Abstract
The etiology and reasons underlying the ethnic disparities in systemic sclerosis (SSc) remain unknown. African Americans are disproportionally affected by SSc and yet are underrepresented in research. The aim of this study was to comprehensively investigate the association of DNA methylation levels with SSc in dermal fibroblasts from patients of African ancestry. Reduced representation bisulfite sequencing (RRBS) was performed on primary dermal fibroblasts from 15 SSc patients and 15 controls of African ancestry, and over 3.8 million CpG sites were tested for differential methylation patterns between cases and controls. The dermal fibroblasts from African American patients exhibited widespread reduced DNA methylation. Differentially methylated CpG sites were most enriched in introns and intergenic regions while depleted in 5′ UTR, promoters, and CpG islands. Seventeen genes and eleven promoters showed significant differential methylation, mostly in non-coding RNA genes and pseudogenes. Gene set enrichment analysis (GSEA) and gene ontology (GO) analyses revealed an enrichment of pathways related to interferon signaling and mesenchymal differentiation. The hypomethylation of DLX5 and TMEM140 was accompanied by these genes’ overexpression in patients but underexpression for lncRNA MGC12916. These data show that differential methylation occurs in dermal fibroblasts from African American patients with SSc and identifies novel coding and non-coding genes.
1. Introduction
Systemic sclerosis (SSc or scleroderma) is a rare, multisystem, connective tissue disease characterized by cutaneous and visceral fibrosis, immune dysregulation, and vasculopathy. Patients are commonly classified into two main subsets, limited cutaneous SSc (lcSSc) and diffuse cutaneous SSc (dcSSc), with dcSSc having a worse prognosis [1]. Relative to individuals of European ancestry, individuals of African ancestry are more likely to develop SSc, to be diagnosed with dcSSc, and to experience higher disease severity, greater morbidity, reduced survival, and earlier death [2,3,4,5,6,7,8]. This higher disease burden in African Americans is not fully explained by differences in socioeconomic status or access to health care [8,9].
The etiology of SSc and the factors underlying its ethnic disparities remain elusive. Genetic and epigenetic studies conducted mostly in individuals of European ancestry uncovered multiple loci associated with SS [10]. A role of DNA methylation in SSc is supported by a X chromosome gene methylation analysis of peripheral blood mononuclear cells [11]; the quantification of global methylation in whole blood [12]; as well as genome-wide DNA methylation analyses of dermal fibroblasts [13], whole blood [14], and CD4+ T cells [15,16]. Different ancestral populations exhibit DNA methylation differences [17,18,19,20,21,22,23,24] that are partially explained by their distinct genetic ancestry, thus environmental factors not captured by genetic ancestry are significant contributors to the variation in methylation [19].
In order to understand the pathogenesis of SSc in patients of African ancestry, we assessed the DNA methylation profiles of dermal fibroblasts from African American patients and controls by RRBS, which has a high sensitivity and specificity to detect changes in DNA methylation in genes, promoters, CpG islands, and repetitive regions [25,26]. We then integrated the data with the gene expression of the top differentially methylated genes from the same subjects. This study is the first to unveil the genome-wide patterns of differential methylation in skin fibroblasts from African American patients with SSc.
2. Materials and Methods
2.1. Subjects
A total of 15 SSc cases and 15 healthy controls were recruited for this study. All the participants were self-reported African American and the patients met the 2013 ACR/EULAR classification criteria for SSc. The cases and controls were age-balanced within 5 years. This study was approved by the Institutional Review Board at the Medical University of South Carolina on 9 April 2014 (application number Pro# 33636). Informed consent was obtained from all the participants. All the research included in this manuscript conforms with the Declaration of Helsinki.
2.2. Primary Dermal Fibroblast Isolation and Culture
Primary dermal fibroblasts were isolated from 3 mm skin biopsies obtained from the involved forearm skin and cultured as described [27]. Cells were cultured for 3 passages, then DNA and RNA were isolated using the DNeasy and RNeasy kits (Qiagen, Germantown, MD, USA) following the manufacturer’s protocols.
2.3. Reduced Representation Bisulfite Sequencing (RRBS)
RRBS was performed using the Ovation® RRBS Methyl-Seq System 1–16 (NuGEN Technologies, Inc., San Carlos, CA, USA), following the manufacturer’s procedure. We generated DNA methylation data for over 5 million CpGs in each sample and between 10× to 40× coverage in CpG sites.
2.4. Genome-Wide DNA Methylation Data Analysis
Alignment and methylation calling were performed using Bismarck v0.16.3 and the GRCh37/hg19 reference genome [28]. Data were filtered, normalized, and analyzed with RnBeads v1.6.1 [29]. A differential methylation analysis was conducted at the CpG, promoter, and gene levels (including RNA, pseudo-, and protein-coding genes) [29]. Genes and promoters were defined by Ensembl (ensembl.org), and CpG islands were defined as the CpG island track of the UCSC Genome Browser (genome.ucsc.edu). As implemented in RnBeads, CpG site p-values were computed using the linear models in the limma package. For gene regions, promoter regions, and CpG islands, the mean of the mean methylation levels for cases and controls across all sites in a region was computed, as well as the following three quantities: the mean difference in means (MDM) across all sites in a region, the mean of quotients in mean methylation across all sites in a region, and a combined p-value calculated from all site p-values in the region [29]. Each gene, promoter, and CpG island was assigned a rank based on these three criteria. A combined rank is computed as the maximum (i.e., worst) value among the three ranks. The smaller the combined rank for a region, the more evidence for differential methylation it exhibits. All genes, promoters, and CpG islands were ranked based on this Combined Score approach implemented in RnBeads [29]. Of note, while a negative MDM represents hypomethylation in cases relative to controls, a positive MDM denotes hypermethylation in cases relative to controls.
2.5. Genomic Annotation Enrichment Analysis
To annotate the position of each CpG to the corresponding genomic location, we used the annotatePeaks.pl program of HOMER v4.9.1 with annotation from the hg19 human genome assembly [30]. CpGs were annotated to promoter, transcription termination site (TTS), exon, intron, 5′ UTR exon, 3′ UTR exon, intergenic, CpG island, repeat elements, and other detailed annotations using default region definitions. HOMER uses annotations based on the UCSC Genome Browser (genome.ucsc.edu). To investigate the distribution of differentially methylated CpGs (DMC) in different genomic locations, all CpGs that met an FDR-adjusted p-value < 0.4 were used to compare their localization in different genomic locations as provided by HOMER’s annotations [30]. Odds ratios (ORs), 95% confidence intervals (CI), and p-values were computed against the general distribution of the 3,870,251 CpGs of our dataset using GraphPad Prism v9.
