Abstract
It has been estimated that 80% of the pre-mRNA undergoes alternative splicing, which exponentially increases the flow of biological information in cellular processes and can be an attractive therapeutic target. It is a crucial mechanism to increase genetic diversity. Disturbed alternative splicing is observed in many disorders, including neuromuscular diseases and carcinomas. Spinal Muscular Atrophy (SMA) is an autosomal recessive neurodegenerative disease. Homozygous deletion in 5q13 (the region coding for the motor neuron survival gene (SMN1)) is responsible for 95% of SMA cases. The nearly identical SMN2 gene does not compensate for SMN loss caused by SMN1 gene mutation due to different splicing of exon 7. A pathologically low level of survival motor neuron protein (SMN) causes degeneration of the anterior horn cells in the spinal cord with associated destruction of α-motor cells and manifested by muscle weakness and loss. Understanding the regulation of the SMN2 pre-mRNA splicing process has allowed for innovative treatment and the introduction of new medicines for SMA. After describing the concept of splicing modulation, this review will cover the progress achieved in this field, by highlighting the breakthrough accomplished recently for the treatment of SMA using the mechanism of alternative splicing.
1. Introduction
Splicing is an essential part of pre-mRNA maturation in a eukaryotic cell. That process consists of excising noncoding intronic sequences from the initial product of gene transcription and ligating remaining exons before translation to protein [1]. Splicing reaction is controlled by the spliceosome, the macromolecular ribonucleoprotein complex. Determination of the beginning and end of the intron, which is marked by the 5′ and 3′ splice sites (5′ss and 3′ss), plays a key role in the splicing mechanism. Specific sequences are recognized by the spliceosome, the macromolecular ribonucleoprotein structure that catalyses splicing [2]. An alternatively spliced gene is a source of multiple mRNA isoforms, which increases coding potential of the eukaryotic genome. Alternative splicing is regulated by the cis-acting splicing regulatory elements (SREs) that recruit trans-acting factors in a sequence-unique manner [3]. Trans-acting RNA binding proteins (RBPs) bound to an intronic or exonic splice enhancer (ISE or ESE) stabilize spliceosome formation and lead to exon recognition and retention. Analogically RBPs bound to intronic or exonic splice silencing motifs (ISS or ESS) preclude the formation of the spliceosome and promote exon removing [4]. The most common RBPs are proteins rich in serine/arginine (SR) rests and the heterogeneous ribonucleoprotein (hRNP). Due to tissue-specific RBPs bounding, final products from the same gene primary transcript can be different depending on the tissue. the tissue-specific RBPs bounding, final products from the same gene primary transcript can be different depending on the tissue. The binding of these proteins is a variable process resulting in diverse combinations of included or excluded introns and exons [5]. SR proteins are responsible for phosphorylation, which regulates their localization and activity [6]. Oxidative stress affects mutations within splicing regulatory sequences or disturbed expression of splice factors, which causes a growing number of diseases and has an emerging role in aging [7,8,9]. Loss of balance in the splicing process influences the development of neurodegenerative diseases, renitis pigmentosa, Prader-Willi syndrome, familial adenomatous polyposis, breast or lung cancer [10,11,12,13,14,15,16]. The aim of this review is to present the novel forms of therapies of spinal muscular atrophy (SMA) based on alternative splicing regulation mechanisms.
2. Spinal Muscular Atrophy (SMA)
SMA is a congenital neurodegenerative disorder with an autosomal recessive inheritance, characterized by loss of motor neurons leading to progressive muscle weakness [17]. Knowledge about SMA has changed considerably since the first reports of patients with this disease, written by Werdnig (1891) [18] and Hofmann (1893) [19]. The SMA incidence is about 1 in 6000 to 11,000, with a carrier frequency of SMN1 mutations from 2 to 3% (1 in 40) in the general population [20,21,22]. In Cuba, a six-year study was conducted to investigate the prevalence of type I SMA in people of different ethnicities. The results of the study suggest that type I SMA is less common in the African American group [23]. According to the statistics of the Polish SMA Foundation, one in 35 inhabitants of Poland carry the SMN1 gene mutation, and the disease phenotype will appear on average in every 7000 children born in Poland [24]. Based on the progression and variability of symptoms, SMA was divided into five types, from congenital lethal (SMA0) to adult onset (SMA4) [25]. The clinical phenotype of SMA is heterogeneous, ranging from severe to mild. It is generally divided into three main subtypes: Type I (also called Werdnig Hoffmann disease), Type II, and Type III (also called Kugelberg Welander disease). However, these phenotypes are viewed more as a continuum rather than as separate subtypes, and further subtypes are sometimes observed at both ends of the spectrum. Type 0 SMA is a very severe form with onset in utero, limited, or missing movements, contractures, and a requirement for assisted mechanical ventilation at birth and death before six months of age, while Type IV SMA is a mild late (adult) form that has a normal life span [20,21].
In most cases this disease develops due to mutations in the gene SMN1 (survival of motor neuron 1), SMN T, telomeric, located on chromosome 5q13.2 [26]. The majority of the patients (92–95%) have a homozygous deletion of SMN1 [20,21]. The intragenic mutations within SMN1 are responsible for the remaining 5% of cases [27]. In some severe cases of SMA, loss of the NAIP (neuronal apoptosis protein inhibitor), GTF2H2A (general transcription factor IIH, p44), and SERF1A (small EDRK-rich factor 1A, H4F5A) genes are also observed [28,29,30,31,32,33]. A study by Ahn, Eun Ji et al. on a group of 33 Korean patients suggests that coexisting deletions of SMN1 and NAIP are connected with earlier onset of symptoms and poor prognosis in SMA patients [34]. The transcription of SMN1 produces a functionally complete mRNA that encodes SMN protein. Significantly fewer SMN proteins come from the SMN2 gene. Only 10–15% of total SMN2 transcripts are full-length mRNA [35]. Thus, SMN2 is identical to SMN1, except for a single C-T substitution in exon 7. This substitution promotes 80 to 85% splicing during transcription and consequent exon 7 deletion [35]. The SMN2 genes are not functionally equivalent. The ability of the SMN2 gene to modify the course of the disease is regulated by epigenetic factors that, through DNA methylation, have the ability to silence the gene. In patients with different types of SMA, differences in methylation levels are observed at positions −296 and −290 in the island 2 CpG of SMN2. A milder disease course correlates with lower methylation levels [36]. It is worth noting that truncated mRNA causes similarly truncated non-functional proteins. Patients with SMA lack SMN1 and therefore they depend on the residual SMN2 production of a functional SMN protein for α function of the motor neuron and subsequent survival [21]. The SMN protein is localized in all eukaryotic cells and has been shown to have a pivotal role in homeostatic cellular pathways in all cells [37]. According to hypotheses, the SMN protein in the cytoplasm was shown to have an important role in the transport of mRNA through axons and transport of containing b-actin ribonucleoprotein complexes. Another hypothesis states that the SMN protein takes part in synthesis of small nuclear RNA (snRNA) and therefore plays a key role in the formation of a spliceosome that removes introns from pre-mRNA into functional mRNA [38,39]. As motor neurons are sensitive to malfunctioning of the spliceosome directly or indirectly through misspliced mRNAs, any damage to motor neurons results in the development of dysfunctions in proteins essential for neuronal function [21].
3. Mechanisms of SMN2 Splicing Regulation Targeted by Therapeutics
In human cells, there are two nearly identical genes responsible for SMN protein production. The presence of two SMN genes is attributed to large tandem chromosomal duplication [40]. In the region of this duplication, on the long arm of chromosome 5 (5q13.2) lie four protein-encoding genes: SMN, NAIP, GTF2H2A, and SERF1A. The duplicated genes are identical to their partner gene (SERF1B), differ in a low number of nucleotides (SMN2) or are pseudogenes (ΨGTF2H2B and ΨNAIPΔ5) [41]. Both SMN genes consist of 10 exons (1,2A,2B,3–6,6B,7,8). It is worth noting that Exon 6b is a new discovery and is generated by exonification of the Alu element in intron 6 [42]. Under certain conditions such as starvation, hypoxia, or oxidative stress, transcription of these genes may proceed differently [8,43]. Factors that regulate SMN levels and modify transcription are tissue-specific [44].
