Abstract
Nasopharyngeal carcinoma (NPC) and alcohol flush syndrome are thought to be strongly influenced by genetic factors and are highly prevalent amongst East Asians. Diminished activity of aldehyde dehydrogenase (ALDH), a major enzyme in the alcohol-metabolizing pathway, causes the flushing syndrome associated with alcoholic consumption. The genetic effect of ALDH isoforms on NPC is unknown. We therefore investigated the association between the genetic polymorphisms of all 19 ALDH isoforms and NPC among 458 patients with NPC and 1672 age- and gender-matched healthy controls in Taiwan. Single-nucleotide polymorphisms (SNPs) located between the 40,000 base pairs upstream and downstream of the 19 ALDH isoform coding regions were collected from two genome-wise association studies conducted in Taiwan and from the Taiwan Biobank. Thirteen SNPs located on ALDH4A1, ALDH18A1, ALDH3B2, ALDH1L2, ALDH1A2, and ALDH2 Glu487Lys (rs671) were associated with NPC susceptibility. Stratification by alcohol status revealed a cumulative risk effect for NPC amongst drinkers and non-drinkers, with odds ratios of 4.89 (95% confidence interval 2.15–11.08) and 3.57 (1.97–6.47), respectively. A synergistic effect was observed between SNPs and alcohol. This study is the first to report associations between genetic variants in 19 ALDH isoforms, their interaction with alcohol consumption and NPC in an East Asian population.
1. Introduction
Nasopharyngeal carcinoma (NPC) is more common in East and Southeast Asia than in Western countries, with more than 70% of cases worldwide originating from this Asian region. Recently reported age-standardized incidence rates range from 3.0 per 100,000 in China to 0.4 per 100,000 in Western countries [1,2]. The remarkable geographical distribution of NPC incidence and family history as a strong risk factor suggests that host genetic susceptibility plays an important role [2,3]. Familial linkage studies, genetic case-control association studies, genome-wide association studies (GWAS) and whole-exome sequencing association studies have identified susceptibility genes/loci related to the risk of NPC, including the HLA genes [4,5], CLPTM1L/TERT [6], MST1R [7] and NIPAL1 genes [3]. Additional host genetic susceptibility factors have remained elusive.
Lifestyle behaviors such as salted fish intake and cigarette smoking significantly increase the risk of NPC in Asian populations [8]. The association between alcohol consumption and NPC risk is inconsistent in many studies [9,10]. Two meta-analyses have indicated that the risk of developing NPC may increase with alcohol consumption; in both meta-analyses, drinking and high-frequency drinking increased the risk of NPC [11,12].
People of East Asian descent have the highest prevalence (35–45%) of aldehyde dehydrogenase 2 (ALDH2) deficiency, which causes the flushing syndrome observed after consumption of alcoholic beverages [13]. The major alcohol-metabolizing enzymes are alcohol dehydrogenase-1B (ADH1B) and ALDH2, and the two most frequently reported polymorphisms, ADH1B Arg47His (rs1229984) and ALDH2 Glu487Lys (rs671), have been shown to alter the effect of alcohol and potentially influence carcinogenesis [14,15]. In humans, the ALDH family consists of 19 members identified through similar amino acid sequences and functions [16]. Several recent studies have shown that ALDH1A1, ALDH2 and ALDH3A1 may be related to different cancers, such as head and neck cancer (HNC) [17], esophageal cancer [18], cholangiocarcinoma [19], and colorectal cancer (CRC) [14]. Elevated ALDH1 activity has been used as a cancer stem cell biomarker of tumor aggressiveness in the invasive front of NPC [20], while a lower expression of ALDH2 has been associated with poor prognoses in breast cancer, lung adenocarcinoma, and HNC squamous cell carcinomas [21].
Although NPC and ALDH2 deficiency occur with high frequencies among East Asians, the genetic effects of ALDH isoforms on NPC remain unknown. Thus, we sought to determine associations between the genetic polymorphisms of 19 ALDH isoforms, [22] their interaction with alcohol consumption and NPC in an Asian population in Taiwan.