2.6. Gene Set Enrichment Analysis (GSEA)
A gene set enrichment analysis GSEA was performed to determine whether a priori defined sets of genes (e.g., pathways) are significantly enriched in the list of genes ranked by their correlation with the disease. The full ranked lists of genes and promoters generated by RnBeads’ Combined Score approach [29] were used as inputs to GSEA Desktop v3.0 [31,32]. The genes were ranked by their differential methylation between cases and controls (hyper- and hypomethylated), and the Reactome Pathway Knowledgebase (reactome.org) [33] was used as the gene set. An enrichment score statistic represents the enrichment of Reactome pathways in genes that are hyper- or hypomethylated in patients, and the significance of the pathway enrichment score is estimated by an empirical phenotype-based permutation test procedure [31,32]. The threshold for statistical significance was defined as FDR ≤ 0.25, as recommended [31,32].
2.7. Gene Ontology (GO) Enrichment Analysis
Enrichment analysis for GO terms associated with the top-ranking differentially methylated genes and promoters was performed using RnBeads v1.6.1 [29]. A GO enrichment analysis of biological process (BP) was conducted separately on each of the 100 hypo- and hypermethylated genes and promoters. The enrichment of GO BP terms associated with the top ranking genes and promoters was determined by a hypergeometric test implemented in RnBeads [29].
2.8. Gene Expression Analysis
Using the available cultured fibroblast samples from the same 15 SSc cases and 14 healthy controls, cDNA was prepared using the Superscript IV First Strand synthesis system (ThermoFisher, Waltham, MA, USA) from 1 μg of isolated RNA. qPCR was performed using the Taqman Real-Time PCR master mix (Applied Biosystems, Foster City, CA, USA). All the samples were run in duplicate using an Applied Biosystems Real-Time PCR System and analyzed using the StepOne Plus Applied Biosystems software. The gene quantification cycle values were normalized to β 2 microglobulin (B2M) expression using the ΔΔCT method to obtain relative cell equivalents. All the primers were purchased from ThermoFisher Scientific (Waltham, MA, USA): CDA (Hs00156401_m1); TMEM140 (Hs00251020_m1); ACKR4 (Hs00664347_s1); DLX5 (Hs01573641_m1); FAM180B (Hs03988397_m1); MGC12916 (Hs04419380_s1); LOC102724927 (Hs04395955_s1); LOC101929882 (Hs04938653_m1); B2M (Hs00187842_m1). Statistical significance was determined using the Mann–Whitney test and defined as p-values ≤ 0.05.
3. Results
3.1. Subject Characteristics
The clinical and demographic characteristics of the volunteer African ancestry SSc patients and healthy controls are summarized in Table 1. Most patients were female with relatively early disease (mean duration of 5 years). Most presented with dcSSc, having more extensive skin disease involving the proximal limbs and trunk and no concomitant rheumatic disease. One patient had the rarer SSc sine scleroderma (ssSSc), which is the total or partial absence of cutaneous manifestations but the presence of internal organ involvement and/or serologic findings consistent with SSc.

Table 1.
Demographic and clinical characteristics of the study participants.
3.2. Differentially Methylated Sites and Genes
Over 3.8 million CpG sites were tested for differential methylation between SSc cases and controls (Supplementary Figure S1). A total of 1180 differentially methylated CpGs (DMCs), which corresponds to 0.03% of all cytosines tested, meet an FDR-adjusted p-value < 0.4. The rationale for the FDR setting was guided by the desire to perform a system-level analysis and include as many CpGs sites as possible, as well as our previous studies demonstrating that this threshold permits a sensitive analysis at a system level of genes that are relevant to the underlying biology of the trait [34,35]. Patients exhibited widespread hypomethylation throughout the genome, with over 85% of CpGs that met an FDR-adjusted p-value < 0.4 showing decreased methylation in the cultured skin fibroblasts from the patients compared to the controls. We first sought to investigate any potential enrichment (or conversely, underrepresentation) of DMCs in defined genomic regions. Among the 1180 DMCs that met an adjusted p-value < 0.4, there was an overrepresentation of DMCs in introns (OR = 1.7, p < 0.0001), intergenic regions (OR = 1.5, p < 0.0001), transcription termination sites (TTS) (OR = 1.5, p = 0.007), and short interspersed nuclear elements (SINE) (OR = 1.2, p = 0.003) (Figure 1 and Supplementary Table S1). Notably, there was a depletion of DMCs in 5′ UTR (OR = 0.2, p < 0.0001), promoters (OR = 0.3, p < 0.0001), and CpG islands (OR = 0.6, p < 0.0001) (Figure 1 and Supplementary Table S1). In most of the genomic regions, the majority of DMCs were hypomethylated in the patients compared to controls. In contrast with other genomic regions, in CpG islands 71% of the DMCs were more methylated in patients than the controls.
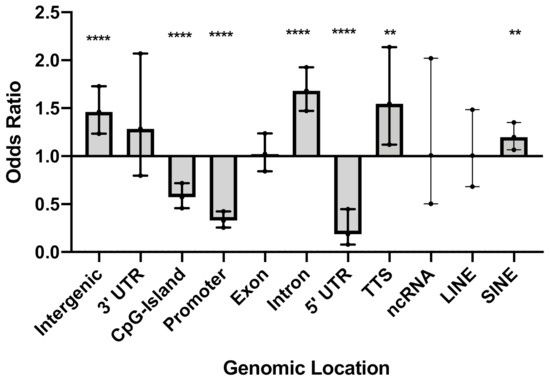
Figure 1.
Genomic location of differentially methylated CpGs (DMC) that met an adjusted p-value < 0.4. Odds ratio (OR), 95% confidence intervals (CI), and p-values were computed against the general distribution of the 3,870,251 CpGs of our dataset using GraphPad Prism v9. Error bars represent the 95% CI. OR indicates the enrichment or depletion of DMCs in each region. Transcription termination site (TTS); non-coding RNA (ncRNA); long interspersed nuclear elements (LINE); short interspersed nuclear elements (SINE). ** p ≤ 0.01, **** p ≤ 0.0001.
