Abstract
Melanoma comprises <5% of cutaneous malignancies, yet it causes a significant proportion of skin cancer-related deaths worldwide. While new therapies for melanoma have been developed, not all patients respond well. Thus, further research is required to better predict patient outcomes. Using long-range nanopore sequencing, RT-qPCR, and RNA sequencing analyses, we examined the transcription of BARD1 splice isoforms in melanoma cell lines and patient tissue samples. Seventy-six BARD1 mRNA variants were identified in total, with several previously characterised isoforms (γ, φ, δ, ε, and η) contributing to a large proportion of the expressed transcripts. In addition, we identified four novel splice events, namely, Δ(E3_E9), ▼(i8), IVS10+131▼46, and IVS10▼176, occurring in various combinations in multiple transcripts. We found that short-read RNA-Seq analyses were limited in their ability to predict isoforms containing multiple non-contiguous splicing events, as compared to long-range nanopore sequencing. These studies suggest that further investigations into the functional significance of the identified BARD1 splice variants in melanoma are warranted.
1. Introduction
Melanoma develops following the malignant transformation of pigment-producing melanocytes and is the fifth most common form of cancer in adults [1]. Importantly, while melanoma is responsible for the majority of skin cancer-related deaths, it accounts for <5% of cutaneous malignancies [2]. Over the past decade, great progress has been made in treating unresectable and metastatic melanomas. Indeed, five-year survival rates have increased from ~5% a decade ago to ~50% for those treated with combination immunotherapies, and ~33% for combination BRAF/MAPK kinase inhibitors [1]. Despite these tremendous advances, not all patients respond well to approved treatments. Moreover, the incidence of cutaneous melanoma is increasing at a startling 4–6% per annum [3]. Thus, there is a great need to further investigate melanoma cell biology to increase our understanding of long-term response and treatment outcomes.
BARD1 (BRCA1-Associated RING Domain 1; 2q34-q35) was initially discovered as a BRCA1-binding partner [4]. The interaction between RING domains within BARD1 and BRCA1 is critical for the tumour suppressive effects of the protein heterodimer, stimulating ubiquitin ligase activity [5]. The identified substrates for ubiquitination by the BARD1–BRCA1 heterodimer include the histone proteins, H2A, and the H2AX variant, γ tubulin, and subunits of the RNA polymerase 2 holoenzyme [6,7,8]. As such, the heterodimer is implicated in the regulation of multiple cellular processes, including nucleosome remodelling, transcription, centrosome formation, and cell cycle progression. Moreover, the heterodimer’s function is crucial for cell survival and regulating DNA damage responses [9]. Under prolonged stress, BARD1 is capable of inducing apoptosis via interactions with TP53 [10].
The role of BARD1 in limiting tumorigenesis has been highlighted in studies of neuroblastoma, where single nucleotide polymorphisms (SNPs), which increase full-length BARD1 mRNA (BARD1-FL) expression, hinder tumour progression [11]. Conversely, SNPs associated with decreased BARD1-FL expression were associated with higher-risk neuroblastoma [12]. Despite the recognition of BARD1’s role in limiting tumorigenesis, oncogenic mutations within BARD1 are infrequent, especially when compared to that of BRCA1 [13]. This observation has spurred much interest into investigating novel splice variants of BARD1. Traditionally, splice variants refer to exon-modifying events at the level of the RNA, while isoforms define the subsequent changes that may be carried through to the final protein structure [14]. In addition to the 11-exon containing BARD1-FL, numerous splice isoforms have been identified thus far, including BARD1-α, -β, -γ, -δ, -ε, -η, and -ω [15,16]. Interestingly, BARD1 splice isoforms, which lack the RING domain, such as BARD1-β and -δ, show effects that are antagonistic to that of BARD1-FL, and have been associated with progression and poor prognosis in multiple cancer types [17,18,19]. Moreover, it has been suggested that increased prevalence of BARD1 isoforms over BARD1-FL promotes tumorigenesis [20].
Currently, investigations into BARD1 splice isoforms in melanoma are not present in the literature. However, due to the recognition of BARD1’s pleiotropic roles in DNA repair processes, in addition to the high mutation burden commonly observed in melanoma, we hypothesised a pro-tumorigenic role for alternate BARD1 isoforms in melanoma [21,22,23,24,25,26].