2. Materials and Methods
2.1. Study Population
This study included 458 NPC cases enrolled in two GWAS studies conducted in Taiwan [4,23], all of whom were recruited from Chang Gung Memorial Hospital (CGMH) between 1983 and 2008. Their pathology records were reviewed for confirmation of NPC diagnosis according to World Health Organization (WHO) pathological classification criteria. Age- and gender-matched healthy controls were randomly selected from subjects without any NPC family history from the Taiwan Biobank (TWB) [24]. After matching, a total of 413 NPC cases and 1672 healthy controls were included in the present study (case:control ratio 1:4). The TWB has collected specimens and associated data (including genetic information) from the general Taiwanese population since 2013 and follows up with subjects every two to four years. The TWB data in this study involved individuals aged 30–70 years who self-reported as being of Taiwanese Han Chinese descent. The study was reviewed and approved by the Institutional Review Broad of Chang Gung Medical Foundation, Taiwan (IRB 103-7224B). Written informed consent was obtained from each study participant at the time of enrollment.
2.2. Data Collection
Survey questionnaires collected information about alcohol consumption, betel quid chewing, and cigarette smoking. Cases were designated alcohol users if they had consumed an alcoholic beverage at least once weekly for six months, betel nut users if they had chewed at least two betel nuts daily for a year, and cigarette smokers if they had smoked daily for at least one year. Among controls, alcohol users were defined as persons who reported drinking more than 150 mL of alcohol per week during the 6 months before the study health examination, betel nut users if they had ever chewed betel nuts daily for one month, and cigarette smokers if they had smoked daily for at least 6 months.
2.3. Genotyping and Imputation
Genotyping of the NPC cohort was performed by Illumina Hap550v3_A (for 277 NPC cases) and Human610-Quad Beadchips (for 181 NPC cases), according to the manufacturer’s protocols (Illumina, Inc., San Diego, CA, USA). The Affymetrix Axiom genome-wide TWB array was used to genotype the TWB cohort. Genotyping and quality control measures involving samples and single nucleotide polymorphisms (SNPs) followed those described in previous studies [4,23]. Since the GWAS results were obtained using three different genotyping platforms, genotype imputations were performed separately in each platform before data combination. Imputations were performed using IMPUTE2 [25] with the 1000 Genomes Project Phase III reference panel (October 2014 release). A total of 78,605 SNPs were identified between the 20,000 base pairs upstream and 20,000 base pairs downstream of 19 ALDH isoform coding regions, based on GENCODE release 38. SNPs with low imputation quality (information < 0.3), call rate < 99%, minor allele frequency < 0.05, and Hardy-Weinberg equilibrium in controls (p < 0.0005) were removed from analysis.
2.4. Statistical Analysis
For the baseline characteristics, continuous data are presented as means with standard deviation, and categorical data are presented as proportions. We used t-tests to compare mean values of continuous variables and chi-squared tests to compare the frequencies of categorical variables between two groups. The association between SNP genotype/cumulative risk alleles and disease status was evaluated using logistic regression while controlling for alcohol use, betel quid chewing, and cigarette smoking to obtain the p values, odds ratios (ORs) and 95% confidence intervals (CIs) in PLINK (version 1.90) [26]. Permutation testing was performed 10,000 times using the PLINK “-mperm 10000” command. All tests were two-sided, and a p value < 0.05 was considered to be statistically significant. Statistical analyses were performed using SPSS software v21.0 for Windows (IBM, Armonk, NY, USA) and R version 3.4.4 (R Core Team, 2018).
3. Results
3.1. Characteristics of the Study Participants
A total of 1245 subjects (249 cases and 996 controls) served as the discovery cohort to search for genetic risk factors associated with NPC, while 840 subjects (164 cases and 676 controls) served as the replication cohort for the identified genetic SNPs (Figure 1). Demographic characteristics of patients and controls are presented in Table 1. Around three-quarters (75%) of the study population were males; mean ages were 47.98 ± 10.03 years in the NPC group and 48.03 ± 10.37 years in the control group. Significantly higher proportions of the NPC group consumed alcohol, chewed betel quid, and smoked cigarettes, compared with the controls (36.8% vs. 15.6%; 23.0% vs. 6.0%; and 48.9% vs. 35.1%, respectively; all p values < 0.001). Around two-thirds of the NPC cases (67.5%) were diagnosed with late-stage (III and IV) disease, as according to the WHO classification (data not shown). Clinical characteristics including alcohol use, betel quid chewing, and cigarette smoking are risk factors for NPC and were included in the subsequent adjusted genetic SNP analysis.
