, M. Krassowski/Complex-Upset. 2020. Available online: https://zenodo.org/record/5762625 (accessed on 21 December 2021).
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc Collection of Microbial Genomes and Metabolic Pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Vreeland, R.H. Mechanisms of Halotolerance in Microorganisms. Crit. Rev. Microbiol. 2009, 14, 311–356. [Google Scholar] [CrossRef]
- Vera-Gargallo, B.; Ventosa, A. Metagenomic Insights into the Phylogenetic and Metabolic Diversity of the Prokaryotic Community Dwelling in Hypersaline Soils from the Odiel Saltmarshes (SW Spain). Genes 2018, 9, 152. [Google Scholar] [CrossRef]
- Youssef, N.H.; Savage-Ashlock, K.N.; McCully, A.L.; Luedtke, B.; Shaw, E.I.; Hoff, W.D.; Elshahed, M.S. Trehalose/2-Sulfotrehalose Biosynthesis and Glycine-Betaine Uptake Are Widely Spread Mechanisms for Osmoadaptation in the Halobacteriales. ISME J. 2014, 8, 636–649. [Google Scholar] [CrossRef]
- Matarredona, L.; Camacho, M.; Zafrilla, B.; Bonete, M.-J.; Esclapez, J. The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules 2020, 10, 1390. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of Adaptation of Microorganisms of the Three Domains of Life to High Salt Concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Becker, E.A.; Seitzer, P.M.; Tritt, A.; Larsen, D.; Krusor, M.; Yao, A.I.; Wu, D.; Madern, D.; Eisen, J.A.; Darling, A.E.; et al. Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-Response. PLOS Genet. 2014, 10, e1004784. [Google Scholar] [CrossRef]
- Kolde, R. pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: http://CRAN.R-project.org/package=pheatmap (accessed on 21 December 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using Average Nucleotide Identity to Improve Taxonomic Assignments in Prokaryotic Genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S rRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Tyson, G.W.; DeLong, E.F. Widespread Known and Novel Phosphonate Utilization Pathways in Marine Bacteria Revealed by Functional Screening and Metagenomic Analyses. Environ. Microbiol. 2010, 12, 222–238. [Google Scholar] [CrossRef]
- Elevi Bardavid, R.; Oren, A. Acid-Shifted Isoelectric Point Profiles of the Proteins in a Hypersaline Microbial Mat: An Adaptation to Life at High Salt Concentrations? Extremophiles 2012, 16, 787–792. [Google Scholar] [CrossRef]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the Marine Roseobacter Lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Giebel, H.-A.; Simon, M. Diversity, Ecology, and Genomics of the Roseobacter Clade: A Short Overview. Arch. Microbiol. 2008, 189, 531–539. [Google Scholar] [CrossRef]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of Moderately Halophilic Aerobic Bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 504–544. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular Signature of Hypersaline Adaptation: Insights from Genome and Proteome Composition of Halophilic Prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef] [PubMed]
- Dyhrman, S.T.; Chappell, P.D.; Haley, S.T.; Moffett, J.W.; Orchard, E.D.; Waterbury, J.B.; Webb, E.A. Phosphonate Utilization by the Globally Important Marine Diazotroph Trichodesmium. Nature 2006, 439, 68–71. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).