Genome Size Dynamics in Marine Ribbon Worms (Nemertea, Spiralia)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nemertean Species Selection
2.2. Genome Size Estimation
2.3. DNA Isolation, PCR, Sequencing, and Phylogenetic Reconstruction
2.4. Data Analyses
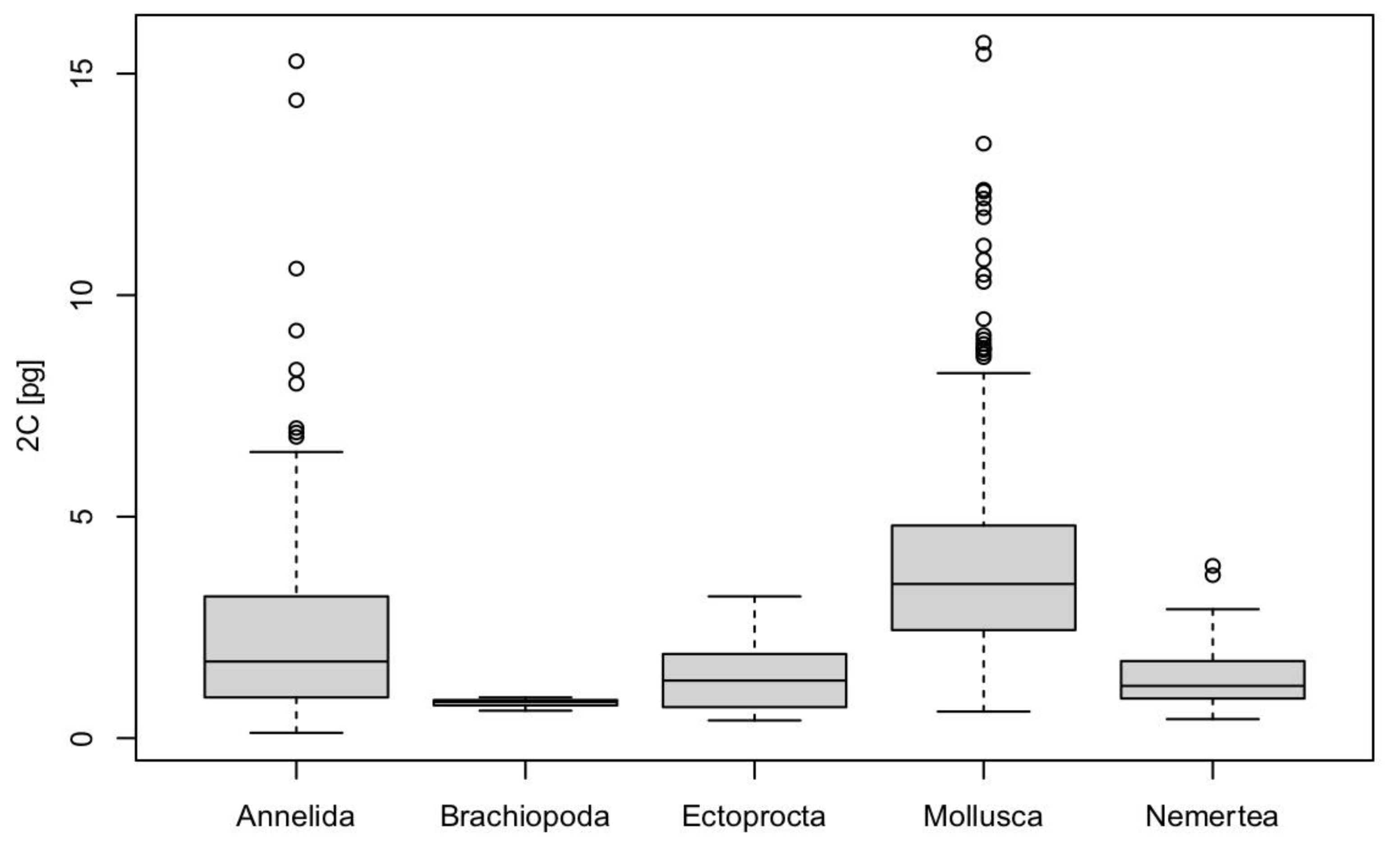
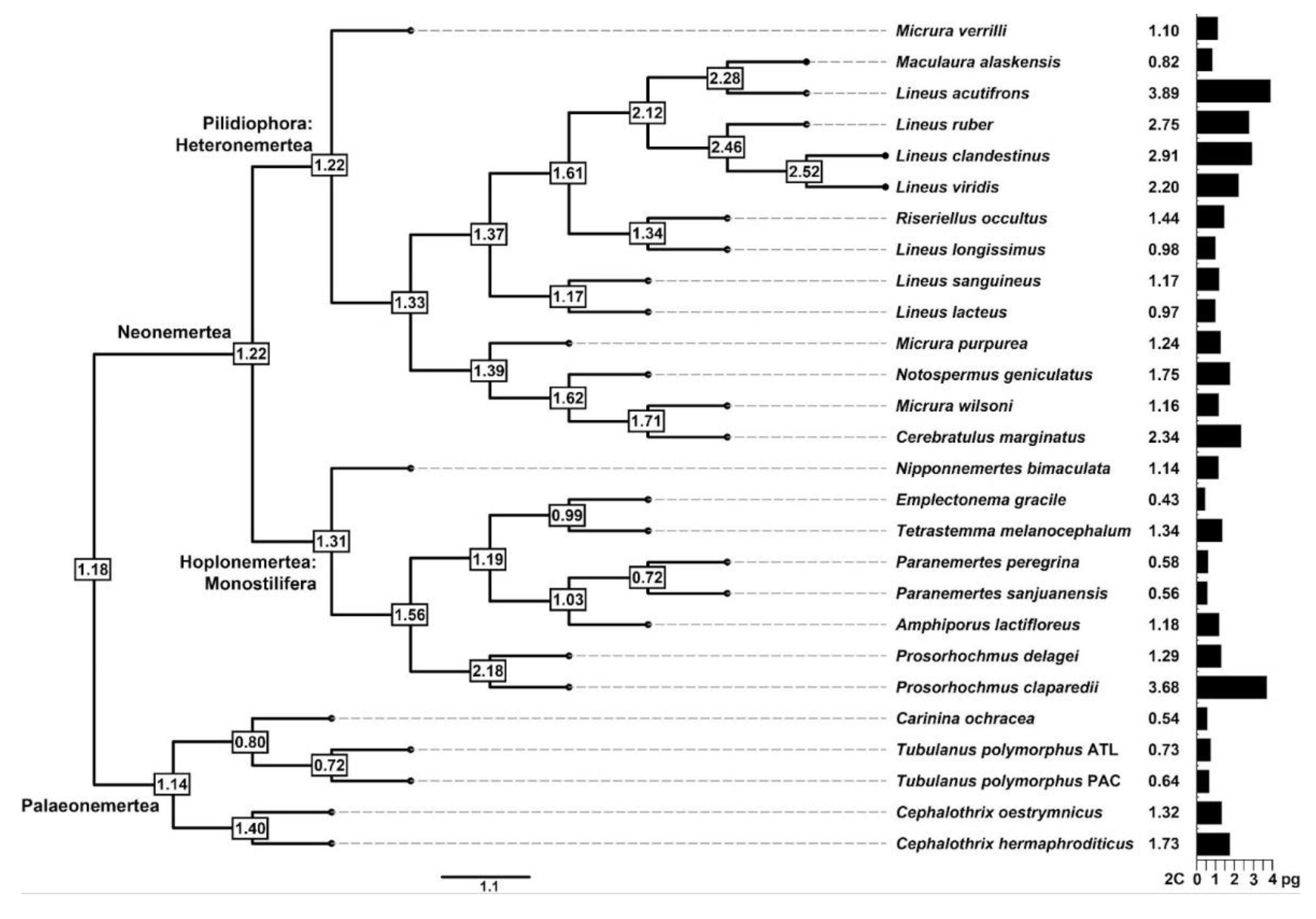
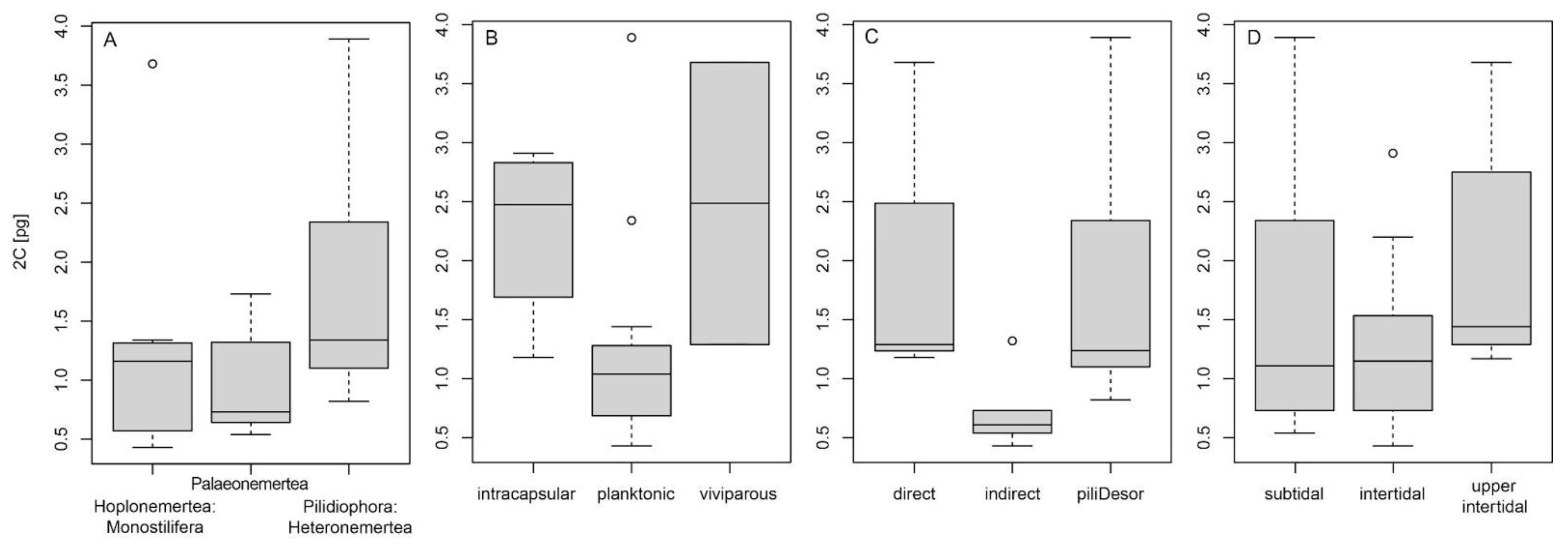
3. Results
4. Discussion
4.1. Genome Size in Nemertea
4.2. Evolutionary Genome Size Dynamics
4.3. Body Size and Genome Size
4.4. Life History and Ecological Traits and Genome Size
4.5. Nemertea and Genomic Biodiversity
4.6. Remarks on Nemertean Taxonomy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.-Q. Animal biodiversity: An update of classification and diversity in 2013. In: Zhang, Z.-Q. (Ed.) Animal Biodiversity: An Outline of Higher-level Classification and Survey of Taxonomic Richness (Addenda 2013). Zootaxa 2013, 3703, 5–11. [Google Scholar] [CrossRef] [Green Version]
- Bleidorn, C. Recent progress in reconstructing lophotrochozoan (spiralian) phylogeny. Org. Divers. Evol. 2019, 19, 557–566. [Google Scholar] [CrossRef]
- Kajihara, H.; Chernyshev, A.V.; Sun, S.-C.; Sundberg, P.; Crandall, F.B. Checklist of Nemertean Genera and Species Published between 1995 and 2007. Species Divers. 2008, 13, 245–274. [Google Scholar] [CrossRef] [Green Version]
- Gittenberger, A.; Schipper, C. Long live Linnaeus, Lineus longissimus (Gunnerus, 1770) (Vermes: Nemertea: Anopla: Heteronemertea: Lineidae), the longest animal worldwide and its relatives occurring in the Netherlands. Zool. Meded. 2008, 82, 59–63. [Google Scholar]
- von Döhren, J.; Bartolomaeus, T. Nemertea. In Guide to the Identification of Marine Meiofauna; Schmidt-Rhaesa, A., Ed.; Pfeil Verlag: Munich, Germany, 2020; pp. 227–238. [Google Scholar]
- Gibson, R. Nemertean genera and species of the world: An annotated checklist of original names and description citations, synonyms, current taxonomic status, habitats and recorded zoogeographic distribution. J. Nat. Hist. 1995, 29, 271–561. [Google Scholar] [CrossRef]
- Maslakova, S.A. The Invention of the Pilidium Larva in an Otherwise Perfectly Good Spiralian Phylum Nemertea. Integr. Comp. Biol. 2010, 50, 734–743. [Google Scholar] [CrossRef]