The key difference between these genes lies in the splicing of exon 7. The amino acids encoded in exon 7 are responsible for SMN stability as they determine the crucial C-terminus of the protein. In the SMN2 gene due to alternative splicing, exon 7 is more often skipped, resulting in more of the truncated, partially functional and unstable SMNΔ7 protein than full-length SMN [45,46]. The primary reason exon 7 is excluded is C-to-T substitution at position 6 of exon 7 (C6U) (Figure 1). Mutation or deletion of the SMN1 gene is a major cause of spinal muscular atrophy, through deficiency of SMN [39]. Restoring exon 7 inclusion has therapeutic benefits proven in mouse models [47].
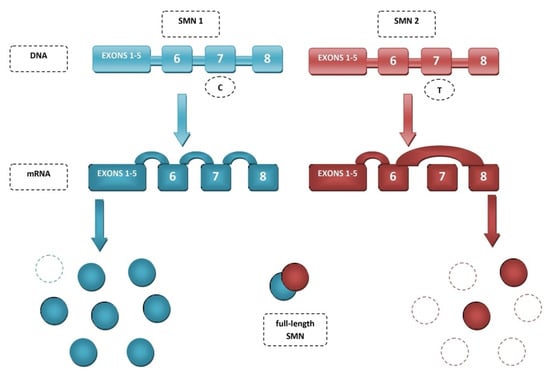
Figure 1.
Splicing of SMN genes.
Mechanisms regulating exon 7 splicing are good potential therapeutic targets. The best described to date splicing factors, binding directly to exon 7 splicing enhancer regions SE1 and SE2 are serine/arginine-rich splicing factor 1 (SRSF1) and transformer 2 protein homolog β (Tra2B) [48]. The best-known negative regulators of exon 7 splicing are heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) [49] and src-associated substrate in mitosis 68 (Sam68) [50]. C6U substitution results in hnRNA A1 or Sam68 being bound to SE1 in place of the positive regulator. Exon 7 exclusion can also occur through binding of hnRNP A1 to SE2 sequences or the intronic silencer sequence N1 (ISS-N1) [51] (Figure 2). Other factors showing altering splicing activity include SRp30c [52], TDP-43 [53], TIA1 [54], hnRNP Q [55], hnRNP G [56].
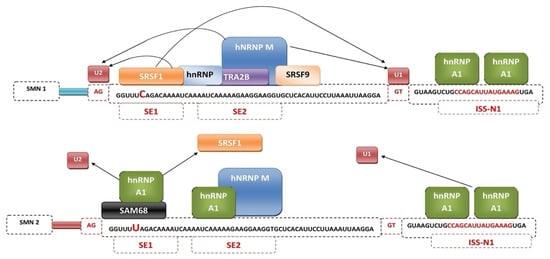
Figure 2.
Mechanism of regulation of SMN1 and SMN2 gene splicing by splicing factors.
Exon 7 skipping may be caused by increased activity of a regulatory sequence located at the 3′ end of exon 7, called terminal stem loop 2 (TSL2). It exhibits inhibitory activity, probably by competing with U1 snRNP for a binding site [57]. The inhibitory effect of TSL2 was confirmed by observing the effect of U40G or A54C substitution on exon 7 splicing. Separately, they disrupted TSL2 by promoting exon 7 incorporation, but combined they reproduced the structure of TSL2 and thus inhibition of exon 7 splicing [58]. Modification of splicing through TSL2 requires further study. It can be a good target for screening small molecules [59].
In 2006, Singh et al. discovered an ISS SMN2 intron-7 in the human SMN1/2 gene, named ISS-N1 [60]. ISS-N1 is a sequence located immediately downstream of the 5′ss of exon 7. It is 15-nt long and binds positions 10 to 24 of intron 7, producing a strongly inhibitory effect on exon 7 inclusion [61]. Interestingly, ISS-N1 deletion reduced the requirement for positive cis-elements in exon 7 inclusions, an effect similar to the A54G mutation [51]. Blocking ISS-N1 with low concentrations of antisense oligonucleotide (ASO) effectively increased SMN protein levels in studies in mouse models or fibroblasts collected from SMA patients. This demonstrates the high availability and binding efficiency of ISS-N1 for this group of compounds [47]. Importantly, modifying alternative splicing of SMN2 by targeting ISS-N1 is already used by the ASO drug nusinersen (Spinraza) in the first approved therapy to treat SMA [62].
A more specific target for ASO than ISS-N1 appeared to be the GC-rich sequence (GCRS), which spans from the 7th to 14th position of intron 7 overlapping ISS-N1. Studies on SMA type I patient cells and severe SMA mouse models demonstrated the efficacy of 8-mer ASO binding to GCRS, which not only elevated SMN levels but also increased the levels of Gemin 2 and Gemin 8 factors involved in snRNP biogenesis and Tra2-β1 and hnRNP Q, responsible for proper RNA splicing [63,64].
GCRS participates in the formation of a 5′ strand of a unique RNA structure called the internal stem formed by long-distance interactions (ISTL-1). 279-nts divides the two 8-bp ISTL1 strands. The last position of ISTL-1 and the first position of ISS-N1 is the C residue located at the 10th intronic position (10C) [65]. Interestingly, an experiment was conducted by targeting ISS-N1 with ASOs of equal length (14-mer). F-14 binding to the first nucleotides of ISS-N1, including 10C, promoted exon 7 inclusion, while L-14 targeting the terminal positions of ISS-N1 without 10C had the opposite effect, promoting exon 7 exclusion [66]. ISTL-1 has been shown to negatively regulate exon 7 splicing independently of snRNP A1. Destabilization of ISTL-1 induces abolition of long-distance interaction (LDI) mediated by C10. ASO-mediated sequestration of the 3′ strand of ISTL-1 and upstream sequences form ISS-N2, which results in correction of alternative splicing and restoration of full-length SMN secretion [66,67].
Sequences flanking exon 7 such as Element 1 located in intron 6 also appear to be promising therapeutic targets. Element 1 is a cis-element that is an extended inhibitory sequence located upstream of the 3′ss exon 7 [68]. The effect of promoting full-length SMN expression was demonstrated by targeting Element 1 with morpholino ASOs in mouse models [69].
4. Antisense Oligonucleotides
A novel approach to the therapy of SMA and other genetically determined diseases is represented by the use of ASO [70]. ASOs are short (about 15–30 nucleotides in length), single-stranded molecules of chemically modified nucleic acids or nucleotide analogs that, on the basis of complementarity, recognize and bind target sequences in RNA through Watson-Crick base pairing [71,72]. Depending on the binding site, ASOs affect transcript inactivation or splicing, leading to changes in exon content [73]. ASOs are designed to pair bases and form a steric block for binding splicing factors to pre-mRNAs. RNA alters the recognition of splicing sites by the spliceosome, leading to a change in the normal splicing of the target transcript [72]. Modified sequence-dependent ASOs can appropriately lead to the exclusion or inclusion of an exon that would have been excised, as is the case in SMA [72].
This relies on exon 7 appearing in the mature SMN2 transcript. It is necessary to block the action of the intron folding silencer. Antisense oligonucleotides recognize, on the basis of nitrogenous base complementarity, precisely this SMN2 pre-mRNA fragment and sterically block its recognition by appropriate proteins. This prevents the formation of a complex that would inhibit detection of the exon/intron boundary. Subsequently, the split between exon 7 and intron 7 is detected, resulting in the incorporation of exon 7 into the mature transcript [74]. The use of appropriate ASOs to treat spinal muscular atrophy allows exon 7 to be incorporated into the transcript of the SMN2 gene.