Herein, we present the first study assessing the expression of BARD1 mRNA splice isoforms in melanoma cell lines and archival formalin-fixed paraffin-embedded (FFPE) tissue samples. The results show that previously identified BARD1 mRNAs encoding isoforms, as well as novel splice isoforms, can be detected using both long-range nanopore sequencing, in addition to short-read and targeted approaches, including RNA-Seq and RT-qPCR in both melanoma cell lines and tissue samples. Due to the nature of the read lengths provided by nanopore sequencing, the technique provides the most comprehensive analyses of splice isoforms. Comparison of the aforementioned RNA sequencing technologies are reviewed elsewhere [27,28,29].
2. Materials and Methods
2.1. NZM and Melanocyte Cell Lines
The 12 New Zealand melanoma (NZM) cell lines used for this study were generated from metastatic melanoma, as described previously [30]. Written informed consent was obtained from all patients under the Auckland Area Health Board Ethics Committee guidelines. The NZM cell lines were maintained in α-MEM media (Gibco, ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% FCS (Moregate, Bulimba, QLD, Australia) and 1% penicillin–streptomycin (Gibco, ThermoFisher Scientific, Waltham, MA, USA). Cell cultures were passaged twice at 80% confluency prior to harvesting for RNA extraction using the appropriate kit (QIAGEN, Germantown, MD, USA).
The melanocyte cell lines used as a non-malignant control were commercial cell lines isolated from human epidermal melanocytes of neonates (HEMnLP) or adult (HEMaLP) tissue. The melanocyte cell line Mel-ST (immortalised using SV40ER and hTERT) was used for nanopore sequencing.
The RNA was extracted using an RNeasy Mini Kit (QIAGEN, Germantown, MD, USA). The quantity and quality of isolated RNA was determined using a NanoPhotometer (Implen, Munich, Germany) and a Qubit Fluorometer (ThermoFisher Scientific, Waltham, MA, USA). For reverse transcription, 20 ng/µL (totalling 200 ng per reaction) of total RNA was used, according to the manufacturer’s instructions (High-Capacity cDNA RT kit, ThermoFisher Scientific). The NZM cell lines had been authenticated by the Auckland Cancer Society Research Centre.
2.2. Archival Formalin-Fixed Paraffin-Embedded (FFPE) Tissue Blocks
From the biobank at the Department of Tumour Pathology, Maria Sklodowska-Curie National Research Institute of Oncology, Krakow, Poland, 18 metastatic melanoma tissue sample blocks were selected. In order to minimise sample heterogeneity, tissue macro-dissection was performed. RNA was extracted using the FormaPure XL Total Kit (Beckman Coulter, Brea, CA, USA). The quantity and quality of isolated RNA was determined using a NanoPhotometer (Implen, Munich, Germany) and a Qubit Fluorometer (ThermoFisher Scientific, Waltham, MA, USA). cDNA synthesis was performed as described above. An overview of the respective techniques utilised for each sample is displayed in Supplementary Table S1.
The study was approved by the bioethics committee—ethical board approval number KB/430-74/20. This was retrospective non-interventional study and all the data analysed were collected as part of a routine clinical practice for diagnosis and treatment. All of the patients signed an informed consent form for treatment as per the standard operating procedures in our hospitals. In addition, the patients were diagnosed and treated following the national guidelines and policies.
2.3. Nanopore and Sanger Sequencing
Long-read sequencing (nanopore) was performed in three melanoma cell lines (NZM3, NZM6, and NZM15) and a non-malignant melanocyte cell line (Mel-ST). The complete BARD1 sequence was amplified, as previously described [31]. Subsequent to gel electrophoresis (on 0.8% agarose gel), the PCR product was purified using Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA). The libraries were prepared using the Direct cDNA Native Barcoding Kit (SQK-DCS109 with EXP-NBD104), and ~22 fmol of each library were pooled. To identify high-confidence BARD1 isoforms, the FLAIR pipeline was used to align nanopore reads using splice aware options to human genome (hg19, www.gencodegene.org) (accessed on 24 December 2020). The --stringent option was used to return high-confidence isoforms that had at least three supporting reads (in at least one cell line) that spanned >80% of a given isoform with 25 bp in the 5′ and 3′ exons [32].