- Maslakova, S.A.; Hiebert, T.C. From trochophore to pilidium and back again-a larva’s journey. Int. J. Dev. Biol. 2014, 58, 585–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thollesson, M.; Norenburg, J.L. Ribbon worm relationships: A phylogeny of the phylum Nemertea. Proc. R. Soc. B Boil. Sci. 2003, 270, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Andrade, S.C.S.; Montenegro, H.; Strand, M.; Schwartz, M.L.; Kajihara, H.; Norenburg, J.L.; Turbeville, J.M.; Sundberg, P.; Giribet, G. A Transcriptomic Approach to Ribbon Worm Systematics (Nemertea): Resolving the Pilidiophora Problem. Mol. Biol. Evol. 2014, 31, 3206–3215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, S.; Strand, M.; Schwartz, M.L.; Chen, H.; Kajihara, H.; Von Döhren, J.; Sun, S.; Junoy, J.; Thiel, M.; Norenburg, J.L.; et al. Disentangling ribbon worm relationships: Multi-locus analysis supports traditional classification of the phylum Nemertea. Cladistics 2011, 28, 141–159. [Google Scholar] [CrossRef]
- Kvist, S.; Laumer, C.E.; Junoy, J.; Giribet, G. New insights into the phylogeny, systematics and DNA barcoding of Nemertea. Invertebr. Syst. 2014, 28, 287–308. [Google Scholar] [CrossRef] [Green Version]
- Thiel, M.; Junoy, J. Mating behavior of nemerteans: Present knowledge and future directions. J. Nat. Hist. 2006, 40, 1021–1034. [Google Scholar] [CrossRef]
- von Döhren, J. Nemertea. Evolutionary Developmental Biology of Invertebrates Vol. 2: Lophotrochozoa (Spiralia); Wanninger, A., Ed.; Springer: Wien, Austria, 2015; pp. 155–192. [Google Scholar]
- Zattara, E.E.; Fernández-Álvarez, F.A.; Hiebert, T.C.; Bely, A.E.; Norenburg, J.L. A phylum-wide survey reveals multiple independent gains of head regeneration in Nemertea. Proc. R. Soc. B Boil. Sci. 2019, 286, 20182524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Döhren, J. The fate of the larval epidermis in the Desor-larva of Lineus viridis (Pilidiophora, Nemertea) displays a historically constrained functional shift from planktotrophy to lecithotrophy. Zoomorphology 2011, 130, 189–196. [Google Scholar] [CrossRef]