The first drug approved for the treatment of SMA was nusinersen (Spinraza TM) [75]. Its discovery took place in 2010 [76]. It is an antisense oligonucleotide that binds to the splicing inhibitory sequence of intron 7. Nusinersen is an 18-mer oligonucleotide in which the sugar-phosphate backbone has been chemically modified [77]. Nusinersen complementary hybridizes to ISS-N1 to block hnRNP recruitment, resulting in the inclusion of exon 7 incorporation into the SMN2 transcript, resulting in higher levels of fully functional SMN protein [78] (Figure 3). This protein is associated with SMA. As the amount of SMN protein increases, the degeneration of motor neurons stops and the disease progresses [78]. This influences a patient’s longer survival, better motor function and faster achievement of milestones. Nusinersen does not cross the blood-brain barrier and therefore requires intrathecal administration. The half-life of the drug is 163 days, and doses must be repeated throughout life [79]. Monitoring in patients of thrombocyte count, prothrombin time, partial thromboplastin time, and urinalysis results is necessary during nusinersen therapy because it can lead to thrombocytopenia and coagulation disorders, and is nephrotoxic [80]. It can be used to treat all types of SMA [81]. Another breakthrough in the treatment of SMA was the December 2016 approval by the American Food and Drug Administration of the medicine nusinersen (SpinrazaTM), also known as ISIS-SMNRx or ISIS [82]. If a patient does not reach an advanced stage of muscle atrophy, appropriate physiotherapy and multidisciplinary care in combination with nusinersen can produce a significant improvement in the condition of the treated patient [83,84] (Table 1).
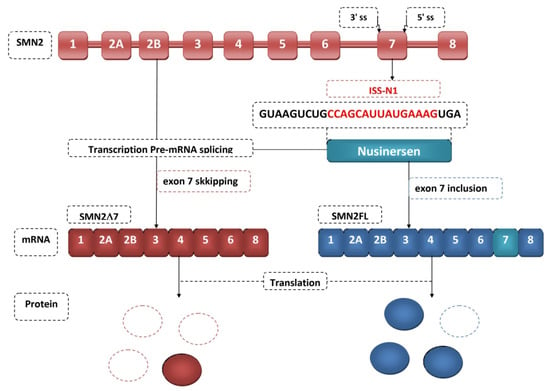
Figure 3.
Nusinersen therapeutic mechanism.

Table 1.
Comparison of potential and the newest targets for ASOs in SMA therapy.
5. Small Molecules
A project led by PTC-Roche (PTC Therapeutics, South Plainfield, New Jersey and Hoffmann-La Roche, Basel, Switzerland) to identify an orally available molecule to treat SMA began about a decade ago. Both groups identified small molecules and reported three orally delivered compounds, namely SMN-C1 (isocoumarin), SMN-C2 (coumarin), and SMN-C3 (pyridopyrimidinone derivative); each promoted exon 7 inclusion from SMN2 [92]. Small molecules can exhibit high selectivity, affecting the modulation of RNA folding of only one or a few genes, among the many thousands of genes expressed in cells [93]. Most drugs are inhibitors of enzyme proteins or receptors. It is worth noting that it is possible to obtain modulators of interactions in RNA-RNA and RNA-protein complexes [94]. Risdiplam is being developed by Roche, PTC Therapeutics Inc and the SMA Foundation for the treatment of SMA. In August 2020, the European Medicines Agency (EMA) approved the use of risdiplam to treat patients with the SMN1 gene mutation [95]. This experimental drug manifests high selectivity for modulation of RNA folding against the SMN2 transcript. It affects the alternative splicing of a small pool of other genes, such as FOXM1, MADD or STRN3 [94,95]. Risdiplam is not a substrate for the transport protein MDR1, and thus crosses the blood-brain barrier well. It is properly distributed in the CNS and peripheral tissues of mice, rats, and monkeys after single or repeated oral or intraperitoneal administration. Risdiplam also increased levels of functional SMN protein in the CNS and peripheral tissues of mouse models of SMA [96].
Risdiplam is a highly potent SMN2 splicing modifier that increases exon 7 inclusion in SMN2 mRNA transcripts in in vitro assays and in transgenic mouse models of SMA [95,96,97]. Risdiplam binds to the SMN2 transcript at two sites—the exonic splicing enhancer 2 (ESE2) in exon 7 and 5′ss of intron 7, thereby dislocating hnRNPG and enhancing 5′ss recognition and binding by U1snRNP. This results in exon 7 not being excised from the transcript and the full SMN protein being able to be synthesized [94,98]. Risdiplam can also increase the binding of far upstream element binding protein 1 (FUBP1) and KH-type splicing regulatory protein (KHSRP) splicing modulators to the SMN2 pre-mRNA complex, activating SMN2 splicing [99]. Some of the first preclinical studies have shown that risdiplam can reach the central nervous system and peripheral organs in vivo and can lead to significant increases in SMN protein levels in blood, brain, and muscle, with increased survival in various mouse models of SMA [100,101]. The advantage of this drug is the oral route of administration [102]. Preclinical studies allow for hypothesizing the possibility of a therapeutic effect also in tissues other than the nervous system [102]. This is particularly important because numerous studies in human and animal models indicate that SMA may be considered a multisystem disorder with involvement of the neuromuscular junction, gastrointestinal tract, cardiovascular system, and lung and liver tissues [103,104]. According to the Food and Drug Administration (FDA), on 7 August 2020 risdiplam was approved for the treatment of spinal muscular atrophy in adults and children 2 months of age and older [105]. A recent study analyzing the administration of risdiplam to infants from 1–7 months of age (type 1 SMA) has led to increased expression of functional SMN protein in the blood [100].
Branaplam is another small molecule, administered orally, that modulates SMN2 splicing with high specificity. It is currently in Phase 2 clinical trials [101,106]. It has been shown to modulate splicing, increase full-length SMN protein levels, and increase survival in a mouse model of severe SMA [107]. The mechanism of action is similar to risdiplam.
To the best of our knowledge, two more molecules PK4C9 and TEC-1, according to recent reports, increase exon 7 SMN2 inclusion with high specificity [59,101,107]. TEC-1 permeabilizes the central nervous system and confers therapeutic efficacy in a mouse model of SMA [59,108]. PCK4C9 targeting the TSL2 tri-loop appeared to cover the “3′-cluster,” a negative element identified by in vivo selection of the entire exon 7 [109,110] (Table 2).

Table 2.
Comparison of potential and the newest targets for small molecules in SMA therapy.
6. Future Prospects
The advancement of SMA therapies has allowed many patients to survive and improve their lives. Current drugs focus on replacing the SMN1 gene (onasemnogene abeparvovec) or changing SMN2 splicing (nusinersen, risdiplam). Work is currently underway on a complementary treatment independent of SMN. This applies, for example, to neuroprotective drugs, nerve connection stabilizers, myostatin inhibitors, or activators of muscle function [106,119,120,121] (Figure 4). Many studies also emphasize the importance of early diagnosis and treatment implementation, even presymptomatically. Efforts should be made to develop effective neonatal screening for SMA and to update treatment regimens due to the evolving phenotype of the disease [122,123,124]. Currently, the most important modifier of SMA is the SMN2 copy number; however, it has been noticed that patients with the same SMN2 copy number show a difference in the disease phenotype. Work is currently underway to find new biomarkers of disease evolution [125,126,127]. It is also important to provide multidisciplinary care for treated children [128,129].
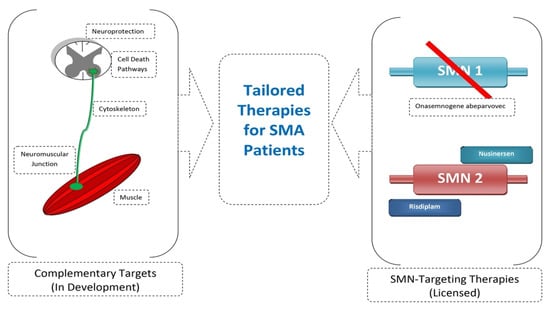
Figure 4.
Combined SMN-independent and SMN-dependent therapy as future direction in SMA therapy.