The presence of Δ(E3_E9, ▼(i8), IVS10▼176, and IVS10+131▼46 was validated using Sanger sequencing. All of the primers used for amplification and the subsequent Sanger sequencing are displayed in Table S2.
2.4. RT-qPCR Data
RT-qPCR was performed on 12 New Zealand melanoma cell lines and 18 archival melanoma FFPE tissues samples. To amplify the six previously reported BARD1 isoforms, named β, δ, γ, ε, η, and φ, as well as four novel splice events (Δ(E3_E9), ▼(i8), IVS10▼176, and IVS10+131▼46) we used primers targeting specific exon–exon junctions (β, δ, γ, ε, η, φ, and Δ(E3_E9)) or, alternatively, exon–intron junctions ▼(i8), IVS10▼176 and IVS10+131▼46). The primer sequences used for RT-qPCR analysis are listed in Table S2. Real-time quantitative PCR was then performed on a LightCycler 480 instrument (Roche) using TB green Ex Taq master mix (TaKaRa, San Jose, CA, USA) and 5 nM of the appropriate primer pairs. All samples were performed in triplicate with amplification over 50 cycles, which were analysed using qbase+ software (version 2.7.11; http://www.biogazelle.com/qbaseplus (accessed on 24 December 2020); Biogazelle, Ghent, Belgium). To calculate the ΔCt values, the raw Ct values were normalised using TATA-binding protein (TBP), ubiquitin C (UBC), and phosphoglycerate kinase 1 (PGK1) as reference genes.
2.5. RNA-seq Data
RNA-Seq data generated from a sample of human melanocytes, two melanocyte cell lines (HEMnLP, HEMaLP), and three melanoma cell lines were used for the RNA-Seq analysis. The melanoma cell line data were generated from the same three cell lines as those used to generate the nanopore data (NZM 3, 6, and 15); however, the melanocyte data were from different cell lines. Reads were adaptor trimmed using cleanadaptors and aligned to the human reference genome GRCh37 using the alignment tool STAR [33]. Assembly was performed with the GENCODE v29 gene annotations and Stringtie [34]. The transcripts per kilobase million (TPM) expression counts for the annotated BARD1 isoforms were obtained from the Stringtie gtf file. For the detection of novel isoforms, the BAM files generated by STAR were visualised in IGV to identify whether junction reads were detected to support novel splicing events and retained introns; however, this analysis was not quantitative.
More information on the screening protocols can be obtained from the corresponding author upon request.
2.6. Sequencing Data Availability
Nanopore sequencing data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher upon request.
3. Results
3.1. Long-Range Nanopore Sequencing
Herein, we present, for the first time, the expression of alternative BARD1 splice isoforms in various melanoma samples. Using long-range PCR and nanopore sequencing, we identified a set of mRNAs corresponding to 76 different BARD1 splice variants in total (Table S3), with six variant transcripts being expressed predominately (Figure 1A). Within these six, we identified a deletion encompassing exons 3 to 9 of BARD1 (Δ(E3_E9); r.216_1903del), resulting in a frame shift, creating a premature stop codon (p.Glu646Ter).
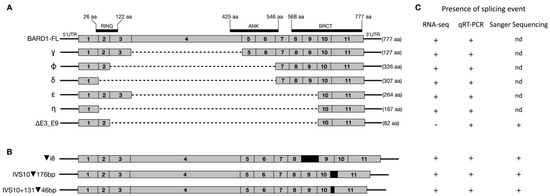
Figure 1.
Schematic diagram of BARD1 and identified isoforms (selected). (A) Structure of full-length BARD1 (BARD1-FL) and six abundantly expressed alternative isoforms (γ, φ, δ, ε, η, and Δ(E3_E9)). (B) Structure of identified splicing events. (C) Presence of identified isoforms as detected using various sequencing techniques. (nd)- not done.