- Gregory, T.R. Coincidence, coevolution, or causation? DNA content, cellsize, and the C-value enigma. Biol. Rev. 2007, 76, 65–101. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Origins of Genome Complexity. Science 2003, 302, 1401–1404. [Google Scholar] [CrossRef] [Green Version]
- Petrov, D.A. Mutational Equilibrium Model of Genome Size Evolution. Theor. Popul. Biol. 2002, 61, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T. Insertion–deletion biases and the evolution of genome size. Gene 2004, 324, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Arriaza, J.R.L.; Mueller, R.L. Slow DNA Loss in the Gigantic Genomes of Salamanders. Genome Biol. Evol. 2012, 4, 1340–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapusta, A.; Suh, A.; Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA 2017, 114, E1460–E1469. [Google Scholar] [CrossRef] [Green Version]
- Gregory, T. The Bigger the C-Value, the Larger the Cell: Genome Size and Red Blood Cell Size in Vertebrates. Blood Cells Mol. Dis. 2001, 27, 830–843. [Google Scholar] [CrossRef] [Green Version]
- Gregory, T.R.; Johnston, J.S. Genome size diversity in the family Drosophilidae. Heredity 2008, 101, 228–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, C.B.; MacKenzie, S.A.; Gregory, T.R. Genome size and wing parameters in passerine birds. Proc. R. Soc. B Boil. Sci. 2008, 276, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Wright, N.A.; Gregory, T.R.; Witt, C. Metabolic ‘engines’ of flight drive genome size reduction in birds. Proc. R. Soc. B Boil. Sci. 2014, 281, 20132780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Liu, K.; Sun, S. Karyotype analysis of four nemertean species. Chin. J. Oceanol. Limnol. 2009, 27, 748–752. [Google Scholar] [CrossRef]
- Ament-Velásquez, S.L.; Figuet, E.; Ballenghien, M.; Zattara, E.E.; Norenburg, J.; Fernández-Álvarez, F.; Bierne, J.; Bierne, N.; Galtier, N. Population genomics of sexual and asexual lineages in fissiparous ribbon worms (Lineus, Nemertea): Hybridization, polyploidy and the Meselson effect. Mol. Ecol. 2016, 25, 3356–3369. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, K.L.; Hiebert, T.C.; Jeffery, N.W.; Gregory, T.R. First estimates of genome size in ribbon worms (phylum Nemertea) using flow cytometry and Feulgen image analysis densitometry. Can. J. Zool. 2014, 92, 847–851. [Google Scholar] [CrossRef]