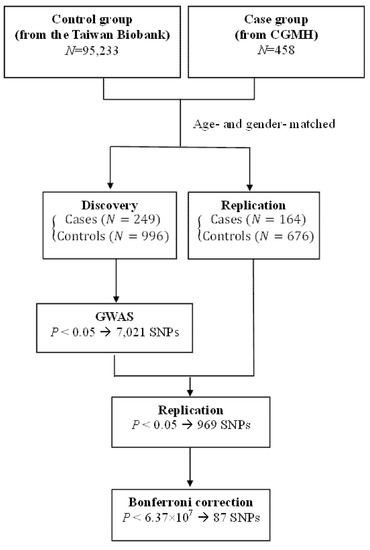
Figure 1.
Flow chart of the study design.

Table 1.
Demographics of the study population.
3.2. ALDH Isoforms and Candidate SNPs Confer Susceptibility for NPC
We determined the association between the genetic polymorphisms of 19 ALDH isoforms and the risk of NPC in Taiwan Chinese. Multivariate logistic regression analysis adjusted for alcohol drinking, betel quid chewing, and cigarette smoking identified 12 SNPs on ALDH4A1, ALDH18A1, ALDH3B2, ALDH1L2, and ALDH1A2 that were significantly associated with an increased risk for NPC (all p values < 0.05, Table 2). In this study, we used the permutation test, a robust but computationally intensive alternative to the conservative Bonferroni correction for correcting multiple testing [27]. Although none of the SNPs remained significant after Bonferroni correction (0.05/78,605), 6 SNPs located in ALDH4A1, ALDH18A1 and ALDH3B2 passed a 10,000 random shuffled permutation test (p perm < 0.05). In particular, one SNP (rs7534676) located in ALDH4A1 had a significant permutation p value of <0.01 (Table 2).

Table 2.
Associations between ALDH gene polymorphisms and nasopharyngeal carcinoma risk.
This study also investigated the two most frequently reported gene polymorphisms related to alcohol metabolism, ADH1B Arg47His rs1229984 and ALDH2 Glu487Lys rs671. An association was observed between the rs671 polymorphism in ALDH2 and NPC risk. The adjusted OR was 1.23 (95% CI = 1.03–1.48, p = 0.00225) when increased by one A allele. No association was observed between the rs1229984 polymorphism in ADH1B and NPC risk. After adjusting for potential confounders, the OR was 0.97 (95% CI = 0.78–1.22, p = 0.0801) (Table 2).
3.3. Cumulative Risk Effect of 13 SNPs on NPC Susceptibility
Total risk allele counts for the 13 SNPs that we have identified were calculated for each subject (range 13–26; median 23). In the multivariate logistic regression model, alcohol use, betel nut chewing and the cumulative risk allele were all independent risk factors for NPC. ORs were 2.61 (95% CI = 1.60–4.26, p < 0.001) for alcohol use and 2.63 (1.40–4.94, p = 0.003) for betel nut chewing. Study subjects with more than 23 risk alleles had a significantly higher risk of NPC (OR = 3.98; 95% CI = 2.45–6.46, p < 0.001) compared with subjects with fewer than 23 risk alleles (Table 3).

Table 3.
Cumulative risk effect of 13 SNPs on nasopharyngeal carcinoma susceptibility in multivariate logistic regression model.
3.4. ALDH Genes Confer Susceptibility for NPC after Stratification for Alcohol Use
To investigate the confounding effect of alcohol use on NPC, associations between SNPs and NPC were stratified by alcohol consumption. Among subjects who did not consume alcohol, the homozygous risk allele for most SNPs (except rs1229984) increased the risk of NPC (p < 0.05). Among alcohol users, the homozygous risk alleles for rs7534676, rs7554974, rs7518631, rs7518631, rs72936453, rs1711068, rs76655136, rs1975431, and rs28829404 increased the risk of NPC (p < 0.05). For ALDH2 rs671, the risk of NPC was higher for study subjects with the AA/AG alleles compared with subjects with the GG allele, whether alcohol was consumed (OR = 1.47; 95% CI = 0.95–2.27, p = 0.082) or not (1.27; 0.97–1.67, p = 0.087). A cumulative risk allele effect for NPC was observed with alcohol consumption: the risk was lower for subjects not using alcohol (OR = 3.57; 95% CI = 1.97–6.47, p < 0.001) than for those who were (4.89; 2.15–11.08, p < 0.001) (Table 4).