The Combined Score approach implemented in RnBeads v1.6.1 [29] was used to independently identify differentially methylated genes, promoters, and CpG islands. Of note, RnBeads uses the definitions of genes and promoters from Ensembl and CpG islands from the UCSC Genome Browser. A total of 197 (out of 30,771) genes, 112 (out of 29,720) promoters, and 97 (out of 24,117) CpG islands were identified and ranked using this approach. The gene and promoter regions identified are shown in Table 2. A total of 9 CpG islands, 17 genes (including RNA, pseudo- and protein-coding genes), and 11 promoters showed significant differential methylation levels between cases and controls at the gene level. The top differentially methylated genes constitute mostly non-coding RNA genes (42%), followed by pseudogenes (27%) and then protein-coding genes (19%) (Table 2). Among the protein-coding genes, cytidine deaminase (CDA), a marker of monocyte/macrophage differentiation [36], is involved in innate immunity pathways. Atypical chemokine receptor 4 (ACKR4) is involved in chemokine signaling [37]. Distal-less homeobox 5 (DLX5) is a transcription factor involved in bone development and the morphogenesis of connective tissue [38]. The functions of the remaining genes are currently unknown.

Table 2.
Gene and promoter regions below the Combined Score cutoff.
3.3. Gene Set Enrichment Analysis (GSEA)
To gain insight into the most differentially methylated genes and promoters, GSEA [31,32] was conducted to predict biologically relevant Reactome pathways [33]. Table 3 lists all Reactome pathways with an FDR ≤ 0.25, as recommended by GSEA [32]. This analysis highlighted an immune pathway (immunoregulatory interactions between a lymphoid and a non-lymphoid cell) to be overrepresented in the set of hypermethylated genes, while metabolism pathways (glucuronidation, chondroitin sulfate dermatan sulfate metabolism) showed enrichment among hypomethylated genes. Pathways involved in cell development (the regulation of β cell development and gene expression) and cell signaling (gap junction trafficking, the activation of kainate receptors upon glutamate binding, G β:γ signaling), were also enriched among hypomethylated genes.

Table 3.
Summary of gene set enrichment analysis results.
3.4. Gene Ontology (GO) Enrichment Analysis
To further aid in the interpretation of the differentially methylated genes and promoters, we performed an enrichment analysis for GO terms associated with the top-ranking genes and promoters in Table 4. Multiple development and morphogenesis, immune, and metabolic-related terms show enrichment. Meanwhile, hypomethylated genes are enriched for GO terms associated with interferon (IFN) signaling (type I IFN signaling pathway, p = 8.0 × 10−4; response to type I IFN, p = 8.0 × 10−4) and hypermethylated genes are enriched for GO terms associated with mesenchyme and epithelial development and cell differentiation (epithelial to mesenchymal transition, p = 1.0 × 10−4; nephron tubule formation, p = 1.0 × 10−4; mesenchymal cell differentiation p = 1.0 × 10−4) (Table 4).

Table 4.
Enriched GO terms (p ≤ 0.005) among hypo- and hypermethylated regions.
3.5. Comparison of DNA Methylation with Previous Reports in Dermal Tissues
The 28 genes and promoters identified in this study (Table 2) were compared to results from published genome-wide DNA methylation [13] and gene expression studies [39,40,41,42,43,44,45,46,47,48,49] in cultured dermal fibroblasts or skin biopsies. Of our top genes and promoters, two CpGs in distal-less homeobox 5 (DLX5) were reported as hypermethylated in skin fibroblasts from dcSSc patients [13], which is consistent with our results.
When compared to gene expression profiling studies in cultured dermal fibroblasts or skin biopsies, DLX5 was reported to be under-expressed in patients with SSc [43], while transmembrane protein 140 (TMEM140) was reported to be overexpressed in patients with SSc [43] and correlated with the modified Rodnan skin thickness score (mRSS) in dcSSc patients [47].
When compared to the genes with compelling evidence of genetic association with SSc [10], none of our top 28 genes has been previously reported. Of note, these genome-wide DNA methylation [13] and gene expression studies [39,40,41,42,43,44,45,46,47,48,49] in skin-related tissues, as well as genetic association studies [10], were all performed in individuals of mostly European ancestry.
3.6. Gene Expression of Differentially Methylated Genes
To evaluate the functional effects of DNA methylation on gene expression in our sample of African American subjects, we performed qPCR on the five protein-coding genes (CDA, TMEM140, ACKR4, DLX5, FAM180B) and three long non-coding (lnc) RNA genes (MGC12916, LOC102724927, LOC101929882). These genes were chosen based on their known functions, an increased number of CpG sites detected (>30), and/or detectable transcripts from primary dermal fibroblasts using the RNA isolation/purification technique outlined in the methods section. Of the eight gene transcripts quantified, DLX5, TMEM140, and MCG12916, showed significant differential expression in cases compared to controls (Figure 2). Although these three genes showed hypermethylation, both the DLX5 and TMEM140 steady-state transcript levels were increased, while the MCG12916 steady-state transcript levels were decreased in patients compared to controls (Figure 2a–c).
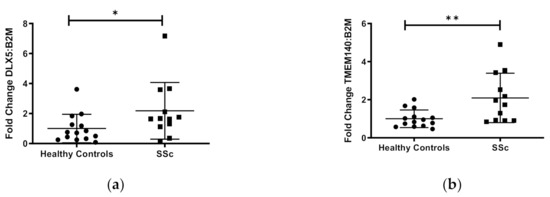
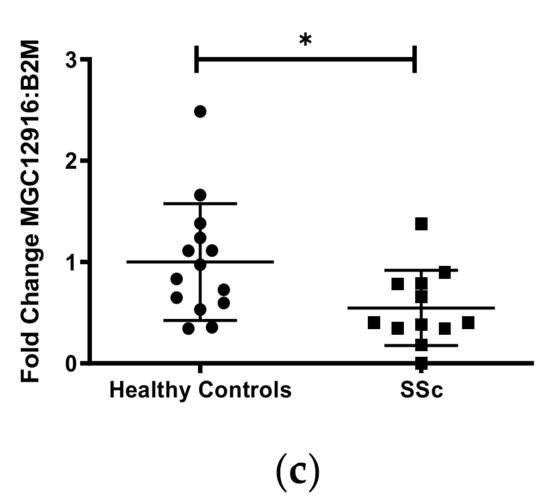
Figure 2.