- Bartolomaeus, T.; Von Döhren, J. Comparative morphology and evolution of the nephridia in Nemertea. J. Nat. Hist. 2010, 44, 2255–2286. [Google Scholar] [CrossRef]
- Stricker, S.A. Phylum Nemertea. In Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast; Strathmann, M.F., Ed.; University of Washington Press: Seattle, WA, USA, 1987; pp. 129–137. [Google Scholar]
- Maslakova, S.A.; von Döhren, J. Larval development with transitory epidermis in Paranemertes peregrina and other hoplonemerteans. Biol. Bull. 2009, 216, 273–292. [Google Scholar] [CrossRef] [Green Version]
- Hiebert, T.C.; Maslakova, S.A. Larval Development of Two N. E. Pacific Pilidiophoran Nemerteans (Heteronemertea; Lineidae). Biol. Bull. 2015, 229, 265–275. [Google Scholar] [CrossRef]
- Hiebert, T.C.; Maslakova, S. Integrative Taxonomy of the Micrura alaskensisCoe, 1901 Species Complex (Nemertea: Heteronemertea), with Descriptions of a New GenusMaculauragen. nov. and Four New Species from the NE Pacific. Zool. Sci. 2015, 32, 615–637. [Google Scholar] [CrossRef] [Green Version]
- Stricker, S.A. The Morphology of Paranemertes sanjuanensis sp.n. (Nemertea, Monostilifera) from Washington, U.S.A. Zool. Scr. 1982, 11, 107–115. [Google Scholar] [CrossRef]
- Riser, N.W. New Zealand nemertines from kelp holdfasts: Heteronemertinea II. Notospermus geniculatus (Delle Chiaje, 1828) n. comb. New Zealand J. Zool. 1991, 18, 427–438. [Google Scholar] [CrossRef]
- Junoy, J.; Gibson, R. A new species of Procephalothrix (Anopla, Archinemertea) from North-Western Spain (Nemertea). Zool. Anz. 1991, 226, 185–194. [Google Scholar]
- Sundberg, P.; Chernyshev, A.V.; Kajihara, H.; Kånneby, T.; Strand, M. Character-matrix based descriptions of two new nemertean (Nemertea) species. Zool. J. Linn. Soc. 2009, 157, 264–294. [Google Scholar] [CrossRef] [Green Version]
- Beckers, P.; Bartolomaeus, T.; Von Döhren, J. Observations and Experiments on the Biology and Life History of Riseriellus occultus (Heteronemertea: Lineidae). Zool. Sci. 2015, 32, 531–546. [Google Scholar] [CrossRef]
- Von Döhren, J. First record on the development of the larva of the basally branching nemertean species Carinina ochracea (Palaeonemertea). Helgol. Mar. Res. 2016, 70, 141. [Google Scholar] [CrossRef] [Green Version]