Table 4.
Associations between ALDH genes and nasopharyngeal carcinoma susceptibility after stratification for alcohol use.
3.5. The Effects of Interaction between Alcohol Consumption and SNPs on the Risk of NPC
An investigation into the effects of interaction between alcohol consumption and SNPs on the risk of NPC revealed that the risk increases with either the presence of a risk allele or alcohol consumption. For the rs671 polymorphism, the NPC risk was significantly increased among AA/AG carriers who were not consuming alcohol or GG carriers who were consuming alcohol (OR = 1.63; 95% CI = 1.27–2.10, p < 0.001), and AA/AG carriers who were consuming alcohol (4.55; 3.02–6.84, p < 0.001), compared with carriers of the GG genotype who did not consume alcohol (Table 5).

Table 5.
Effects of interactions between SNPs and alcohol consumption on nasopharyngeal carcinoma risk.
4. Discussion
To the best of our knowledge, this study is the first to investigate the association between genetic variants in 19 ALDH isoform polymorphisms and the risk of NPC in an East Asian population residing in Taiwan. Besides the known alcohol metabolism genetic variant, rs671, we identified 12 SNPs located on the ALDH4A1, ALDH18A1, ALDH3B2, ALDH1L2, and ALDH1A2 genes from the ALDH multigene family that were associated with an elevated NPC risk.
ALDHs are a family of intracellular enzymes that are involved in aldehyde metabolism, cellular detoxification, differentiation, and cancer drug [28,29]. Several isoforms of the ALDH1 family (ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH1L1, and ALDH1L2) are used as cancer stem cell markers in a variety of cancers [29,30,31]. Strong correlations between ALDH1 expression in the invasive tumor front of NPC, epithelial-mesenchymal transition (EMT) and tumor aggressiveness suggest that ALDH1 expression in the invasive front of NPC could be a useful prognostic marker for NPC patients [20]. RNA sequencing data from The Cancer Genome Atlas (TCGA) database have revealed downregulated ALDH1A2 and ALDH1L1 expression in esophageal squamous cell carcinoma and HNC squamous cell carcinoma [21]. Meta-analysis results found that lower ALDH1A1 and ALDH1L1 expression was associated with poorer overall survival and poorer progression-free survival in cancer patients [21]. In our study, SNPs located on the ALDH1L2 and ALDH1A2 genes were associated with the risk of developing NPC. Decreased levels of ALDH1A1, ALDH1A2, ALDH1A3, and ALDH1L1 expression were observed in 5 pairwise samples of nasopharynx squamous cell carcinoma (the results are not shown).
Polymorphisms in genes responsible for the alcohol metabolism pathways can affect the amount of acetaldehyde and reactive oxygen species generated during the metabolic process, and thus alter the effects of alcohol and potentially influence carcinogenesis [14,15]. ADH1B Arg47His (rs1229984) and ALDH2 Glu487Lys (rs671) are the most frequently reported genetic polymorphisms related to alcohol metabolism. Both variants are not only related to alcohol metabolism but also to cancer risk. A 40-fold decrease in ADH1B activity has been observed in ADH1B His/His individuals [32], while a loss of ALDH2 enzyme activity has been observed in individuals with the ALDH2 Lys/Lys phenotype [14,33]. Many studies have demonstrated that the genetic effect of ADH1B and ALDH2 increase the risk of different types of cancers [34]. However, SNP rs1229884 in the ADH1B gene was not significantly associated with NPC in our Han Chinese patients in Taiwan, which is consistent with the results from previous meta-analyses showing that the ALDH2 polymorphism, but not the ADH1B polymorphism, significantly increases the risk of CRC in East Asians [14,35].