7. Conclusions
The use of the molecular basis of SMA by drugs such as nusinersen, Zolgensma, and risdiplam has brought significant benefits to patients with this fatal disease. Modification of exon 7 alternative splicing turns out to be a key mechanism and target for further research. Current research proves that this therapeutic strategy can effectively increase the level of the SMN protein and, as a result, reduce the course of the disease. Early diagnosis and initiation of treatment in the patient allow for the extension of lifespan and the achievement of milestones. Work is underway on the implementation of other compounds in SMA therapy that bind to factors involved in the regulation of splicing.
Author Contributions
Conceptualization, J.L. and M.L.; methodology, J.L., G.Z. and M.L.; the acquisition of the literature for the manuscript J.L. and G.Z.; writing—original draft preparation, J.L., G.Z. and M.L.; writing—review and editing, J.L., G.Z., P.G. and M.L.; visualization, G.Z.; supervision, P.G. and M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We acknowledge support from the Medical University of Lublin for Open Access Publishing. We acknowledge Maria Ochal for help with preparing the final version manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Faustino, N.A.; Cooper, T.A. Pre-MRNA Splicing and Human Disease. Genes Dev. 2003, 17, 419–437. [Google Scholar] [CrossRef]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke Off. J. Int. Stroke Soc. 2018, 13, 612–632. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Systematical Identification of Splicing Regulatory Cis-Elements and Cognate Trans-Factors. Methods 2014, 65, 350–358. [Google Scholar] [CrossRef]
- Black, A.J.; Gamarra, J.R.; Giudice, J. More than a Messenger: Alternative Splicing as a Therapeutic Target. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194395. [Google Scholar] [CrossRef]
- Montes, M.; Sanford, B.L.; Comiskey, D.F.; Chandler, D.S. RNA Splicing and Disease: Animal Models to Therapies. Trends Genet. TIG 2019, 35, 68–87. [Google Scholar] [CrossRef]
- Long, Y.; Sou, W.H.; Yung, K.W.Y.; Liu, H.; Wan, S.W.C.; Li, Q.; Zeng, C.; Law, C.O.K.; Chan, G.H.C.; Lau, T.C.K.; et al. Distinct Mechanisms Govern the Phosphorylation of Different SR Protein Splicing Factors. J. Biol. Chem. 2019, 294, 1312–1327. [Google Scholar] [CrossRef]
- Cooper, T.A.; Wan, L.; Dreyfuss, G. RNA and Disease. Cell 2009, 136, 777–793. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Singh, N.N.; Ottesen, E.W.; Sivanesan, S.; Shishimorova, M.; Singh, R.N. Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene. PLoS ONE 2016, 11, e0154390. [Google Scholar] [CrossRef] [PubMed]
- Deschênes, M.; Chabot, B. The Emerging Role of Alternative Splicing in Senescence and Aging. Aging Cell 2017, 16, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Apicco, D.J.; Zhang, C.; Maziuk, B.; Jiang, L.; Ballance, H.I.; Boudeau, S.; Ung, C.; Li, H.; Wolozin, B. Dysregulation of RNA Splicing in Tauopathies. Cell Rep. 2019, 29, 4377–4388.e4. [Google Scholar] [CrossRef]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted Alternative Splicing for Genes Implicated in Splicing and Ciliogenesis Causes PRPF31 Retinitis Pigmentosa. Nat. Commun. 2018, 9, 4234. [Google Scholar] [CrossRef]
- Perrone, B.; La Cognata, V.; Sprovieri, T.; Ungaro, C.; Conforti, F.L.; Andò, S.; Cavallaro, S. Alternative Splicing of ALS Genes: Misregulation and Potential Therapies. Cell. Mol. Neurobiol. 2020, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Aretz, S.; Uhlhaas, S.; Sun, Y.; Pagenstecher, C.; Mangold, E.; Caspari, R.; Möslein, G.; Schulmann, K.; Propping, P.; Friedl, W. Familial Adenomatous Polyposis: Aberrant Splicing Due to Missense or Silent Mutations in the APC Gene. Hum. Mutat. 2004, 24, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Coulson, R.L.; Powell, W.T.; Yasui, D.H.; Dileep, G.; Resnick, J.; LaSalle, J.M. Prader-Willi Locus Snord116 RNA Processing Requires an Active Endogenous Allele and Neuron-Specific Splicing by Rbfox3/NeuN. Hum. Mol. Genet. 2018, 27, 4051–4060. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Zhang, W.; Chen, D.; Wang, Y. Aberrant Alternative Splicing in Breast Cancer. J. Mol. Cell Biol. 2019, 11, 920–929. [Google Scholar] [CrossRef]
- Papatsirou, M.; Adamopoulos, P.G.; Artemaki, P.I.; Georganti, V.P.; Scorilas, A.; Vassilacopoulou, D.; Kontos, C.K. Next-Generation Sequencing Reveals Alternative L-DOPA Decarboxylase (DDC) Splice Variants Bearing Novel Exons, in Human Hepatocellular and Lung Cancer Cells. Gene 2021, 768, 145262. [Google Scholar] [CrossRef]
- Chen, T.-H. New and Developing Therapies in Spinal Muscular Atrophy: From Genotype to Phenotype to Treatment and Where Do We Stand? Int. J. Mol. Sci. 2020, 21, 3297. [Google Scholar] [CrossRef]
- Werdnig, G. Two Early Infantile Hereditary Cases of Progressive Muscular Atrophy Simulating Dystrophy, but on a Neural Basis. 1891. Arch. Neurol. 1971, 25, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J. Ueber chronische spinale Muskelatrophie im Kindesalter, auf familiärer Basis. Dtsch. Z. Nervenheilkd. 1893, 3, 427–470. [Google Scholar] [CrossRef]
- Verhaart, I.E.C.; Robertson, A.; Wilson, I.J.; Aartsma-Rus, A.; Cameron, S.; Jones, C.C.; Cook, S.F.; Lochmüller, H. Prevalence, Incidence and Carrier Frequency of 5q–Linked Spinal Muscular Atrophy—A Literature Review. Orphanet J. Rare Dis. 2017, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Burr, P.; Reddivari, A.K.R. Spinal Muscle Atrophy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Crawford, T.O.; Pardo, C.A. The Neurobiology of Childhood Spinal Muscular Atrophy. Neurobiol. Dis. 1996, 3, 97–110. [Google Scholar] [CrossRef]
- Zaldívar, T.; Montejo, Y.; Acevedo, A.M.; Guerra, R.; Vargas, J.; Garofalo, N.; Alvarez, R.; Alvarez, M.A.; Hardiman, O. Evidence of Reduced Frequency of Spinal Muscular Atrophy Type I in the Cuban Population. Neurology 2005, 65, 636–638. [Google Scholar] [CrossRef]
- Jak Częste Jest SMA? Fund SMA. Available online: https://www.fsma.pl/rdzeniowy-zanik-miesni/jak-czeste-jest-sma/ (accessed on 24 May 2021).