Moreover, nanopore sequencing analyses identified three different splicing events: (1) retention of intron 8 (▼(i8); r.1810_1811ins562), (2) inclusion of 175 bp from intron 10 (IVS10▼176; r.2001_2002ins2001+1_2001_176), and (3) activation of intronic exon within intron 10 (IVS10+131▼46; r.2001_2002ins2001+131_2001+176) (Figure 1B). Interestingly, all three splicing events were identified in several different transcripts, co-occurring with various other splicing arrangements, e.g., (▼(i8) was observed in four independent transcripts: row numbers 40, 44, 58, and 59. Similarly, IVS10▼176 and IVS10+131▼46 were present in 13 and six transcripts, respectively (Table S3). The presence of all of the above isoforms was confirmed with Sanger sequencing, which revealed characteristics of exon–exon or exon–intron junctions (Figure 1C and Figure S1).
Among the identified transcripts, semi-quantitative analysis of the mean expression levels was highest for BARD1-ε and BARD1-η at 21.8% (range 20.6–22.6%) and 21.3% (range 19.9–23.5%), respectively (Figure 2). In non-malignant melanocytes, ε and η accounted for 16.9% and 28.9% of the expressed transcripts, respectively. BARD1-δ was more abundantly expressed in melanocytes, at 20.7%, than in NZM melanoma cell lines (mean expression 16.7%). BARD1-φ had a decreased representation within the melanocyte cell line (9% of total transcripts expressed), compared to the mean expression level of 18.3% (range 13.7–21.2%) in the NZM melanoma cell lines. Surprisingly, considering that amplification of full-length transcripts was efficiently achieved, the BARD1-FL was not the predominantly expressed transcript, contributing only 5.3% of total transcripts in melanocytes. Although, a possible PCR bias cannot be excluded via preferential amplification of shorter PCR products. Similarly, the mean expression was 4.7% (range 1–8.3%) in the NZM cell lines. The novel variant, Δ(E3_E9), was amongst the highest expressed transcripts present in both the melanoma and melanocyte cell lines with a frequency of 3.6% (range 2–4.5%) and 3.7%, respectively.
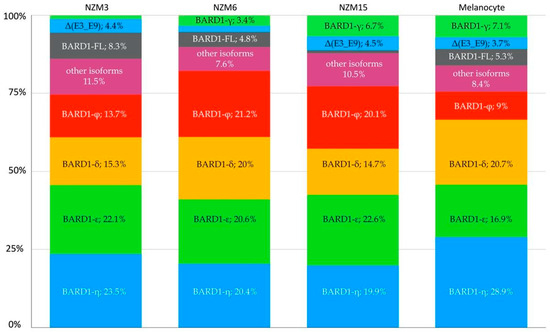
Figure 2.
Proportion of BARD1 transcript isoforms quantified using nanopore sequencing. The first three bars correspond to the New Zealand melanoma (NZM) cell lines, and the last bar corresponds to the non-malignant (melanocyte) control.
3.2. RT-qPCR
Next, by using RT-qPCR, we successfully validated the nanopore sequencing results and quantified all analysed transcripts, both in the cell lines and the archival (FFPE) melanoma samples (Figure S2. BARD1-ɣ was the highest expressed alternative isoform in the FFPE samples (mean normalised value 2.15), followed by BARD1-η, BARD1-π, and BARD1-β, accounting for, respectively, 1.72, 1.58, and 1.53. BARD1-δ was the lowest expressed transcript in the archival material (mean normalised value 0.27). In the NZM cell lines, BARD1-β, BARD1-ɣ, and BARD1-η were similarly expressed, with a mean normalised value of 1.41 to 1.43. In contrast to the nanopore sequencing results, BARD1-δ had the lowest expression level (0.135). In the non-malignant melanocyte control, the highest expression was observed for BARD1-η (3.00) and the lowest for BARD1-δ (0.005). Among the novel isoforms, Δ(E3_E9) was the highest expressed in the analysed FFPE samples (mean normalised value 0.597, range 0.05 to 0.75) (Figure S3).
3.3. RNA-Seq
Lastly, to assess whether RNA-Seq was consistent with the other methods used, we analysed RNA-Seq datasets that were generated from the same melanoma cell lines used for nanopore sequencing and RT-qPCR. To enable quantification of the annotated BARD1 isoform transcripts, the data were aligned in STAR and then assembly was performed using Stringtie.