- Gibson, R.; Knight-Jones, E.W. Flatworms and Ribbon Worms. In Handbook of the Marine Fauna of North-West Europe; Hayward, P.J., Ryland, J.S., Eds.; Oxford University Press: Oxford, UK, 2017; pp. 133–164. [Google Scholar]
- Krämer, D.; Schmidt, C.; Podsiadlowski, L.; Beckers, P.; Horn, L.; Von Döhren, J. Unravelling the Lineus ruber/viridis species complex (Nemertea, Heteronemertea). Zool. Scr. 2016, 46, 111–126. [Google Scholar] [CrossRef]
- Von Döhren, J.; Bartolomaeus, T. Unexpected ultrastructure of an eye in Spiralia: The larval ocelli of Procephalothrix oestrymnicus (Nemertea). Zoomorphology 2018, 137, 241–248. [Google Scholar] [CrossRef]
- Sagorny, C.; Wesseler, C.; Krämer, D.; Von Döhren, J. Assessing the diversity and distribution of Cephalothrix species (Nemertea: Palaeonemertea) in European waters by comparing different species delimitation methods. J. Zool. Syst. Evol. Res. 2019, 57, 497–519. [Google Scholar] [CrossRef]
- Greilhuber, J. The Origin, Evolution and Proposed Stabilization of the Terms ‘Genome Size’ and ‘C-Value’ to Describe Nuclear DNA Contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Otto, F. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In Methods in Cell Biology; Vol. 33 Flow Cytometry; Darzynkiewicz, Z., Crissman, H.A., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; pp. 105–110. [Google Scholar]
- Dolezel, J.; Doleželová, M.; Novák, F.J. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol. Plant. 1994, 36, 351–357. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Lucretti, S.; Meister, A.; Lysák, M.A.; Nardi, L.; Obermayer, R. Plant Genome Size Estimation by Flow Cytometry: Inter-laboratory Comparison. Ann. Bot. 1998, 82, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Doležel, J.; Bartoš, J.; Voglmayr, H.; Greilhuber, J. Letter to the editor. Cytometry 2003, 51, 127–128. [Google Scholar] [CrossRef]
- Luo, Y.-J.; Kanda, M.; Koyanagi, R.; Hisata, K.; Akiyama, T.; Sakamoto, H.; Sakamoto, T.; Satoh, N. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat. Ecol. Evol. 2017, 2, 141–151. [Google Scholar] [CrossRef]
- Gregory, T.R. Animal Genome Size Database. Available online: http://www.genomesize.com (accessed on 25 August 2011).