- Jędrzejowska, M.; Kostera-Pruszczyk, A. Spinal Muscular Atrophy—New Therapies, New Challenges. Neurol. Neurochir. Pol. 2020, 54, 8–13. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M. Identification and Characterization of a Spinal Muscular Atrophy-Determining Gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Alías, L.; Bernal, S.; Fuentes-Prior, P.; Barceló, M.J.; Also, E.; Martínez-Hernández, R.; Rodríguez-Alvarez, F.J.; Martín, Y.; Aller, E.; Grau, E.; et al. Mutation Update of Spinal Muscular Atrophy in Spain: Molecular Characterization of 745 Unrelated Patients and Identification of Four Novel Mutations in the SMN1 Gene. Hum. Genet. 2009, 125, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Wirth, B.; Hahnen, E.; Morgan, K.; DiDonato, C.J.; Dadze, A.; Rudnik-Schöneborn, S.; Simard, L.R.; Zerres, K.; Burghes, A.H. Allelic Association and Deletions in Autosomal Recessive Proximal Spinal Muscular Atrophy: Association of Marker Genotype with Disease Severity and Candidate CDNAs. Hum. Mol. Genet. 1995, 4, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Burlet, P.; Bürglen, L.; Clermont, O.; Lefebvre, S.; Viollet, L.; Munnich, A.; Melki, J. Large Scale Deletions of the 5q13 Region Are Specific to Werdnig-Hoffmann Disease. J. Med. Genet. 1996, 33, 281–283. [Google Scholar] [CrossRef]
- Velasco, E.; Valero, C.; Valero, A.; Moreno, F.; Hernández-Chico, C. Molecular Analysis of the SMN and NAIP Genes in Spanish Spinal Muscular Atrophy (SMA) Families and Correlation between Number of Copies of CBCD541 and SMA Phenotype. Hum. Mol. Genet. 1996, 5, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.R.; Owen, N.; Talbot, K.; Patel, S.; Muntoni, F.; Ignatius, J.; Dubowitz, V.; Davies, K.E. Gene Deletions in Spinal Muscular Atrophy. J. Med. Genet. 1996, 33, 93–96. [Google Scholar] [CrossRef]
- Bürglen, L.; Seroz, T.; Miniou, P.; Lefebvre, S.; Burlet, P.; Munnich, A.; Pequignot, E.V.; Egly, J.M.; Melki, J. The Gene Encoding P44, a Subunit of the Transcription Factor TFIIH, Is Involved in Large-Scale Deletions Associated with Werdnig-Hoffmann Disease. Am. J. Hum. Genet. 1997, 60, 72–79. [Google Scholar]
- Carter, T.A.; Bönnemann, C.G.; Wang, C.H.; Obici, S.; Parano, E.; De Fatima Bonaldo, M.; Ross, B.M.; Penchaszadeh, G.K.; Mackenzie, A.; Soares, M.B.; et al. A Multicopy Transcription-Repair Gene, BTF2p44, Maps to the SMA Region and Demonstrates SMA Associated Deletions. Hum. Mol. Genet. 1997, 6, 229–236. [Google Scholar] [CrossRef][Green Version]
- Ahn, E.J.; Yum, M.S.; Kim, E.H.; Yoo, H.W.; Lee, B.H.; Kim, G.H.; Ko, T.S. Genotype-Phenotype Correlation of SMN1 and NAIP Deletions in Korean Patients with Spinal Muscular Atrophy. J. Clin. Neurol. 2017, 13, 27–31. [Google Scholar] [CrossRef]
- Kolb, S.J.; Kissel, J.T. Spinal Muscular Atrophy: A Timely Review. Arch. Neurol. 2011, 68, 979–984. [Google Scholar] [CrossRef]
- Hauke, J.; Riessland, M.; Lunke, S.; Eyüpoglu, I.Y.; Blümcke, I.; El-Osta, A.; Wirth, B.; Hahnen, E. Survival Motor Neuron Gene 2 Silencing by DNA Methylation Correlates with Spinal Muscular Atrophy Disease Severity and Can Be Bypassed by Histone Deacetylase Inhibition. Hum. Mol. Genet. 2009, 18, 304–317. [Google Scholar] [CrossRef]
- Kolb, S.J.; Battle, D.J.; Dreyfuss, G. Molecular Functions of the SMN Complex. J. Child Neurol. 2007, 22, 990–994. [Google Scholar] [CrossRef]
- D’Amico, A.; Mercuri, E.; Tiziano, F.D.; Bertini, E. Spinal Muscular Atrophy. Orphanet J. Rare Dis. 2011, 6, 71. [Google Scholar] [CrossRef]
- Bowerman, M.; Becker, C.G.; Yáñez-Muñoz, R.J.; Ning, K.; Wood, M.J.A.; Gillingwater, T.H.; Talbot, K. Therapeutic Strategies for Spinal Muscular Atrophy: SMN and Beyond. Dis. Model. Mech. 2017, 10, 943–954. [Google Scholar] [CrossRef]
- Stabley, D.L.; Harris, A.W.; Holbrook, J.; Chubbs, N.J.; Lozo, K.W.; Crawford, T.O.; Swoboda, K.J.; Funanage, V.L.; Wang, W.; Mackenzie, W.; et al. SMN1 and SMN2 Copy Numbers in Cell Lines Derived from Patients with Spinal Muscular Atrophy as Measured by Array Digital PCR. Mol. Genet. Genom. Med. 2015, 3, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Butchbach, M.E.R. Copy Number Variations in the Survival Motor Neuron Genes: Implications for Spinal Muscular Atrophy and Other Neurodegenerative Diseases. Front. Mol. Biosci. 2016, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, E.W.; Seo, J.; Singh, N.N.; Singh, R.N. A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements. Front. Microbiol. 2017, 8, 2252. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.E.; Cunningham, D.; Chandler, D.S. SMN Regulation in SMA and in Response to Stress: New Paradigms and Therapeutic Possibilities. Hum. Genet. 2017, 136, 1173–1191. [Google Scholar] [CrossRef]
- Groen, E.J.N.; Perenthaler, E.; Courtney, N.L.; Jordan, C.Y.; Shorrock, H.K.; van der Hoorn, D.; Huang, Y.-T.; Murray, L.M.; Viero, G.; Gillingwater, T.H. Temporal and Tissue-Specific Variability of SMN Protein Levels in Mouse Models of Spinal Muscular Atrophy. Hum. Mol. Genet. 2018, 27, 2851–2862. [Google Scholar] [CrossRef]
- Singh, R.N.; Seo, J.; Singh, N.N. RNA in Spinal Muscular Atrophy: Therapeutic Implications of Targeting. Expert Opin. Ther. Targets 2020, 24, 731–743. [Google Scholar] [CrossRef]
- Cho, S.; Dreyfuss, G. A Degron Created by SMN2 Exon 7 Skipping Is a Principal Contributor to Spinal Muscular Atrophy Severity. Genes Dev. 2010, 24, 438–442. [Google Scholar] [CrossRef]
- Berciano, M.T.; Puente-Bedia, A.; Medina-Samamé, A.; Rodríguez-Rey, J.C.; Calderó, J.; Lafarga, M.; Tapia, O. Nusinersen Ameliorates Motor Function and Prevents Motoneuron Cajal Body Disassembly and Abnormal Poly(A) RNA Distribution in a SMA Mouse Model. Sci. Rep. 2020, 10, 10738. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, Y.; Lorson, C.L.; Stamm, S.; Androphy, E.J.; Wirth, B. Htra2-Beta 1 Stimulates an Exonic Splicing Enhancer and Can Restore Full-Length SMN Expression to Survival Motor Neuron 2 (SMN2). Proc. Natl. Acad. Sci. USA 2000, 97, 9618–9623. [Google Scholar] [CrossRef] [PubMed]
- Wee, C.D.; Havens, M.A.; Jodelka, F.M.; Hastings, M.L. Targeting SR Proteins Improves SMN Expression in Spinal Muscular Atrophy Cells. PLoS ONE 2014, 9, e115205. [Google Scholar] [CrossRef] [PubMed]
- Pedrotti, S.; Bielli, P.; Paronetto, M.P.; Ciccosanti, F.; Fimia, G.M.; Stamm, S.; Manley, J.L.; Sette, C. The Splicing Regulator Sam68 Binds to a Novel Exonic Splicing Silencer and Functions in SMN2 Alternative Splicing in Spinal Muscular Atrophy. EMBO J. 2010, 29, 1235–1247. [Google Scholar] [CrossRef]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the Discovery of ISS-N1 Led to the First Medical Therapy for Spinal Muscular Atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef]