Interestingly, we observed that the proportion of each isoform transcript detected by Stringtie was not consistent with the nanopore sequencing data. The full-length BARD1 isoform transcript was the most abundantly expressed, as reported by the software in all of the analysed RNA-Seq datasets (Table S4). As the RNA-Seq analysis only enabled detection of the annotated isoform mRNAs, we also visualised the BAM files in IGV to establish whether there were any reads to support novel splice variants. No junction reads were detected in any of the datasets to support the Δ(E3_E9) isoform. We did identify reads in the RNA-Seq data that supported the retention of intron 8 and 10; however, we were unable to assign these to a specific transcript.
4. Discussion
We carried out the first detailed analysis of alternative BARD1 transcription in melanoma. To do this, we used a combination of nanopore sequencing, RNA sequencing, and RT-qPCR analyses on RNA from three melanoma cell lines and a melanocyte cell line. Nanopore sequencing enabled end-to-end sequencing across the entire length of PCR-amplified BARD1 transcripts. Conversely, as RNA-Seq covers relatively short RNA fragments, only limited information can be derived about transcript structure. Moreover, using the nanopore sequencing of BARD1 mRNA transcripts in melanoma, we identified the full-length BARD1 isoform, along with five other major alternatively spliced transcripts. We additionally sequenced four novel BARD1 splicing events in the melanoma cell lines, several of which we are reporting for the first time.
Alternative BARD1 transcripts contribute to the genesis of multiple BARD1 isoforms, including α, β, κ, γ, φ, δ, ε, η, and ω. While multiple isoforms are expressed across various cancers, the relative expression levels of specific isoforms have been observed to vary [16]. Among these isoforms, the BARD1-β and BARD1-δ isoforms have attracted significant attention in cancer genomics due to their tumorigenic characteristics and ability to confer resistance to apoptosis [18]. In melanoma cell lines, however, nanopore sequencing identified the expression level of BARD1-β comprised <1% of total BARD1 transcripts (Table S3). In contrast, the expression levels of the BARD1-δ transcript comprised between 14.7% and 20% of the BARD1 transcripts observed. The upregulation of the BARD1-δ isoform expression in cancer has been associated with mitochondrial regulation in response to stress, but not with apoptosis stimulation or altering cell membrane permeability [35].
The other major isoform transcripts detected in melanoma using nanopore sequencing and RNA-Seq included BARD1-γ, BARD1-φ, BARD1-ε, and BARD1-η. However, the expression levels of the BARD1-FL transcripts were very low in all NZM melanoma cell lines, as measured by nanopore sequencing. Interestingly, the proportion of BARD1-FL expressed in our analyses was not concordant with the earlier study [31], despite using the same primer set, although we cannot explain this at present. Structural analyses of the BARD1 isoforms δ, ε, and η predict an absence of the ANK-BRCT binding domain, or part of the BRCT region. These motifs are crucial sites for binding with several tumour suppressor proteins, including TP53 [36,37,38]. BARD1–TP53 complex formation is fundamental for stimulating nuclear export and mitochondrial localisation, and is subsequently required to induce apoptosis [39]. The induction of TP53-mediated apoptosis via BARD1 occurs irrespective of the serine-15 phosphorylation status of TP53 [36,40]. Moreover, BARD1-δ antagonises BARD1-FL. As such, increased levels of BARD1-δ expression relative to BARD1-FL may regulate or inhibit tumour suppressor functions in melanoma cell lines.
Reduced expression of BARD1 isoforms containing the functional RING domain may result in decreased BRCA1–BARD1 interactions [41]. Dimerisation of BRCA1 and BARD1 is required to stimulate their shared ubiquitination activity and is required for the heterodimer tumour suppressor functions [42]. Taken together, the relatively high expression of BARD1-δ, BARD1-ε, and BARD1-η, compared to BARD1-FL in melanoma cells, suggests a role for these transcripts in uncontrolled melanoma cell proliferation and mitochondrial regulation in response to stress. However, despite support for a pro-tumour role of these isoforms in the literature, significant differences were not observed between our melanocyte and melanoma data; thus, further investigations are warranted.