- Goldberg, R.B.; Crain, W.R.; Ruderman, J.V.; Moore, G.P.; Barnett, T.R.; Higgins, R.C.; Gelfand, R.A.; Galau, G.A.; Britten, R.J.; Davidson, E.H. DNA sequence organization in the genomes of five marine invertebrates. Chromosoma 1975, 51, 225–251. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Boil. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venables, W.N.; Ripley, B.D. Package MASS. Available online: http://www.r-project.org (accessed on 17 October 2012).
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2011, 3, 217–223. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Jhwueng, D.-C.; Boettiger, C.; O’Meara, B. Modeling stabilizing selection: Expanding The Ornstein-Uhlenbeck model of adaptive evolution. Evolution 2012, 66, 2369–2383. [Google Scholar] [CrossRef]
- Grafen, A. The phylogenetic regression. Philos. Trans. R. Soc. B Biol. Sci. 1989, 326, 119–157. [Google Scholar] [CrossRef]
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. Caper: Comparative analyses of phylogenetics and evolution in R. R Packag version 0.5. Proc. R. Soc. B. 2012, 2, 458. [Google Scholar]
- Maslakova, S.A.; Norenburg, J.L. Revision of the smiling worms, genus Prosorhochmus Keferstein, 1862, and description of a new species, Prosorhochmus belizeanus sp. nov. (Prosorhochmidae, Hoplonemertea, Nemertea) from Florida and Belize. J. Nat. Hist. 2008, 42, 1219–1260. [Google Scholar] [CrossRef]
- von Döhren, J.; University of Bonn, Bonn, Germany. Personal communication, 2021.
- Du, Y.-P.; Bi, Y.; Zhang, M.-F.; Yang, F.-P.; Jia, G.-X.; Zhang, X.-H. Genome Size Diversity in Lilium (Liliaceae) Is Correlated with Karyotype and Environmental Traits. Front. Plant Sci. 2017, 8, 1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, B.A.; Mitton, J.B. The Relationship Between Genome Size and Genetic Variation. Am. Nat. 1980, 116, 850–861. [Google Scholar] [CrossRef]
- Gregory, T.R.; Hebert, P.; Kolasa, J. Evolutionary implications of the relationship between genome size and body size in flatworms and copepods. Heredity 2000, 84, 201–208. [Google Scholar] [CrossRef]
- Jeffery, N.W.; Ellis, E.A.; Oakley, T.H.; Gregory, T.R. The Genome Sizes of Ostracod Crustaceans Correlate with Body Size and Evolutionary History, but not Environment. J. Hered. 2017, 108, 701–706. [Google Scholar] [CrossRef] [Green Version]
- Wyngaard, G.A.; Rasch, E.M. Patterns of genome size in the copepoda. Hydrobiologia 2000, 417, 43–56. [Google Scholar] [CrossRef]
- Wyngaard, G.A.; Rasch, E.M.; Manning, N.M.; Gasser, K.; Domangue, R. The relationship between genome size, development rate, and body size in copepods. Hydrobiologia 2005, 532, 123–137. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Skeletal DNA and the Evolution of Genome Size. Annu. Rev. Biophys. Bioeng. 1982, 11, 273–302. [Google Scholar] [CrossRef] [PubMed]
- Glazier, D. Genome Size Covaries More Positively with Propagule Size than Adult Size: New Insights into an Old Problem. Biology 2021, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 1–12. [Google Scholar] [CrossRef]
- Talla, V.; Suh, A.; Kalsoom, F.; Dincă, V.; Vila, R.; Friberg, M.; Wiklund, C.; Backström, N. Rapid Increase in Genome Size as a Consequence of Transposable Element Hyperactivity in Wood-White (Leptidea) Butterflies. Genome Biol. Evol. 2017, 9, 2491–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naville, M.; Henriet, S.; Warren, I.; Sumic, S.; Reeve, M.; Volff, J.-N.; Chourrout, D. Massive Changes of Genome Size Driven by Expansions of Non-autonomous Transposable Elements. Curr. Biol. 2019, 29, 1161–1168. [Google Scholar] [CrossRef] [Green Version]