- Simard, M.J.; Chabot, B. SRp30c Is a Repressor of 3’ Splice Site Utilization. Mol. Cell. Biol. 2002, 22, 4001–4010. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; König, J.; Hortobágyi, T.; Nishimura, A.L.; Zupunski, V.; et al. Characterizing the RNA Targets and Position-Dependent Splicing Regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Garzia, A.; Mazzola, M.; Gerstberger, S.; Molina, H.; Tuschl, T. The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol. Cell 2018, 69, 622–635.e6. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-H.; Chang, J.-G.; Lu, R.-M.; Peng, T.-Y.; Tarn, W.-Y. The RNA Binding Protein HnRNP Q Modulates the Utilization of Exon 7 in the Survival Motor Neuron 2 (SMN2) Gene. Mol. Cell. Biol. 2008, 28, 6929–6938. [Google Scholar] [CrossRef] [PubMed]
- Moursy, A.; Allain, F.H.-T.; Cléry, A. Characterization of the RNA Recognition Mode of HnRNP G Extends Its Role in SMN2 Splicing Regulation. Nucleic Acids Res. 2014, 42, 6659–6672. [Google Scholar] [CrossRef]
- Tang, Z.; Zhao, J.; Pearson, Z.J.; Boskovic, Z.V.; Wang, J. RNA-Targeting Splicing Modifiers: Drug Development and Screening Assays. Molecules 2021, 26, 2263. [Google Scholar] [CrossRef]
- Singh, N.N.; Singh, R.N.; Androphy, E.J. Modulating Role of RNA Structure in Alternative Splicing of a Critical Exon in the Spinal Muscular Atrophy Genes. Nucleic Acids Res. 2007, 35, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lopez, A.; Tessaro, F.; Jonker, H.R.A.; Wacker, A.; Richter, C.; Comte, A.; Berntenis, N.; Schmucki, R.; Hatje, K.; Petermann, O.; et al. Targeting RNA Structure in SMN2 Reverses Spinal Muscular Atrophy Molecular Phenotypes. Nat. Commun. 2018, 9, 2032. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Singh, N.N.; Androphy, E.J.; Singh, R.N. Splicing of a Critical Exon of Human Survival Motor Neuron Is Regulated by a Unique Silencer Element Located in the Last Intron. Mol. Cell. Biol. 2006, 26, 1333–1346. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv. Neurobiol. 2018, 20, 31–61. [Google Scholar] [CrossRef]
- Frederiksen, S.B.; Holm, L.L.; Larsen, M.R.; Doktor, T.K.; Andersen, H.S.; Hastings, M.L.; Hua, Y.; Krainer, A.R.; Andresen, B.S. Identification of SRSF10 as a Regulator of SMN2 ISS-N1. Hum. Mutat. 2021, 42, 246–260. [Google Scholar] [CrossRef]
- Singh, N.N.; Shishimorova, M.; Cao, L.C.; Gangwani, L.; Singh, R.N. A Short Antisense Oligonucleotide Masking a Unique Intronic Motif Prevents Skipping of a Critical Exon in Spinal Muscular Atrophy. RNA Biol. 2009, 6, 341–350. [Google Scholar] [CrossRef]
- Keil, J.M.; Seo, J.; Howell, M.D.; Hsu, W.H.; Singh, R.N.; DiDonato, C.J. A Short Antisense Oligonucleotide Ameliorates Symptoms of Severe Mouse Models of Spinal Muscular Atrophy. Mol. Ther. Nucleic Acids 2014, 3, e174. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Singh, R.N. How RNA Structure Dictates the Usage of a Critical Exon of Spinal Muscular Atrophy Gene. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194403. [Google Scholar] [CrossRef]
- Singh, N.N.; Hollinger, K.; Bhattacharya, D.; Singh, R.N. An Antisense Microwalk Reveals Critical Role of an Intronic Position Linked to a Unique Long-Distance Interaction in Pre-MRNA Splicing. RNA 2010, 16, 1167–1181. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Lawler, M.N.; Ottesen, E.W.; Upreti, D.; Kaczynski, J.R.; Singh, R.N. An Intronic Structure Enabled by a Long-Distance Interaction Serves as a Novel Target for Splicing Correction in Spinal Muscular Atrophy. Nucleic Acids Res. 2013, 41, 8144–8165. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, H.; Miyaso, H.; Okumura, M.; Kurisu, J.; Imaizumi, K. Identification of a Cis-Acting Element for the Regulation of SMN Exon 7 Splicing. J. Biol. Chem. 2002, 277, 23271–23277. [Google Scholar] [CrossRef] [PubMed]
- Osman, E.Y.; Washington, C.W.; Kaifer, K.A.; Mazzasette, C.; Patitucci, T.N.; Florea, K.M.; Simon, M.E.; Ko, C.-P.; Ebert, A.D.; Lorson, C.L. Optimization of Morpholino Antisense Oligonucleotides Targeting the Intronic Repressor Element1 in Spinal Muscular Atrophy. Mol. Ther. 2016, 24, 1592–1601. [Google Scholar] [CrossRef]
- Godfrey, C.; Desviat, L.R.; Smedsrød, B.; Piétri-Rouxel, F.; Denti, M.A.; Disterer, P.; Lorain, S.; Nogales-Gadea, G.; Sardone, V.; Anwar, R.; et al. Delivery Is Key: Lessons Learnt from Developing Splice-Switching Antisense Therapies. EMBO Mol. Med. 2017, 9, 545–557. [Google Scholar] [CrossRef]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense Oligonucleotides. Neurol. Genet. 2019, 5, e323. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Hastings, M.L. Splice-Switching Antisense Oligonucleotides as Therapeutic Drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, A.; Śliwa, A.; Żarowski, M.; Jankowska, A. Molecular Basis and Therapy of Spinal Muscular Atrophy. Child Neurol. 2019, 27, 39–46. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Warych, A.; Szoszkiewicz, M. Spinal Muscular Atrophy—Onasemnogene Abeparvovec and Other Therapeutic Options. Farm. Pol. 2020, 76, 10–17. [Google Scholar] [CrossRef]
- Jedrzejowska, M.; Milewski, M.; Zimowski, J.; Borkowska, J.; Kostera-Pruszczyk, A.; Sielska, D.; Jurek, M.; Hausmanowa-Petrusewicz, I. Phenotype Modifiers of Spinal Muscular Atrophy: The Number of SMN2 Gene Copies, Deletion in the NAIP Gene and Probably Gender Influence the Course of the Disease. Acta Biochim. Pol. 2009, 56, 103–108. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense Correction of SMN2 Splicing in the CNS Rescues Necrosis in a Type III SMA Mouse Model. Genes Dev. 2010, 24, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Gidaro, T.; Servais, L. Nusinersen Treatment of Spinal Muscular Atrophy: Current Knowledge and Existing Gaps. Dev. Med. Child Neurol. 2019, 61, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Li, Q. Nusinersen as a Therapeutic Agent for Spinal Muscular Atrophy. Yonsei Med. J. 2020, 61, 273–283. [Google Scholar] [CrossRef]
- Dabbous, O.; Maru, B.; Jansen, J.P.; Lorenzi, M.; Cloutier, M.; Guérin, A.; Pivneva, I.; Wu, E.Q.; Arjunji, R.; Feltner, D.; et al. Survival, Motor Function, and Motor Milestones: Comparison of AVXS-101 Relative to Nusinersen for the Treatment of Infants with Spinal Muscular Atrophy Type 1. Adv. Ther. 2019, 36, 1164–1176. [Google Scholar] [CrossRef]
- Spinraza (Nusinersen); Biogen Inc.: Cambridge, MA, USA, 2018; Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/209531lbl.pdf (accessed on 7 July 2021).