Transcript levels of three differentially expressed genes in cultured skin fibroblasts. Among eight genes that were chosen for analysis, three genes demonstrated significantly differentiated expression in African ancestry SSc patients compared to controls: (a) DLX5, (b) TMEM140, and (c) MGC12916. Participant classification is detailed on the x-axis, while gene transcript-level fold change is expressed on the y-axis. Figure is representative of 14 healthy controls and 15 SSc patients. * p ≤ 0.05, ** p ≤ 0.01.
4. Discussion
This is the first study investigating patterns of differential methylation in primary skin fibroblasts from African American patients with SSc. We found widespread reduced DNA methylation in patients compared with healthy controls, consistent with what has been previously reported in skin fibroblasts from SSc patients of mostly European ancestry [13], and peripheral blood from Black South African patients with SSc [12].
Our findings show novel, top differentially methylated genes constituting mostly non-coding RNA genes and pseudogenes, with the function of most genes currently unknown. Only three protein-coding genes were amongst the top results: CDA and ACKR4 with known roles in immune pathways, and DLX5 with roles in cell development and proliferation. DLX5 was previously reported as hypermethylated in skin fibroblasts from dcSSc patients [13], which is consistent with our results. However, the previous study analyzed DNA methylation using the HumanMethylation450K array. Our study is based on RRBS, which tested eight times more CpGs than those present on the HumanMethylation450K array used in the previous genome-wide study of skin fibroblasts [13]. Thus, these methods are not directly comparable. Because the array contains only 2% of the CpGs we tested [50], minimal overlap can be expected. In addition, extensive differences in DNA methylation are known to exist between individuals of African and European ancestry [17,18,19,20,21,22,23,24], due to both variation in genetic ancestry and environmental factors [19], with Africans showing a higher DNA methylation than Europeans [20]. These differences help explain the new findings and minimal overlap with previous reports.
DLX5, TMEM140, and MCG12916 exhibited concomitant differential gene expression in the same primary dermal fibroblasts among the differentially methylated genes. While these genes exhibited hypermethylation, DLX5 and TMEM140 showed overexpression, while MCG12916 showed downregulation in the same individuals. This is not surprising, as the correlation between DNA methylation and gene expression is positive or negative and is tissue or context specific, in that the local DNA sequence and genomic features largely account for local patterns of methylation [51,52,53]. There is great variation in the quantitative impact of DNA methylation on gene expression among different cell types, with both positive and negative correlations between expression levels and CpG methylation levels [20,54,55,56,57]. Thus, the variable correlation between methylation patterns and gene expression is well established. Our results show that the hypomethylation of CpGs was prominent in all regions but CpGs islands, where DMCs were hypermethylated. DMC sites were enriched in introns, intergenic regions, TTS, and SINE, while depleted in 5′ UTR, promoters, and CpG islands. The overrepresentation of DMCs in introns has been previously reported in whole blood and neutrophils from systemic lupus erythematosus (SLE) patients [58,59], and in CD4+ T cells from SSc patients [15], while their overrepresentation in intergenic regions has also been found in CD4+ T cells from SSc patients [16]. Similarly, the underrepresentation of DMCs in promoters, CpG islands, and 5′ UTR has been previously reported in whole blood and neutrophils from SLE patients [58,59], and that of CpG islands is also found in CD4+ T cells from SSc patients [16]. Because CpG sites preferentially located in enhancers are reported to mediate gene expression, not in the promoters, this further supports a modest role of promoters in epigenetic regulatory mechanisms [20].
Interestingly, despite the differences in tissue and patient characteristics, TMEM140 was reported as overexpressed in skin biopsy specimens from patients with SSc [43] and correlated with the mRSS in dcSSc patients [60], which corroborates our findings. TMEM140 was identified as an IFN-inducible gene in cells infected with the human T-lymphocytic virus [61]. Since IFN signaling occurs in SSc [62,63,64,65] and is confirmed in this report, the overexpression of TMEM140 in AA SSc dermal fibroblasts may be in response to the IFN signature observed in SSc.
On the other hand, DLX5 was reported as under-expressed in skin biopsy specimens from patients with SSc [43]. The different outcomes of gene expression for DLX5 between the experiments could be the result of measuring gene expression in one cell type vs. across multiple cell types in skin biopsies, as well as underlying ancestral differences in gene expression. The inhibition of DLX5 in a uremic model of renal fibrosis causes a decreased expression of Notch receptors, ligands, and target genes [66]. Because Notch signaling is active in skin fibroblasts isolated from SSc patients and contributes to fibrosis in animal models [67,68], DLX5 may also regulate fibrosis through Notch signaling in SSc.
Although multiple lncRNAs have been reported as dysregulated in SSc patient tissues [69], to our knowledge this is the first report that MGC12916 has differential gene methylation and expression in primary dermal fibroblasts from African American patients with SSc.
To elucidate the underlying biological processes associated with SSc, GSEA and GO enrichment analyses were conducted. Among the hypomethylated regions, both GSEA and GO enrichment analyses showed the enrichment of immune pathways, with the GO analysis showing an enrichment in type I IFN signaling. Patients with SSc have excessive IFN and an IFN signature that correlates to early and more severe disease [62,63,64,65]. IFN is also pathogenic in SSc, since exogenous exposure to IFNα or IFNβ leads to its development [70,71,72,73]. The IFN regulatory factor 7 promoter (IRF7) is hypomethylated in SSc peripheral blood mononuclear cells [74], supporting the link of IFN signaling and gene hypomethylation in SSc.
Among hypermethylated regions, GSEA showed an enrichment of metabolism, cell development, and cell signaling pathways, and a GO enrichment analysis revealed an enrichment in specific pathways related to mesenchyme and epithelial development and cell differentiation. The top enriched GO term among hypermethylated regions, endothelial-mesenchymal cell transition (EMT), is consistent with the current hypothesis that EMT likely influences SSc disease characteristics, including endothelial cell dysfunction, dermal fibrosis, and interstitial lung disease [75,76]. To our knowledge, this is the first reported association between gene and promoter hypermethylation and mesenchymal cell differentiation in SSc. Thus, our GESA and GO analyses correlate with previous data regarding known pathways in SSc.