Nanopore and Sanger sequencing has identified two other BARD1 splice variants of interest in melanoma: BARD1 Δ(E3_E9) and IVS10+131▼46 (Figure 1). These splicing events have previously been identified in lymphoblastoid cell lines [31]. Splice variant Δ(E3_E9) was also detected by RT-qPCR in the FFPE melanoma samples, but was unable to be detected in the RNA-Seq data. There are inherent limitations to the use of RNA-Seq for the identification of novel splice variants. This is primarily due to the reliance of assembly tools on annotations for accurate transcript assembly. As the novel Δ(E3_E9) splice variant is not within the Gencode annotations, Stringtie was unable to detect this isoform. We attempted to identify junction reads between exons 3 and 9 using IGV. However, none were detected using this approach and thus we were unable to confirm the presence of this splicing event within the RNA-Seq data. This emphasises a significant limitation of RNA-Seq data in the detection of splice variants. The relatively short reads generated by RNA-Seq experiments make assignment to specific transcripts difficult, particularly when dealing with a gene that shows complex splicing, such as BARD1. Additionally, this may have led to an overestimation of the full-length isoform expression in the RNA-Seq data when compared to nanopore sequencing (Tables S3 and S4). Stringtie takes reads that have aligned to BARD1 and then performs transcript assembly, generating a count for each annotated isoform. Therefore, reads derived from the novel splice isoforms detected by nanopore sequencing may have been assigned to the full-length BARD1 by Stringtie, due to the lack of annotations for these transcripts. Similarly, this may be used to explain the non-concordance of the results observed for other well documented isoforms, e.g., Gamma (Δ(E4)), as this splice event is present in multiple independent transcripts. Unfortunately, we were unable to validate this possible occurrence in the RNA-Seq data via RT-qPCR, as there are no unique sequences within the BARD1-FL isoform such that it could be confidently differentiated from other annotated isoforms.
The role of these isoforms in human biology is largely unknown. In previous studies, the roles of Δ(E3_E6) and Δ(E3_E7) in cell proliferation have been described, with the ΔE3 isoform using an alternative open reading frame to encode functional proteins [43]. In contrast to Walker et al., [31], the IVS10+131▼46 was also present in several different transcripts (co-occurring with other splicing alterations). This might suggest its potential importance in melanoma biology. Similarly, the novel splicing events, named ▼(i8) and IVS10▼176 (which comprise less than 1% of BARD1 transcripts), were also noted in several different transcripts. A frame shift in the BARD1 open reading frame (ORF) resulted both from insertion of intron 8 sequences for the ▼(i8) splice event and the inclusion of 175 bp from intron 10 in the IVS10▼176 splice event. Such frame shift indels could lead to mRNA degradation, owing to nonsense-mediated mRNA decay or the production of truncated proteins [44]. Further work is required to investigate the functional significance of these variants.
5. Conclusions
In conclusion, we identified multiple alternative transcripts corresponding to known BARD1 isoforms, expressed at much higher levels than BARD1-FL in both melanoma and melanocytes. While these results observed in melanoma could be expected from the literature, the same observation in the control melanocytes was unexpected. The identified variants encode proteins lacking the domains required for BRCA1 interaction and tumour suppressor functions. These results therefore do not support a pro-tumorigenic role of BARD1 isoforms in melanoma, although further investigation will be required. Important limitations of the current study include the degree of variation observed between the different sequencing methods. The apparent lack of expression of BARD1 isoforms using RNA-Seq may be attributed to short reads being falsely assigned to BARD1-FL. Nevertheless, using nanopore sequencing technology, we unexpectedly found that what are usually rare and potentially antagonistic isoforms of BARD1 transcription are in fact expressed at relatively high levels in melanoma and melanocytes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/2/320/s1, Figure S1: Validation of Nanopore results by Sanger sequencing, Figure S2: Relative expression of predominant BARD1 isoforms in melanoma tissues, melanoma cell lines and melanocytes, Figure S3: Relative expression of novel BARD1 splice events in melanoma tissues, melanoma cell lines and melanocytes, Table S1: An evaluation of samples included in each technique, Table S2: Primer sequences used in the study, Table S3: BARD1 transcript variants identified using long-range Nanopore sequencing, Table S4: Expression of annotated BARD1 transcripts from RNA-Seq data.