- Rao, V.K.; Kapp, D.; Schroth, M. Gene Therapy for Spinal Muscular Atrophy: An Emerging Treatment Option for a Devastating Disease. J. Manag. Care Spec. Pharm. 2018, 24, S3–S16. [Google Scholar] [CrossRef]
- Wood, M.J.A.; Talbot, K.; Bowerman, M. Spinal Muscular Atrophy: Antisense Oligonucleotide Therapy Opens the Door to an Integrated Therapeutic Landscape. Hum. Mol. Genet. 2017, 26, R151–R159. [Google Scholar] [CrossRef]
- Foead, A.; Yeo, W.Y.; Vishnumukkala, T.; Larvin, M. Rehabilitation in Spinal Muscular Atrophy. J. Int. Soc. Phys. Rehabil. Med. 2019, 2, 62. [Google Scholar] [CrossRef]
- De Wel, B.; Goosens, V.; Sobota, A.; Van Camp, E.; Geukens, E.; Van Kerschaver, G.; Jagut, M.; Claes, K.; Claeys, K.G. Nusinersen Treatment Significantly Improves Hand Grip Strength, Hand Motor Function and MRC Sum Scores in Adult Patients with Spinal Muscular Atrophy Types 3 and 4. J. Neurol. 2021, 268, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Lee, B.M.; Singh, R.N. Splicing Regulation in Spinal Muscular Atrophy by an RNA Structure Formed by Long-Distance Interactions. Ann. N. Y. Acad. Sci. 2015, 1341, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Chiriboga, C.A.; Swoboda, K.J.; Darras, B.T.; Iannaccone, S.T.; Montes, J.; De Vivo, D.C.; Norris, D.A.; Bennett, C.F.; Bishop, K.M. Results from a Phase 1 Study of Nusinersen (ISIS-SMNRx) in Children with Spinal Muscular Atrophy. Neurology 2016, 86, 890–897. [Google Scholar] [CrossRef]
- Howell, M.D.; Ottesen, E.W.; Singh, N.N.; Anderson, R.L.; Singh, R.N. Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1328–1341. [Google Scholar] [CrossRef]
- Touznik, A.; Maruyama, R.; Hosoki, K.; Echigoya, Y.; Yokota, T. LNA/DNA Mixmer-Based Antisense Oligonucleotides Correct Alternative Splicing of the SMN2 Gene and Restore SMN Protein Expression in Type 1 SMA Fibroblasts. Sci. Rep. 2017, 7, 3672. [Google Scholar] [CrossRef] [PubMed]
- Flynn, L.L.; Mitrpant, C.; Pitout, I.L.; Fletcher, S.; Wilton, S.D. Antisense Oligonucleotide-Mediated Terminal Intron Retention of the SMN2 Transcript. Mol. Ther. Nucleic Acids 2018, 11, 91–102. [Google Scholar] [CrossRef]
- Mercuri, E.; Darras, B.T.; Chiriboga, C.A.; Day, J.W.; Campbell, C.; Connolly, A.M.; Iannaccone, S.T.; Kirschner, J.; Kuntz, N.L.; Saito, K.; et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2018, 378, 625–635. [Google Scholar] [CrossRef]
- De Vivo, D.C.; Bertini, E.; Swoboda, K.J.; Hwu, W.-L.; Crawford, T.O.; Finkel, R.S.; Kirschner, J.; Kuntz, N.L.; Parsons, J.A.; Ryan, M.M.; et al. Nusinersen Initiated in Infants during the Presymptomatic Stage of Spinal Muscular Atrophy: Interim Efficacy and Safety Results from the Phase 2 NURTURE Study. Neuromuscul. Disord. 2019, 29, 842–856. [Google Scholar] [CrossRef]
- Naryshkin, N.A.; Weetall, M.; Dakka, A.; Narasimhan, J.; Zhao, X.; Feng, Z.; Ling, K.K.Y.; Karp, G.M.; Qi, H.; Woll, M.G.; et al. Motor Neuron Disease. SMN2 Splicing Modifiers Improve Motor Function and Longevity in Mice with Spinal Muscular Atrophy. Science 2014, 345, 688–693. [Google Scholar] [CrossRef]
- Taladriz-Sender, A.; Campbell, E.; Burley, G.A. Splice-Switching Small Molecules: A New Therapeutic Approach to Modulate Gene Expression. Methods 2019, 167, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Sivaramakrishnan, M.; McCarthy, K.D.; Campagne, S.; Huber, S.; Meier, S.; Augustin, A.; Heckel, T.; Meistermann, H.; Hug, M.N.; Birrer, P.; et al. Binding to SMN2 Pre-MRNA-Protein Complex Elicits Specificity for Small Molecule Splicing Modifiers. Nat. Commun. 2017, 8, 1476. [Google Scholar] [CrossRef]
- Dhillon, S. Risdiplam: First Approval. Drugs 2020, 80, 1853–1858. [Google Scholar] [CrossRef]
- Poirier, A.; Weetall, M.; Heinig, K.; Bucheli, F.; Schoenlein, K.; Alsenz, J.; Bassett, S.; Ullah, M.; Senn, C.; Ratni, H.; et al. Risdiplam Distributes and Increases SMN Protein in Both the Central Nervous System and Peripheral Organs. Pharmacol. Res. Perspect. 2018, 6, e00447. [Google Scholar] [CrossRef] [PubMed]
- Ratni, H.; Ebeling, M.; Baird, J.; Bendels, S.; Bylund, J.; Chen, K.S.; Denk, N.; Feng, Z.; Green, L.; Guerard, M.; et al. Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 ( SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem. 2018, 61, 6501–6517. [Google Scholar] [CrossRef] [PubMed]
- Paluszczak, J. Therapeutic Targeting of Alternative Splicing. Farm. Pol. 2019, 75, 605–616. [Google Scholar] [CrossRef]
- Wang, J.; Schultz, P.G.; Johnson, K.A. Mechanistic Studies of a Small-Molecule Modulator of SMN2 Splicing. Proc. Natl. Acad. Sci. USA 2018, 115, E4604–E4612. [Google Scholar] [CrossRef]
- Baranello, G.; Darras, B.T.; Day, J.W.; Deconinck, N.; Klein, A.; Masson, R.; Mercuri, E.; Rose, K.; El-Khairi, M.; Gerber, M.; et al. Risdiplam in Type 1 Spinal Muscular Atrophy. N. Engl. J. Med. 2021, 384, 915–923. [Google Scholar] [CrossRef]
- Singh, R.N.; Ottesen, E.W.; Singh, N.N. The First Orally Deliverable Small Molecule for the Treatment of Spinal Muscular Atrophy. Neurosci. Insights 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Messina, S.; Sframeli, M. New Treatments in Spinal Muscular Atrophy: Positive Results and New Challenges. J. Clin. Med. 2020, 9, 2222. [Google Scholar] [CrossRef] [PubMed]
- Yeo, C.J.J.; Darras, B.T. Overturning the Paradigm of Spinal Muscular Atrophy as Just a Motor Neuron Disease. Pediatr. Neurol. 2020, 109, 12–19. [Google Scholar] [CrossRef]
- Hamilton, G.; Gillingwater, T.H. Spinal Muscular Atrophy: Going beyond the Motor Neuron. Trends Mol. Med. 2013, 19, 40–50. [Google Scholar] [CrossRef]
- Evrysdi (Risdiplam) FDA Approval History. Available online: https://www.drugs.com/history/evrysdi.html (accessed on 7 July 2021).
- Novartis Pharmaceuticals. An Open Label Multi-Part First-in-Human Study of Oral LMI070 in Infants with Type 1 Spinal Muscular Atrophy. Available online: https://clinicaltrials.gov/ct2/show/NCT02268552 (accessed on 28 June 2021).