There are limitations to this study. First, although we are the first to analyze patterns of DNA methylation in dermal fibroblasts in African Americans, the sample size is modest. Nevertheless, with 15 SSc patients it is comparable to previous genome-wide DNA methylation analyses focused on skin fibroblasts (n = 12 SSc patients) [13], whole blood (n = 27 SSc patients) [14], and CD4+ T cells (n = 9 patients) [15], which included primarily individuals of European ancestry. Second, SSc is a rare disease with a prevalence of only 49,000 US adults [77], and there is currently no existing cohort or repository of samples from African American patients that can be leveraged to replicate and validate our results. Future studies expanded to multiple centers are needed. Third, the comparison of our results to those previously reported in European ancestry patients is hindered by differences in the analytic methods. Future studies including diverse individuals with measures of genetic ancestry as well as self-reported race, ethnicity, and other social and environmental exposures will ensure the validity and relevance of these findings for patients of all backgrounds. Fourth, and inherent to all epigenomic studies, we cannot exclude the possibility of reverse causation, or whether the DNA methylation changes are a cause or an effect of SSc. Future longitudinal studies, as well as studies comparing skin fibroblasts between affected and unaffected skin in patients and between affected and unaffected relatives will help to elucidate the role of DNA methylation in disease etiology. Fifth, it is possible that the DNA methylation changes are due to genetic variation. We lack genotypic data on these samples, but note that none of the top differentially methylated genes has been previously reported to be associated with SSc. We recognize that it is difficult to account for all lifestyle factors that could affect DNA methylation (i.e., diet, physical activity, body weight, smoking, medications, etc.) [78]. Our samples were balanced relative to smoking and age, so their confounding effects are minimized. Finally, we do not know the role of DLX5, TMEM140, and MCG12916 in SSc, but future gene silencing studies to inhibit the expression of these genes in primary dermal SSc fibroblasts will help elucidate their function in this cell type. In spite of these limitations, these findings identify novel loci in SSc and highlight candidate genes for further research.
5. Conclusions
This first genome-wide DNA methylation study of skin fibroblasts from SSc patients of African ancestry identified novel differentially methylated sites and genes. These include sites with evidence of altered methylation in protein-coding, lncRNA, and pseudogenes and concomitant differential expression in MGC12916, DLX5, and TMEM140. Although this cross-sectional study cannot separate causality from response to disease, it identifies DNA methylation alterations in genes and pathways that are important in SSc, showing that distinct DNA methylation changes underlie SSc in African Americans. These findings provide a foundation for further research to determine the functional consequences of the differentially methylated loci. Given the reversible nature of epigenetic marks, these loci might represent attractive targets for the treatment or prevention of autoimmune- and/or fibrotic-related diseases.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/2/129/s1: Table S1: Enrichment of DMC in different genomic locations. Figure S1. Manhattan plot of association of differential DNA methylation with SSc.
Author Contributions
Conceptualization, P.S.R.; Recruitment, J.C.O.; Data Curation, J.F., I.A., G.S.B., R.C.W. and D.B.F.; Data Analysis, W.d.S., E.S.H., P.S.R., G.H., D.B.F. and C.F.-B.; Writing—Original Draft Preparation, D.B.F., K.L.D. and P.S.R.; Writing—Review and Editing, P.S.R., D.B.F., J.C.O., J.F., I.A., G.S.B., R.C.W., W.d.S., E.S.H., G.H., K.L.D. and C.F.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the US National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) under Awards Number T32 AR050958 (DBF), K01 AR067280 (PSR), P30 AR072582 (PSR), R03 AR065801 (PSR), P60 AR062755 (PSR, GSB, JCO), K24 AR060297 (CFB), the US National Institute on Drug Abuse of the NIH under Awards Number U01 DA045300 and U54 MD010706-CHH (GH), the US National Cancer Institute of the NIH under Award Number P30 CA138313 (RCW), the National Center for Advancing Translational Sciences of the NIH under Award Numbers KL2 TR001452 and UL1 TR001450 (DBF, PSR), the MUSC Center for Genomic Medicine (PSR, GH), the Scleroderma Foundation (PSR), and start-up funding from Queens University Belfast (GH).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board at the Medical University of South Carolina (Date: 9 April 2014. Application number Pro# 33636).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are openly available in the NCBI Gene Expression Omnibus (GEO) repository (accession code GSE150592). All the data are also available from the authors on request.
Acknowledgments
The authors would like to acknowledge the participants, Edwin Smith and Richard Silver for performing the skin biopsies, and Carol Lambourne for coordinating participant recruitment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Silver, R.M. Clinical aspects of systemic sclerosis (scleroderma). Ann. Rheum. Dis. 1991, 50 (Suppl. 4), 854–861. [Google Scholar] [CrossRef]
- Mayes, M.D.; Lacey, J.V., Jr.; Beebe-Dimmer, J.; Gillespie, B.W.; Cooper, B.; Laing, T.J.; Schottenfeld, D. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheumatol. 2003, 48, 2246–2255. [Google Scholar] [CrossRef]