Other research has reported that heavy alcohol consumption can increase the risk of certain cancer types, including HNC cancers and NPC [11]. In studies involving East Asian populations, the presence of genetic polymorphisms in ADH1B (rs1229984) and ALDH2 (rs671), as well as alcohol consumption, individually or in combination [13], increase the risk of breast cancer [36], HNC [17], and esophageal cancer [18,35]. Moreover, research has shown that alcohol consumption affects two major folate-metabolizing enzymes, ALDH1L1 and ALDH1L2, with a possible synergistic effect on carcinogenesis [37,38]. In this study, SNPs rs671 located on ALDH2 and rs10778364 located on ALDH1L2 were significantly associated with an increased risk for NPC, with or without alcohol consumption. We also observed a synergistic effect between SNPs and alcohol consumption. These findings indicate that not only alcohol plays a role in the risk of NPC, but that the genetic effects of ALDH2 and ALDH1L2 are also important for NPC risk.
Inconsistent associations for alcohol consumption, betel nut chewing, and tobacco smoking have been recorded in previous studies [8,12]. This inconsistency may be due to differences in study populations, NPC subtypes, or definitions of lifestyle behaviors. A significant association between alcohol intake and NPC risk was observed in this study and other research [39,40,41], while several studies have observed a lack of association between alcohol and NPC risk [10,42,43]. We also observed that betel nut chewing was significantly associated with NPC risk. Although three previous studies found no such association [44], a positive association has been reported between betel nut chewing and NPC risk in NPC high-risk families in Taiwan [45]. A modestly increased risk of NPC associated with tobacco smoking has been reported in southern China [46], which is consistent with our study.
This study is apparently the first to discuss associations between the genetic variants of 19 ALDH isoforms and NPC. However, some limitations in this study must be noted. First, due to the low frequency of alcohol consumption and low frequency of risk alleles, the numbers in each subgroup for SNPs and alcohol interactions are small and the statistical power is limited. Second, recall bias may exist, since the information about alcohol, betel quid chewing, and cigarette smoking was collected by self-reported questionnaires. Third, selection bias may exist, since the NPC cases and controls were enrolled under different projects (a hospital for the NPC cases, whereas controls were recruited from communities throughout Taiwan). Fourth, different measurement scales used for alcohol consumption, betel nut chewing, and cigarette smoking in these two projects may have led to misclassification.
5. Conclusions
In conclusion, our data demonstrate that the risk of NPC is increased in the presence of genetic variants of different ALDH isoforms. The potential of using genetic variants of ALDH as biomarkers to help to identify potential screening populations for NPC awaits future investigations.
Author Contributions
Conceptualization, H.-H.C., C.-H.C. and W.-L.L.; Methodology, W.-L.L. and W.-H.S.; Formal Analysis, F.-C.C. and Y.-W.C.; Writing—Original Draft Preparation, W.-L.L.; Writing—Review & Editing, W.-L.L., C.-H.C., W.-H.S. and K.-P.C.; Resources, K.-P.C. and H.-H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by China Medical University Hospital (DMR-109-197, CMU108-MF-83 and CMU109-MF-38), the Ministry of Science and Technology (MOST 108-2314-B-182-026, MOST 108-2314-B-182A-108-MY3), Chang Gung Memorial Hospital, Taiwan (BMRPC18, CMRPG3J1253, CMRPG3C1913, CMRPD1H0351, CMRPD1J0111), and also by the “Chinese Medicine Research Center, China Medical University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (CMRC-CMA-6). We acknowledge the support given to C-H.C. by NIH award AAA11147 made to Prof. Daria Mochly-Rosen in Stanford University. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Chang Gung Medical Foundation, Taiwan (IRB number 103-7224B; date of approval for the original version, 13 February 2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors. The data are not publicly available due to ethical considerations.