Author Contributions
Conceptualisation, M.R. and M.R.E.; methodology, G.R. and L.C.W.; formal analysis, L.I.M., M.R., G.A.R.W., and C.F.L.-S.; investigation, L.I.M. and M.R.; resources, J.M., A.H.-L., B.C.-S., and J.R.; data curation, M.R., J.M., and G.A.R.W.; writing—original draft preparation, R.M.P., L.I.M. and S.M.H.; writing—review and editing, M.R., G.A.R.W., C.F.L.-S., E.C.M., G.R., L.C.W., S.M.H., and M.R.E.; funding acquisition, M.R.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Health Research Council of New Zealand and the New Zealand Institute for Cancer Research Trust.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committee of Maria Sklodowska-Curie National Research Institute of Oncology, Cracow, Poland (number KB/430-74/20).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data presented in this study, including nanopore sequencing data, is available upon request to the corresponding author.
Acknowledgments
The authors acknowledge Euan Rodger and Suzan Almomani for technical support and helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jenkins, R.W.; Fisher, D.E. Treatment of Advanced Melanoma in 2020 and Beyond. J. Investig. Dermatol. 2020, 1–9. [Google Scholar] [CrossRef]
- Bolick, N.L.; Geller, A.C. Epidemiology of Melanoma. Hematol. Oncol. Clin. N. Am. 2021, 35, 57–72. [Google Scholar] [CrossRef]
- Godar, D.E. Worldwide Increasing Incidences of Cutaneous Malignant Melanoma. J. Skin Cancer 2011, 2011, 858425. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.C.W.; Hwang, L.Y.; Bowcock, A.M.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-derived Mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef]
- Chen, A.; Kleiman, F.E.; Manley, J.L.; Ouchi, T. Autoubiquitination of the BRCA1·BARD1 RING ubiquitin ligase. J. Biol. Chem. 2002, 277, 22085–22092. [Google Scholar] [CrossRef]
- Starita, L.M.; Machida, Y.; Sankaran, S.; Elias, J.E.; Griffin, K.; Schlegel, B.P.; Gygi, S.P.; Parvin, J.D. BRCA1-Dependent Ubiquitination of γ-Tubulin Regulates Centrosome Number. Mol. Cell. Biol. 2004, 24, 8457–8466. [Google Scholar] [CrossRef]
- Starita, L.M.; Horwitz, A.A.; Keogh, M.C.; Ishioka, C.; Parvin, J.D.; Chiba, N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J. Biol. Chem. 2005, 280, 24498–24505. [Google Scholar] [CrossRef]
- Morris, J.R.; Solomon, E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 2004, 13, 807–817. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.; Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 2001, 8, 1255–1266. [Google Scholar] [CrossRef]
- Cimmino, F.; Avitabile, M.; Diskin, S.J.; Vaksman, Z.; Pignataro, P.; Formicola, D.; Cardinale, A.; Testori, A.; Koster, J.; de Torres, C.; et al. Fine mapping of 2q35 high-risk neuroblastoma locus reveals independent functional risk variants and suggests full-length BARD1 as tumor-suppressor. Int. J. Cancer 2018, 143, 2828–2837. [Google Scholar] [CrossRef]
- Capasso, M.; Diskin, S.J.; Totaro, F.; Longo, L.; de Mariano, M.; Russo, R.; Cimmino, F.; Hakonarson, H.; Tonini, G.P.; Devoto, M.; et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis 2013, 34, 605–611. [Google Scholar] [CrossRef]
- Chen, J.; Weiss, W.A. Alternative splicing in cancer: Implications for biology and therapy. Oncogene 2015, 34, 1–14. [Google Scholar] [CrossRef]
- Li, H.D.; Menon, R.; Omenn, G.S.; Guan, Y. The emerging era of genomic data integration for analyzing splice isoform function. Trends Genet. 2014, 30, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Watters, A.K.; Seltzer, E.S.; Mackenzie, D.J.; Young, M.; Muratori, J.; Hussein, R.; Sodoma, A.M.; To, J.; Singh, M. The Effects of Genetic and Epigenetic Alterations of Non-Gynecological Cancers. Genes (Basel) 2020, 11, 829. [Google Scholar] [CrossRef] [PubMed]
- Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016, 72, 1–17. [Google Scholar] [CrossRef]
- Li, L.; Cohen, M.; Wu, J.Y.; Sow, M.H.; Nikolic, B.; Bischof, P.; Irminger-Finger, I. Identification of BARD1 splice-isoforms involved in human trophoblast invasion. Int. J. Biochem. Cell Biol. 2007, 39, 1659–1672. [Google Scholar] [CrossRef]