- Mendoza, F.; Derk, C.T. Systemic sclerosis mortality in the United States: 1999-2002 implications for patient care. J. Clin. Rheumatol. Pract. Rep. Rheum. Musculoskelet. Dis. 2007, 13, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Laing, T.J.; Gillespie, B.W.; Toth, M.B.; Mayes, M.D.; Gallavan, R.H., Jr.; Burns, C.J.; Johanns, J.R.; Cooper, B.C.; Keroack, B.J.; Wasko, M.C.; et al. Racial differences in scleroderma among women in Michigan. Arthritis Rheumatol. 1997, 40, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Greidinger, E.L.; Flaherty, K.T.; White, B.; Rosen, A.; Wigley, F.M.; Wise, R.A. African-American race and antibodies to topoisomerase I are associated with increased severity of scleroderma lung disease. Chest 1998, 114, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, E.; Furst, D.E. Systemic sclerosis mortality in the United States: 1979–1998. Eur. J. Epidemiol. 2005, 20, 855–861. [Google Scholar] [CrossRef] [PubMed]
- McNearney, T.A.; Reveille, J.D.; Fischbach, M.; Friedman, A.W.; Lisse, J.R.; Goel, N.; Tan, F.K.; Zhou, X.; Ahn, C.; Feghali-Bostwick, C.A.; et al. Pulmonary involvement in systemic sclerosis: Associations with genetic, serologic, sociodemographic, and behavioral factors. Arthritis Rheumatol. 2007, 57, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Nietert, P.J.; Mitchell, H.C.; Bolster, M.B.; Shaftman, S.R.; Tilley, B.C.; Silver, R.M. Racial variation in clinical and immunological manifestations of systemic sclerosis. J. Rheumatol. 2006, 33, 263–268. [Google Scholar]
- Morgan, N.D.; Shah, A.A.; Mayes, M.D.; Domsic, R.T.; Medsger, T.A., Jr.; Steen, V.D.; Varga, J.; Carns, M.; Ramos, P.S.; Silver, R.M.; et al. Clinical and serological features of systemic sclerosis in a multicenter African American cohort: Analysis of the genome research in African American scleroderma patients clinical database. Medicine 2017, 96, e8980. [Google Scholar] [CrossRef]
- Angiolilli, C.; Marut, W.; van der Kroef, M.; Chouri, E.; Reedquist, K.A.; Radstake, T. New insights into the genetics and epigenetics of systemic sclerosis. Nat. Rev. Rheumatol. 2018, 14, 657–673. [Google Scholar] [CrossRef]
- Selmi, C.; Feghali-Bostwick, C.A.; Lleo, A.; Lombardi, S.A.; De Santis, M.; Cavaciocchi, F.; Zammataro, L.; Mitchell, M.M.; Lasalle, J.M.; Medsger, T., Jr.; et al. X chromosome gene methylation in peripheral lymphocytes from monozygotic twins discordant for scleroderma. Clin. Exp. Immunol. 2012, 169, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Matatiele, P.; Tikly, M.; Tarr, G.; Gulumian, M. DNA methylation similarities in genes of black South Africans with systemic lupus erythematosus and systemic sclerosis. J. Biomed. Sci. 2015, 22, 34. [Google Scholar] [CrossRef] [PubMed]
- Altorok, N.; Tsou, P.S.; Coit, P.; Khanna, D.; Sawalha, A.H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 2015, 74, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.S.; Zimmerman, K.D.; Haddad, S.; Langefeld, C.D.; Medsger, T.A., Jr.; Feghali-Bostwick, C.A. Integrative analysis of DNA methylation in discordant twins unveils distinct architectures of systemic sclerosis subsets. Clin. Epigenetics 2019, 11, 58. [Google Scholar] [CrossRef]
- Lu, T.; Klein, K.O.; Colmegna, I.; Lora, M.; Greenwood, C.M.T.; Hudson, M. Whole-genome bisulfite sequencing in systemic sclerosis provides novel targets to understand disease pathogenesis. BMC Med. Genom. 2019, 12, 144. [Google Scholar] [CrossRef]
- Li, T.; Ortiz-Fernandez, L.; Andres-Leon, E.; Ciudad, L.; Javierre, B.M.; Lopez-Isac, E.; Guillen-Del-Castillo, A.; Simeon-Aznar, C.P.; Ballestar, E.; Martin, J. Epigenomics and transcriptomics of systemic sclerosis CD4+ T cells reveal long-range dysregulation of key inflammatory pathways mediated by disease-associated susceptibility loci. Genome Med. 2020, 12, 81. [Google Scholar] [CrossRef]
- Michels, K.B.; Binder, A.M.; Dedeurwaerder, S.; Epstein, C.B.; Greally, J.M.; Gut, I.; Houseman, E.A.; Izzi, B.; Kelsey, K.T.; Meissner, A.; et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat. Methods 2013, 10, 949–955. [Google Scholar] [CrossRef]
- Barfield, R.T.; Almli, L.M.; Kilaru, V.; Smith, A.K.; Mercer, K.B.; Duncan, R.; Klengel, T.; Mehta, D.; Binder, E.B.; Epstein, M.P.; et al. Accounting for population stratification in DNA methylation studies. Genet. Epidemiol. 2014, 38, 231–241. [Google Scholar] [CrossRef]
- Galanter, J.M.; Gignoux, C.R.; Oh, S.S.; Torgerson, D.; Pino-Yanes, M.; Thakur, N.; Eng, C.; Hu, D.; Huntsman, S.; Farber, H.J.; et al. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife 2017, 6, e20532. [Google Scholar] [CrossRef]
- Husquin, L.T.; Rotival, M.; Fagny, M.; Quach, H.; Zidane, N.; McEwen, L.M.; MacIsaac, J.L.; Kobor, M.S.; Aschard, H.; Patin, E.; et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 2018, 19, 222. [Google Scholar] [CrossRef]
- Quach, H.; Rotival, M.; Pothlichet, J.; Loh, Y.E.; Dannemann, M.; Zidane, N.; Laval, G.; Patin, E.; Harmant, C.; Lopez, M.; et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell 2016, 167, 643–656.e17. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, S.; Carja, O.; Fagny, M.; Patin, E.; Myrick, J.W.; McEwen, L.M.; Mah, S.M.; Kobor, M.S.; Froment, A.; Feldman, M.W.; et al. Trends in DNA Methylation with Age Replicate across Diverse Human Populations. Genetics 2017, 206, 1659–1674. [Google Scholar] [CrossRef] [PubMed]
- Fagny, M.; Patin, E.; MacIsaac, J.L.; Rotival, M.; Flutre, T.; Jones, M.J.; Siddle, K.J.; Quach, H.; Harmant, C.; McEwen, L.M.; et al. The epigenomic landscape of African rainforest hunter-gatherers and farmers. Nat. Commun. 2015, 6, 10047. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA methylation contributes to natural human variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef]
- Gu, H.; Bock, C.; Mikkelsen, T.S.; Jager, N.; Smith, Z.D.; Tomazou, E.; Gnirke, A.; Lander, E.S.; Meissner, A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods 2010, 7, 133–136. [Google Scholar] [CrossRef]