Acknowledgments
The authors thank all the members of the Cancer Center at Chang Gung Memorial Hospital for their invaluable help. We would like to thank Iona J. MacDonald from China Medical University for her English language revision of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Yu, G.; Hsu, W.-L.; Coghill, A.E.; Yu, K.J.; Wang, C.-P.; Lou, P.-J.; Liu, Z.; Jones, K.; Vogt, A.; Wang, M.; et al. Whole-Exome Sequencing of Nasopharyngeal Carcinoma Families Reveals Novel Variants Potentially Involved in Nasopharyngeal Carcinoma. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Tse, K.-P.; Su, W.-H.; Chang, K.-P.; Tsang, N.-M.; Yu, C.-J.; Tang, P.; See, L.-C.; Hsueh, C.; Yang, M.-L.; Hao, S.-P.; et al. Genome-wide Association Study Reveals Multiple Nasopharyngeal Carcinoma-Associated Loci within the HLA Region at Chromosome 6p21.3. Am. J. Hum. Genet. 2009, 85, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef]
- Bei, J.-X.; Su, W.-H.; Yu, K.; Chin, Y.-M.; Lou, P.-J.; Hsu, W.-L.; McKay, J.D.; Chen, C.-J.; Chang, Y.-S.; Ching-Ching International Nasopharyngeal Carcinoma (NPC) Genetics Working Group. A GWAS Meta-analysis and Replication Study Identifies a Novel Locus within CLPTM1L/TERT Associated with Nasopharyngeal Carcinoma in Individuals of Chinese Ancestry. Cancer Epidemiol. Biomark. Prev. 2016, 25, 188–192. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Zheng, H.; Cheung, A.K.L.; Tang, C.; Ko, J.; Wong, B.W.Y.; Leong, M.M.L.; Sham, P.C.; Cheung, F.; Kwong, D.L.-W.; et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 3317–3322. [Google Scholar] [CrossRef] [Green Version]
- Okekpa, S.; Mydin, R.B.S.M.N.; Mangantig, E.; Azmi, N.S.A.; Zahari, S.N.S.; Kaur, G.; Musa, Y. Nasopharyngeal Carcinoma (NPC) Risk Factors: A Systematic Review and Meta-Analysis of the Association with Lifestyle, Diets, Socioeconomic and Sociodemographic in Asian Region. Asian Pac. J. Cancer Prev. 2019, 20, 3505–3514. [Google Scholar] [CrossRef]
- Polesel, J.; Franceschi, S.; Talamini, R.; Negri, E.; Barzan, L.; Montella, M.; Libra, M.; Vaccher, E.; Franchin, G.; La Vecchia, C.; et al. Tobacco smoking, alcohol drinking, and the risk of different histological types of nasopharyngeal cancer in a low-risk population. Oral Oncol. 2011, 47, 541–545. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, W.; Xie, C.; Wang, B.; Zhang, G.; Zhou, F. Nasopharyngeal carcinoma risk by histologic type in central China: Impact of smoking, alcohol and family history. Int. J. Cancer 2010, 129, 724–732. [Google Scholar] [CrossRef]
- Du, T.; Chen, K.; Zheng, S.; Bao, M.; Huang, Y.; Wu, K. Association Between Alcohol Consumption and Risk of Nasopharyngeal Carcinoma: A Comprehensive Meta-Analysis of Epidemiological Studies. Alcohol. Clin. Exp. Res. 2019, 43, 2262–2273. [Google Scholar] [CrossRef]
- Chen, L.; Gallicchio, L.; Boyd-Lindsley, K.; Tao, X.; Robinson, K.A.; Lam, T.K.; Herman, J.G.; Caulfield, L.E.; Guallar, E.; Alberg, A.J. Alcohol Consumption and the Risk of Nasopharyngeal Carcinoma: A Systematic Review. Nutr. Cancer 2008, 61, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gross, E.R.; Zambelli, V.O.; Small, B.A.; Ferreira, J.C.B.; Chen, C.-H.; Mochly-Rosen, D. A Personalized Medicine Approach for Asian Americans with the Aldehyde Dehydrogenase 2*2 Variant. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 107–127. [Google Scholar] [CrossRef] [Green Version]