- Palacino, J.; Swalley, S.E.; Song, C.; Cheung, A.K.; Shu, L.; Zhang, X.; Van Hoosear, M.; Shin, Y.; Chin, D.N.; Keller, C.G.; et al. SMN2 Splice Modulators Enhance U1-Pre-MRNA Association and Rescue SMA Mice. Nat. Chem. Biol. 2015, 11, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Ando, S.; Suzuki, S.; Okubo, S.; Ohuchi, K.; Takahashi, K.; Nakamura, S.; Shimazawa, M.; Fuji, K.; Hara, H. Discovery of a CNS Penetrant Small Molecule SMN2 Splicing Modulator with Improved Tolerability for Spinal Muscular Atrophy. Sci. Rep. 2020, 10, 17472. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Androphy, E.J.; Singh, R.N. In Vivo Selection Reveals Combinatorial Controls That Define a Critical Exon in the Spinal Muscular Atrophy Genes. RNA 2004, 10, 1291–1305. [Google Scholar] [CrossRef]
- Singh, R.N. Unfolding the Mystery of Alternative Splicing through a Unique Method of in Vivo Selection. Front. Biosci. J. Virtual Libr. 2007, 12, 3263–3272. [Google Scholar] [CrossRef]
- Woll, M.G.; Qi, H.; Turpoff, A.; Zhang, N.; Zhang, X.; Chen, G.; Li, C.; Huang, S.; Yang, T.; Moon, Y.-C.; et al. Discovery and Optimization of Small Molecule Splicing Modifiers of Survival Motor Neuron 2 as a Treatment for Spinal Muscular Atrophy. J. Med. Chem. 2016, 59, 6070–6085. [Google Scholar] [CrossRef]
- Zhao, X.; Feng, Z.; Ling, K.K.Y.; Mollin, A.; Sheedy, J.; Yeh, S.; Petruska, J.; Narasimhan, J.; Dakka, A.; Welch, E.M.; et al. Pharmacokinetics, Pharmacodynamics, and Efficacy of a Small-Molecule SMN2 Splicing Modifier in Mouse Models of Spinal Muscular Atrophy. Hum. Mol. Genet. 2016, 25, 1885–1899. [Google Scholar] [CrossRef]
- Ratni, H.; Karp, G.M.; Weetall, M.; Naryshkin, N.A.; Paushkin, S.V.; Chen, K.S.; McCarthy, K.D.; Qi, H.; Turpoff, A.; Woll, M.G.; et al. Specific Correction of Alternative Survival Motor Neuron 2 Splicing by Small Molecules: Discovery of a Potential Novel Medicine To Treat Spinal Muscular Atrophy. J. Med. Chem. 2016, 59, 6086–6100. [Google Scholar] [CrossRef]
- Pinard, E.; Green, L.; Reutlinger, M.; Weetall, M.; Naryshkin, N.A.; Baird, J.; Chen, K.S.; Paushkin, S.V.; Metzger, F.; Ratni, H. Discovery of a Novel Class of Survival Motor Neuron 2 Splicing Modifiers for the Treatment of Spinal Muscular Atrophy. J. Med. Chem. 2017, 60, 4444–4457. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Hurley, B.; Kerrigan, R.; Shu, L.; Chin, D.N.; Shen, Y.; O’Brien, G.; Sung, M.J.; Hou, Y.; Axford, J.; et al. Discovery of Small Molecule Splicing Modulators of Survival Motor Neuron-2 (SMN2) for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem. 2018, 61, 11021–11036. [Google Scholar] [CrossRef]
- Kletzl, H.; Marquet, A.; Günther, A.; Tang, W.; Heuberger, J.; Groeneveld, G.J.; Birkhoff, W.; Mercuri, E.; Lochmüller, H.; Wood, C.; et al. The Oral Splicing Modifier RG7800 Increases Full Length Survival of Motor Neuron 2 MRNA and Survival of Motor Neuron Protein: Results from Trials in Healthy Adults and Patients with Spinal Muscular Atrophy. Neuromuscul. Disord. NMD 2019, 29, 21–29. [Google Scholar] [CrossRef]
- Sturm, S.; Günther, A.; Jaber, B.; Jordan, P.; Al Kotbi, N.; Parkar, N.; Cleary, Y.; Frances, N.; Bergauer, T.; Heinig, K.; et al. A Phase 1 Healthy Male Volunteer Single Escalating Dose Study of the Pharmacokinetics and Pharmacodynamics of Risdiplam (RG7916, RO7034067), a SMN2 Splicing Modifier. Br. J. Clin. Pharmacol. 2019, 85, 181–193. [Google Scholar] [CrossRef]
- Konieczny, P.; Artero, R. Drosophila SMN2 Minigene Reporter Model Identifies Moxifloxacin as a Candidate Therapy for SMA. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 3021–3036. [Google Scholar] [CrossRef]
- Long, K.K.; O’Shea, K.M.; Khairallah, R.J.; Howell, K.; Paushkin, S.; Chen, K.S.; Cote, S.M.; Webster, M.T.; Stains, J.P.; Treece, E.; et al. Specific Inhibition of Myostatin Activation Is Beneficial in Mouse Models of SMA Therapy. Hum. Mol. Genet. 2019, 28, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Meng, J.; Malerba, A.; Catapano, F.; Sintusek, P.; Jarmin, S.; Feng, L.; Lu-Nguyen, N.; Sun, L.; Mariot, V.; et al. Myostatin Inhibition in Combination with Antisense Oligonucleotide Therapy Improves Outcomes in Spinal Muscular Atrophy. J. Cachexia Sarcopenia Muscle 2020, 11, 768–782. [Google Scholar] [CrossRef] [PubMed]
- Cytokinetics. A Phase 2, Double-Blind, Randomized, Placebo-Controlled, Multiple Dose Study of CK-2127107 in Two Ascending Dose Cohorts of Patients with Spinal Muscular Atrophy. Available online: https://clinicaltrials.gov/ct2/show/NCT02644668 (accessed on 17 August 2021).
- Calucho, M.; Bernal, S.; Alías, L.; March, F.; Venceslá, A.; Rodríguez-Álvarez, F.J.; Aller, E.; Fernández, R.M.; Borrego, S.; Millán, J.M.; et al. Correlation between SMA Type and SMN2 Copy Number Revisited: An Analysis of 625 Unrelated Spanish Patients and a Compilation of 2834 Reported Cases. Neuromuscul. Disord. NMD 2018, 28, 208–215. [Google Scholar] [CrossRef]
- Glascock, J.; Sampson, J.; Haidet-Phillips, A.; Connolly, A.; Darras, B.; Day, J.; Finkel, R.; Howell, R.R.; Klinger, K.; Kuntz, N.; et al. Treatment Algorithm for Infants Diagnosed with Spinal Muscular Atrophy through Newborn Screening. J. Neuromuscul. Dis. 2018, 5, 145–158. [Google Scholar] [CrossRef]
- Tizzano, E.F.; Finkel, R.S. Spinal Muscular Atrophy: A Changing Phenotype beyond the Clinical Trials. Neuromuscul. Disord. NMD 2017, 27, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Oprea, G.E.; Kröber, S.; McWhorter, M.L.; Rossoll, W.; Müller, S.; Krawczak, M.; Bassell, G.J.; Beattie, C.E.; Wirth, B. Plastin 3 Is a Protective Modifier of Autosomal Recessive Spinal Muscular Atrophy. Science 2008, 320, 524–527. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.-H.; Sun, J.; Krainer, A.R.; Hua, Y.; Prior, T.W. A-44G Transition in SMN2 Intron 6 Protects Patients with Spinal Muscular Atrophy. Hum. Mol. Genet. 2017, 26, 2768–2780. [Google Scholar] [CrossRef]
- Darras, B.T.; Crawford, T.O.; Finkel, R.S.; Mercuri, E.; De Vivo, D.C.; Oskoui, M.; Tizzano, E.F.; Ryan, M.M.; Muntoni, F.; Zhao, G.; et al. Neurofilament as a Potential Biomarker for Spinal Muscular Atrophy. Ann. Clin. Transl. Neurol. 2019, 6, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Finkel, R.S.; Muntoni, F.; Wirth, B.; Montes, J.; Main, M.; Mazzone, E.S.; Vitale, M.; Snyder, B.; Quijano-Roy, S.; et al. Diagnosis and Management of Spinal Muscular Atrophy: Part 1: Recommendations for Diagnosis, Rehabilitation, Orthopedic and Nutritional Care. Neuromuscul. Disord. NMD 2018, 28, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Finkel, R.S.; Mercuri, E.; Meyer, O.H.; Simonds, A.K.; Schroth, M.K.; Graham, R.J.; Kirschner, J.; Iannaccone, S.T.; Crawford, T.O.; Woods, S.; et al. Diagnosis and Management of Spinal Muscular Atrophy: Part 2: Pulmonary and Acute Care; Medications, Supplements and Immunizations; Other Organ Systems; and Ethics. Neuromuscul. Disord. NMD 2018, 28, 197–207. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).