- Bock, C.; Tomazou, E.M.; Brinkman, A.B.; Muller, F.; Simmer, F.; Gu, H.; Jager, N.; Gnirke, A.; Stunnenberg, H.G.; Meissner, A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010, 28, 1106–1114. [Google Scholar] [CrossRef]
- Atanelishvili, I.; Shirai, Y.; Akter, T.; Buckner, T.; Noguchi, A.; Silver, R.M.; Bogatkevich, G.S. M10, a caspase cleavage product of the hepatocyte growth factor receptor, interacts with Smad2 and demonstrates antifibrotic properties in vitro and in vivo. Transl. Res. 2016, 170, 99–111. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics (Oxf. Engl.) 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Assenov, Y.; Muller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 2014, 11, 1138–1140. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.C.; Mills, J.N.; Turner-Ivey, B.; Wilson, R.C.; Guest, S.T.; Rutkovsky, A.; Dombkowski, A.; Kappler, C.S.; Hardiman, G.; Ethier, S.P. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol. Oncol. 2016, 10, 850–865. [Google Scholar] [CrossRef]
- Hardiman, G.; Savage, S.J.; Hazard, E.S.; Wilson, R.C.; Courtney, S.M.; Smith, M.T.; Hollis, B.W.; Halbert, C.H.; Gattoni-Celli, S. Systems analysis of the prostate transcriptome in African-American men compared with European-American men. Pharmacogenomics 2016, 17, 1129–1143. [Google Scholar] [CrossRef]
- Kuhn, K.; Bertling, W.M.; Emmrich, F. Cloning of a functional cDNA for human cytidine deaminase (CDD) and its use as a marker of monocyte/macrophage differentiation. Biochem. Biophys. Res. Commun. 1993, 190, 1–7. [Google Scholar] [CrossRef]
- Nibbs, R.J.; Graham, G.J. Immune regulation by atypical chemokine receptors. Nat. Reviews. Immunol. 2013, 13, 815–829. [Google Scholar] [CrossRef]
- Weiss, K.M.; Ruddle, F.H.; Bollekens, J. Dlx and other homeobox genes in the morphological development of the dentition. Connect. Tissue Res. 1995, 32, 35–40. [Google Scholar] [CrossRef]
- Skaug, B.; Khanna, D.; Swindell, W.R.; Hinchcliff, M.E.; Frech, T.M.; Steen, V.D.; Hant, F.N.; Gordon, J.K.; Shah, A.A.; Zhu, L.; et al. Global skin gene expression analysis of early diffuse cutaneous systemic sclerosis shows a prominent innate and adaptive inflammatory profile. Ann. Rheum. Dis. 2020, 79, 379–386. [Google Scholar] [CrossRef]
- Whitfield, M.L.; Finlay, D.R.; Murray, J.I.; Troyanskaya, O.G.; Chi, J.T.; Pergamenschikov, A.; McCalmont, T.H.; Brown, P.O.; Botstein, D.; Connolly, M.K. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc. Natl. Acad. Sci. USA 2003, 100, 12319–12324. [Google Scholar] [CrossRef]
- Gardner, H.; Shearstone, J.R.; Bandaru, R.; Crowell, T.; Lynes, M.; Trojanowska, M.; Pannu, J.; Smith, E.; Jablonska, S.; Blaszczyk, M.; et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheumatol. 2006, 54, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Milano, A.; Pendergrass, S.A.; Sargent, J.L.; George, L.K.; McCalmont, T.H.; Connolly, M.K.; Whitfield, M.L. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE 2008, 3, e2696. [Google Scholar] [CrossRef]
- Assassi, S.; Swindell, W.R.; Wu, M.; Tan, F.D.; Khanna, D.; Furst, D.E.; Tashkin, D.P.; Jahan-Tigh, R.R.; Mayes, M.D.; Gudjonsson, J.E.; et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol. 2015, 67, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Derrett-Smith, E.C.; Martyanov, V.; Chighizola, C.B.; Moinzadeh, P.; Campochiaro, C.; Khan, K.; Wood, T.A.; Meroni, P.L.; Abraham, D.J.; Ong, V.H.; et al. Limited cutaneous systemic sclerosis skin demonstrates distinct molecular subsets separated by a cardiovascular development gene expression signature. Arthritis Res. 2017, 19, 156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pendergrass, S.A.; Lemaire, R.; Francis, I.P.; Mahoney, J.M.; Lafyatis, R.; Whitfield, M.L. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J. Investig. Dermatol. 2012, 132, 1363–1373. [Google Scholar] [CrossRef]
- Johnson, M.E.; Mahoney, J.M.; Taroni, J.; Sargent, J.L.; Marmarelis, E.; Wu, M.R.; Varga, J.; Hinchcliff, M.E.; Whitfield, M.L. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS ONE 2015, 10, e0114017. [Google Scholar] [CrossRef]
- Rice, L.M.; Ziemek, J.; Stratton, E.A.; McLaughlin, S.R.; Padilla, C.M.; Mathes, A.L.; Christmann, R.B.; Stifano, G.; Browning, J.L.; Whitfield, M.L.; et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2015, 67, 3004–3015. [Google Scholar] [CrossRef]
- Stifano, G.; Sornasse, T.; Rice, L.M.; Na, L.; Chen-Harris, H.; Khanna, D.; Jahreis, A.; Zhang, Y.; Siegel, J.; Lafyatis, R. Skin Gene Expression Is Prognostic for the Trajectory of Skin Disease in Patients With Diffuse Cutaneous Systemic Sclerosis. Arthritis Rheumatol. 2018, 70, 912–919. [Google Scholar] [CrossRef]
- Hinchcliff, M.; Huang, C.C.; Wood, T.A.; Matthew Mahoney, J.; Martyanov, V.; Bhattacharyya, S.; Tamaki, Z.; Lee, J.; Carns, M.; Podlusky, S.; et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J. Investig. Dermatol. 2013, 133, 1979–1989. [Google Scholar] [CrossRef]
- Walker, D.L.; Bhagwate, A.V.; Baheti, S.; Smalley, R.L.; Hilker, C.A.; Sun, Z.; Cunningham, J.M. DNA methylation profiling: Comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip. Epigenomics 2015, 7, 1287–1302. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Arcelus, M.; Ongen, H.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Yurovsky, A.; Bryois, J.; Padioleau, I.; Romano, L.; Planchon, A.; et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 2015, 11, e1004958. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Manzo, M.; Baubec, T. Dynamics and Context-Dependent Roles of DNA Methylation. J. Mol. Biol. 2017, 429, 1459–1475. [Google Scholar] [CrossRef]
- Gutierrez-Arcelus, M.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Ongen, H.; Yurovsky, A.; Bryois, J.; Giger, T.; Romano, L.; Planchon, A.; et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2013, 2, e00523. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Luijk, R.; Zhernakova, D.V.; Moed, M.; Deelen, P.; Vermaat, M.; van Iterson, M.; van Dijk, F.; van Galen, M.; Bot, J.; et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017, 49, 131–138. [Google Scholar] [CrossRef]