- Kalendar, R.; Tanskanen, J.; Immonen, S.; Nevo, E.; Schulman, A.H. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 2000, 97, 6603–6607. [Google Scholar] [CrossRef] [Green Version]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Finigan, P.; Tanurdzic, M.; Martienssen, R.A. Origins of Novel Phenotypic Variation in Polyploids. In Polyploidy and Genome Evolution; Springer: Berlin, Heidelberg, 2012; pp. 57–76. [Google Scholar] [CrossRef]
- Lewin, H.A.; Robinson, G.E.; Kress, W.J.; Baker, W.J.; Coddington, J.A.; Crandall, K.A.; Durbin, R.; Edwards, S.V.; Forest, F.; Gilbert, M.; et al. Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. USA 2018, 115, 4325–4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drukewitz, S.H.; Von Reumont, B.M. The Significance of Comparative Genomics in Modern Evolutionary Venomics. Front. Ecol. Evol. 2019, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Göransson, U.; Jacobsson, E.; Strand, M.; Andersson, H.S. The Toxins of Nemertean Worms. Toxins 2019, 11, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Reumont, B.M.; Lüddecke, T.; Timm, T.; Lochnit, G.; Vilcinskas, A.; Von Döhren, J.; Nilsson, M.A. Proteo-Transcriptomic Analysis Identifies Potential Novel Toxins Secreted by the Predatory, Prey-Piercing Ribbon Worm Amphiporus lactifloreus. Mar. Drugs 2020, 18, 407. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-W.; Harkess, A. A guide to sequence your favorite plant genomes. Appl. Plant Sci. 2018, 6, e1030. [Google Scholar] [CrossRef]
- Schwartz, M.L.; Norenburg, J.L. Can we infer heteronemertean phylogeny from available morphological data? Hydrobiologia 2001, 456, 165–174. [Google Scholar] [CrossRef]
- Cornet, R.; Marche-Marchad, I. Inventaire de la faune Marine de Roscoff. In Travaux de la Station Biologique de Roscoff; Presses Universitaires de France: Paris, France, 1955; Volume 7, pp. 1–15. [Google Scholar]
- Gibson, R.; Moore, J. The genus Prosorhochmus Keferstein, 1862 (Hoplonemertea). J. Zool. 1985, 206, 145–162. [Google Scholar] [CrossRef]
- Krämer, D.; University of Bonn, Bonn, Germany. Personal communication, 2017.
| Species | Order:Class | Method | Standard | 2C [pg] | ±SD | 1C [Mbp] | ±SD | Source |
|---|---|---|---|---|---|---|---|---|
| Amphiporus lactifloreus (Johnston, 1828) | Hoplonemertea: Monostilifera | FCM | G. max | 1.18 | 0.01 | 575.78 | 6.13 | this study |
| Carinina ochracea [38] | Palaeonemertea: Tubulaniformes | FCM | G. max | 0.54 | 0.00 | 264.88 | 1.03 | this study |
| Cephalothrix hermaphroditicus Gibson, Sanchez, and Mendez, 1990 | Palaeonemertea: Archinemertea | FCM | G. max | 1.73 | 0.01 | 847.72 | 4.31 | this study |
| Cephalothrix oestrymnicus Junoy and Gibson, 1991 | Palaeonemertea: Archinemertea | FCM | G. max | 1.32 | 0.03 | 646.34 | 12.24 | this study |
| Cerebratulus marginatus Renier, 1804 | Pilidiophora: Heteronemertea | FCM | O. mykiss | 2.34 | 0.06 | 1144.26 | 29.34 | [29] |
| Emplectonema gracile (Johnston, 1837) | Hoplonemertea: Monostilifera | FCM | G. max | 0.43 | 0.01 | 209.75 | 5.42 | this study |
| Lineus acutifrons Southern, 1913 | Pilidiophora: Heteronemertea | FCM | G. max | 3.89 | 0.13 | 1904.45 | 64.20 | this study |
| Lineus clandestinus Krämer, Schmidt, Podsiadlowski, Beckers, Horn and von Döhren, 2016 | Pilidiophora: Heteronemertea | FCM | P. sativum | 2.91 | 0.10 | 1423.91 | 51.16 | this study |
| Lineus lacteus (Rathke, 1843) | Pilidiophora: Heteronemertea | FCM | G. max | 0.97 | 0.02 | 474.88 | 10.66 | this study |
| Lineus longissimus (Gunnerus, 1770) | Pilidiophora: Heteronemertea | FCM | G. max | 0.98 | 0.04 | 477.06 | 17.58 | this study |
| Lineus ruber (Müller, 1774) | Pilidiophora: Heteronemertea | FCM | P. sativum | 2.75 | 0.04 | 1342.41 | 19.59 | this study |