- Bosse, K.R.; Diskin, S.J.; Cole, K.A.; Wood, A.C.; Schnepp, R.W.; Norris, G.; Nguyen, L.B.; Jagannathan, J.; Laquaglia, M.; Winter, C.; et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012, 72, 2068–2078. [Google Scholar] [CrossRef]
- Ozden, O.; Bishehsari, F.; Bauer, J.; Park, S.H.; Jana, A.; Baik, S.H.; Sporn, J.C.; Staudacher, J.J.; Yazici, C.; Krett, N.; et al. Expression of an Oncogenic BARD1 Splice Variant Impairs Homologous Recombination and Predicts Response to PARP-1 Inhibitor Therapy in Colon Cancer. Sci. Rep. 2016, 6, 26273. [Google Scholar] [CrossRef]
- Cimmino, F.; Formicola, D.; Capasso, M. Dualistic role of BARD1 in cancer. Genes (Basel) 2017, 8, 375. [Google Scholar] [CrossRef]
- Baer, R.; Ludwig, T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 2002, 12, 86–91. [Google Scholar] [CrossRef]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016, 23, 647–655. [Google Scholar] [CrossRef]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Laufer, M.; Nandula, S.V.; Modi, A.P.; Wang, S.; Jasin, M.; Murty, V.V.V.S.; Ludwig, T.; Baer, R. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 2007, 282, 34325–34333. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed]
- Kabbarah, O.; Chin, L. Revealing the genomic heterogeneity of melanoma. Cancer Cell 2005, 8, 439–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Kim, J.E.; Stones, C.; Joseph, W.R.; Leung, E.; Finlay, G.J.; Shelling, A.N.; Phillips, W.A.; Shepherd, P.R.; Baguley, B.C. Comparison of growth factor signalling pathway utilisation in cultured normal melanocytes and melanoma cell lines. BMC Cancer 2012, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; Lattimore, V.L.; Kvist, A.; Kleiblova, P.; Zemankova, P.; de Jong, L.; Wiggins, G.A.R.; Hakkaart, C.; Cree, S.L.; Behar, R.; et al. Comprehensive Assessment of BARD1 Messenger Ribonucleic Acid Splicing With Implications for Variant Classification. Front. Genet. 2019, 10, 1139. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Tembe, V.; Henderson, B.R. BARD1 translocation to mitochondria correlates with bax oligomerization, loss of mitochondrial membrane potential, and apoptosis. J. Biol. Chem. 2007, 282, 20513–20522. [Google Scholar] [CrossRef]
- Feki, A.; Jefford, C.E.; Berardi, P.; Wu, J.Y.; Cartier, L.; Krause, K.H.; Irminger-Finger, I. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene 2005, 24, 3726–3736. [Google Scholar] [CrossRef]
- Kleiman, F.E.; Manley, J.L. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science (80-. ) 1999, 285, 1576–1579. [Google Scholar] [CrossRef]
- Kleiman, F.E.; Manley, J.L. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 2001, 104, 743–753. [Google Scholar] [CrossRef]
- Tembe, V.; Martino-Echarri, E.; Marzec, K.A.; Mok, M.T.S.; Brodie, K.M.; Mills, K.; Lei, Y.; DeFazio, A.; Rizos, H.; Kettle, E.; et al. The BARD1 BRCT domain contributes to p53 binding, cytoplasmic and mitochondrial localization, and apoptotic function. Cell. Signal. 2015, 27, 1763–1771. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Soriano, J.V.; Vaudan, G.; Montesano, R.; Sappino, A.P. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 1998, 143, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.; Klevit, R.E. Structure of a BRCA1 – BARD1 heterodimeric RING – RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.A.; Schüchner, S.; Au, W.W.Y.; Fabbro, M.; Henderson, B.R. Nuclear-cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene 2004, 23, 1809–1820. [Google Scholar] [CrossRef]
- Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007, 67, 11876–11885. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, S.; Stone, O.A.; Lessard, S.; Lavertu, A.; Desjardins, J.; Beaudoin, M.; Rivas, M.; Stainier, D.Y.R.; Lettre, G. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS ONE 2017, 12, e0178700. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).