- Bonder, M.J.; Kasela, S.; Kals, M.; Tamm, R.; Lokk, K.; Barragan, I.; Buurman, W.A.; Deelen, P.; Greve, J.W.; Ivanov, M.; et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genom. 2014, 15, 860. [Google Scholar] [CrossRef]
- Grundberg, E.; Meduri, E.; Sandling, J.K.; Hedman, A.K.; Keildson, S.; Buil, A.; Busche, S.; Yuan, W.; Nisbet, J.; Sekowska, M.; et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet. 2013, 93, 876–890. [Google Scholar] [CrossRef]
- Coit, P.; Ortiz-Fernandez, L.; Lewis, E.E.; McCune, W.J.; Maksimowicz-McKinnon, K.; Sawalha, A.H. A longitudinal and transancestral analysis of DNA methylation patterns and disease activity in lupus patients. JCI Insight 2020, 5, e143654. [Google Scholar] [CrossRef]
- Imgenberg-Kreuz, J.; Carlsson Almlof, J.; Leonard, D.; Alexsson, A.; Nordmark, G.; Eloranta, M.L.; Rantapaa-Dahlqvist, S.; Bengtsson, A.A.; Jonsen, A.; Padyukov, L.; et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 736–743. [Google Scholar] [CrossRef]
- Rice, L.M.; Stifano, G.; Ziemek, J.; Lafyatis, R. Local skin gene expression reflects both local and systemic skin disease in patients with systemic sclerosis. Rheumatology (Oxf.) 2016, 55, 377–379. [Google Scholar] [CrossRef][Green Version]
- Refaat, A.; Owis, M.; Abdelhamed, S.; Saiki, I.; Sakurai, H. Retrospective screening of microarray data to identify candidate IFN-inducible genes in a HTLV-1 transformed model. Oncol. Lett. 2018, 15, 4753–4758. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.K.; Zhou, X.; Mayes, M.D.; Gourh, P.; Guo, X.; Marcum, C.; Jin, L.; Arnett, F.C., Jr. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatol. (Oxf.) 2006, 45, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Higgs, B.W.; Liu, Z.; White, B.; Zhu, W.; White, W.I.; Morehouse, C.; Brohawn, P.; Kiener, P.A.; Richman, L.; Fiorentino, D.; et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann. Rheum. Dis. 2011, 70, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Higgs, B.W.; Zhu, W.; Richman, L.; Fiorentino, D.F.; Greenberg, S.A.; Jallal, B.; Yao, Y. Identification of activated cytokine pathways in the blood of systemic lupus erythematosus, myositis, rheumatoid arthritis, and scleroderma patients. Int. J. Rheum. Dis. 2012, 15, 25–35. [Google Scholar] [CrossRef]
- Brkic, Z.; van Bon, L.; Cossu, M.; van Helden-Meeuwsen, C.G.; Vonk, M.C.; Knaapen, H.; van den Berg, W.; Dalm, V.A.; Van Daele, P.L.; Severino, A.; et al. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann. Rheum. Dis. 2016, 75, 1567–1573. [Google Scholar] [CrossRef]
- Wang, X.F.; Zhang, B.H.; Lu, X.Q.; Wang, R.Q. DLX5 gene regulates the Notch signaling pathway to promote glomerulosclerosis and interstitial fibrosis in uremic rats. J. Cell. Physiol. 2019, 234, 21825–21837. [Google Scholar] [CrossRef]
- Dees, C.; Zerr, P.; Tomcik, M.; Beyer, C.; Horn, A.; Akhmetshina, A.; Palumbo, K.; Reich, N.; Zwerina, J.; Sticherling, M.; et al. Inhibition of Notch signaling prevents experimental fibrosis and induces regression of established fibrosis. Arthritis Rheum. 2011, 63, 1396–1404. [Google Scholar] [CrossRef]
- Beyer, C.; Dees, C.; Distler, J.H. Morphogen pathways as molecular targets for the treatment of fibrosis in systemic sclerosis. Arch. Dermatol. Res. 2013, 305, 1–8. [Google Scholar] [CrossRef]
- Ramos, P.S. Epigenetics of scleroderma: Integrating genetic, ethnic, age, and environmental effects. J. Scleroderma Relat. Disord. 2019, 4, 238–250. [Google Scholar] [CrossRef]
- Beretta, L.; Caronni, M.; Vanoli, M.; Scorza, R. Systemic sclerosis after interferon-alfa therapy for myeloproliferative disorders. Br. J. Dermatol. 2002, 147, 385–386. [Google Scholar] [CrossRef]
- Solans, R.; Bosch, J.A.; Esteban, I.; Vilardell, M. Systemic sclerosis developing in association with the use of interferon alpha therapy for chronic viral hepatitis. Clin. Exp. Rheumatol. 2004, 22, 625–628. [Google Scholar] [PubMed]
- Tahara, H.; Kojima, A.; Hirokawa, T.; Oyama, T.; Naganuma, A.; Maruta, S.; Okada, K.; Ban, S.; Yoshida, K.; Takagi, H.; et al. Systemic sclerosis after interferon alphacon-1 therapy for hepatitis C. Intern. Med. 2007, 46, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.; Myles, M.L.; Yacyshyn, E. The development of systemic sclerosis in a female patient with multiple sclerosis following beta interferon treatment. Clin. Rheumatol. 2008, 27, 1467–1468. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, R.; Mahmoudi, M.; Gharibdoost, F.; Kavosi, H.; Dashti, N.; Imeni, V.; Jamshidi, A.; Aslani, S.; Mostafaei, S.; Vodjgani, M. IRF7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int. J. Rheum. Dis. 2017, 20, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Manetti, M.; Romano, E.; Rosa, I.; Guiducci, S.; Bellando-Randone, S.; De Paulis, A.; Ibba-Manneschi, L.; Matucci-Cerinic, M. Endothelial-to-mesenchymal transition contributes to endothelial dysfunction and dermal fibrosis in systemic sclerosis. Ann. Rheum Dis 2017, 76, 924–934. [Google Scholar] [CrossRef]
- Mendoza, F.A.; Piera-Velazquez, S.; Farber, J.L.; Feghali-Bostwick, C.; Jimenez, S.A. Endothelial Cells Expressing Endothelial and Mesenchymal Cell Gene Products in Lung Tissue From Patients with Systemic Sclerosis-Associated Interstitial Lung Disease. Arthritis Rheumatol. 2016, 68, 210–217. [Google Scholar] [CrossRef]
- Helmick, C.G.; Felson, D.T.; Lawrence, R.C.; Gabriel, S.; Hirsch, R.; Kwoh, C.K.; Liang, M.H.; Kremers, H.M.; Mayes, M.D.; Merkel, P.A.; et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheumatol. 2008, 58, 15–25. [Google Scholar] [CrossRef]
- Lim, U.; Song, M.A. Dietary and lifestyle factors of DNA methylation. Methods Mol. Biol. 2012, 863, 359–376. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).