| Lineus sanguineus (Rathke, 1799) | Pilidiophora: Heteronemertea | FCM | G. max | 1.17 | 0.03 | 572.93 | 13.05 | this study |
| Lineus viridis (Müller, 1774) | Pilidiophora: Heteronemertea | FCM | P. sativum | 2.20 | 0.05 | 1074.02 | 25.35 | this study |
| Maculaura alaskensis (Coe, 1901) (published as Micura alaskensis) | Pilidiophora: Heteronemertea | FCM | O. mykiss | 0.82 | NA | 400.98 | NA | [29] |
| Micrura purpurea (Dalyell, 1853) | Pilidiophora: Heteronemertea | FCM | G. max | 1.24 | 0.02 | 606.21 | 9.81 | this study |
| Micrura verrilli Coe, 1901 | Pilidiophora: Heteronemertea | FCM | O. mykiss | 1.10 | NA | 537.90 | NA | [29] |
| Micrura wilsoni (Coe, 1904) | Pilidiophora: Heteronemertea | FCM | O. mykiss | 1.16 | NA | 567.24 | NA | [29] |
| Nipponnemertes bimaculata (Coe, 1901) (published as N. bimaculatus) | Hoplonemertea: Monostilifera | FCM | O. mykiss | 1.14 | 0.02 | 557.46 | 9.78 | [29] |
| Notospermus geniculatus (Delle Chiaje, 1828) | Pilidiophora: Heteronemertea | k-mer | NA | 1.75 | NA | 859.00 | NA | [50] |
| Paranemertes peregrina Coe, 1901 | Hoplonemertea: Monostilifera | FCM | O. mykiss | 0.58 | 0.00 | 283.62 | 0.00 | [29] |
| Paranemertes sanjuanensis Stricker, 1982 | Hoplonemertea: Monostilifera | FCM | O. mykiss | 0.56 | 0.16 | 273.84 | 78.24 | [29] |
| Prosorhochmus claparedii Keferstein, 1862 | Hoplonemertea: Monostilifera | FCM | G. max | 3.68 | 0.10 | 1801.70 | 50.97 | this study |
| Prosorhochmus delagei Oxner, 1907 | Hoplonemertea: Monostilifera | FCM | P. sativum | 1.29 | 0.02 | 631.60 | 11.34 | this study |
| Riseriellus occultus Rogers, Junoy, Gibson, and Thorpe, 1993 | Pilidiophora: Heteronemertea | FCM | G. max | 1.44 | 0.02 | 706.15 | 10.34 | this study |
| Tetrastemma melanocephalum (Johnston, 1837) | Hoplonemertea: Monostilifera | FCM | G. max | 1.34 | 0.04 | 657.40 | 18.19 | this study |
| Tubulanus polymorphus Renier, 1804 (Atlantic) | Palaeonemertea: Tubulaniformes | FCM | G. max | 0.73 | 0.02 | 358.47 | 11.80 | this study |
| Tubulanus polymorphus Renier, 1804 (Pacific) | Palaeonemertea: Tubulaniformes | FCM | O. mykiss | 0.64 | 0.08 | 312.96 | 39.12 | [29] |
| Genomic Character/Model | Groups | α | σ2 | θ (S.E.) |
|---|---|---|---|---|
| dev1/OUMV | intracapsular | 0.349 | 0.004 | 2.973 (1.150) |
| planktonic | 0.349 | 0.039 | 0.975 (0.118) | |
| viviparous | 0.349 | 0.513 | 5.609 (2.978) | |
| dev2/OU1 | non-feeding | 0.219 | 0.165 | 1.106 (0.267) |
| feeding | 0.219 | 0.165 | 1.106 (0.267) | |
| dev3/BMS | direct | N/A | 0.631 | 1.314 (0.418) |
| indirect | N/A | 0.006 | 1.314 (0.418) | |
| pilidium/Desor larva | N/A | 0.539 | 1.314 (0.418) | |
| habitat/OUMV | subtidal | 0.305 | 0.191 | 0.437 (1.394) |
| intertidal | 0.305 | 0.025 | 1.096 (0.097) | |
| upper intertidal | 0.305 | 0.207 | 3.615 (1.093) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paule, J.; von Döhren, J.; Sagorny, C.; Nilsson, M.A. Genome Size Dynamics in Marine Ribbon Worms (Nemertea, Spiralia). Genes 2021, 12, 1347. https://doi.org/10.3390/genes12091347
Paule J, von Döhren J, Sagorny C, Nilsson MA. Genome Size Dynamics in Marine Ribbon Worms (Nemertea, Spiralia). Genes. 2021; 12(9):1347. https://doi.org/10.3390/genes12091347
Chicago/Turabian StylePaule, Juraj, Jörn von Döhren, Christina Sagorny, and Maria A. Nilsson. 2021. "Genome Size Dynamics in Marine Ribbon Worms (Nemertea, Spiralia)" Genes 12, no. 9: 1347. https://doi.org/10.3390/genes12091347
APA StylePaule, J., von Döhren, J., Sagorny, C., & Nilsson, M. A. (2021). Genome Size Dynamics in Marine Ribbon Worms (Nemertea, Spiralia). Genes, 12(9), 1347. https://doi.org/10.3390/genes12091347