- Crous-Bou, M.; Rennert, G.; Cuadras, D.; Salazar, R.; Cordero, D.; Rennert, H.S.; Lejbkowicz, F.; Kopelovich, L.; Lipkin, S.M.; Gruber, S.B.; et al. Polymorphisms in Alcohol Metabolism Genes ADH1B and ALDH2, Alcohol Consumption and Colorectal Cancer. PLoS ONE 2013, 8, e80158. [Google Scholar] [CrossRef] [Green Version]
- Seitz, H.K.; Stickel, F. Acetaldehyde as an underestimated risk factor for cancer development: Role of genetics in ethanol metabolism. Genes Nutr. 2010, 5, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Perez-Miller, S.J.; Hurley, T.D. Coenzyme Isomerization is Integral to Catalysis in Aldehyde Dehydrogenase†,‡. Biochemistry 2003, 42, 7100–7109. [Google Scholar] [CrossRef]
- Tsai, S.-T.; Wong, T.-Y.; Ou, C.-Y.; Fang, S.-Y.; Chen, K.-C.; Hsiao, J.-R.; Huang, C.-C.; Lee, W.-T.; Lo, H.-I.; Huang, J.-S.; et al. The interplay between alcohol consumption, oral hygiene, ALDH2 and ADH1B in the risk of head and neck cancer. Int. J. Cancer 2014, 135, 2424–2436. [Google Scholar] [CrossRef]
- Brooks, P.J.; Enoch, M.-A.; Goldman, D.; Li, T.-K.; Yokoyama, A. The Alcohol Flushing Response: An Unrecognized Risk Factor for Esophageal Cancer from Alcohol Consumption. PLoS Med. 2009, 6, e1000050. [Google Scholar] [CrossRef]
- Chen, M.-H.; Weng, J.-J.; Cheng, C.-T.; Wu, R.-C.; Huang, S.-C.; Wu, C.-E.; Chung, Y.-H.; Liu, C.-Y.; Chang, M.-H.; Chiang, K.-C.; et al. ALDH1A3, the Major Aldehyde Dehydrogenase Isoform in Human Cholangiocarcinoma Cells, Affects Prognosis and Gemcitabine Resistance in Cholangiocarcinoma Patients. Clin. Cancer Res. 2016, 22, 4225–4235. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.-R.; Yao, K.-T. Cancer stem cell characteristics, ALDH1 expression in the invasive front of nasopharyngeal carcinoma. Virchows Arch. 2014, 464, 35–43. [Google Scholar] [CrossRef]
- Chang, P.M.-H.; Chen, C.-H.; Yeh, C.-C.; Lu, H.-J.; Liu, T.-T.; Chen, M.-H.; Liu, C.-Y.; Wu, A.T.H.; Yang, M.-H.; Tai, S.-K.; et al. Transcriptome analysis and prognosis of ALDH isoforms in human cancer. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Vasiliou, V.; Nebert, D.W. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum. Genom. 2005, 2, 138–143. [Google Scholar] [CrossRef] [Green Version]
- Su, W.-H.; Shugart, Y.Y.; Chang, K.-P.; Tsang, N.-M.; Tse, K.-P.; Chang, Y.-S. How Genome-Wide SNP-SNP Interactions Relate to Nasopharyngeal Carcinoma Susceptibility. PLoS ONE 2013, 8, e83034. [Google Scholar] [CrossRef]
- Wei, C.-Y.; Yang, J.-H.; Yeh, E.-C.; Tsai, M.-F.; Kao, H.-J.; Lo, C.-Z.; Chang, L.-P.; Lin, W.-J.; Hsieh, F.-J.; Belsare, S.; et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom. Med. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Howie, B.N.; Donnelly, P.; Marchini, J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.D.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sham, P.C.; Purcell, S.M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 2014, 15, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Moreb, J.; Schweder, M.; Suresh, A.; Zucali, J.R. Overexpression of the human aldehyde dehydrogenase class I results in in-creased resistance to 4-hydroperoxycyclophosphamide. Cancer Gene Ther. 1996, 3, 24–30. [Google Scholar]
- Yang, C.; Wang, X.; Liao, X.; Han, C.; Yu, T.; Qin, W.; Zhu, G.; Su, H.; Yu, L.; Liu, X.; et al. Aldehyde dehydrogenase 1 (ALDH1) isoform expression and potential clinical implications in hepatocellular carcinoma. PLoS ONE 2017, 12, e0182208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikawa, S.; Konno, M.; Hamabe, A.; Hasegawa, S.; Kano, Y.; Ohta, K.; Fukusumi, T.; Sakai, D.; Kudo, T.; Haraguchi, N.; et al. Aldehyde dehydrogenasehigh gastric cancer stem cells are resistant to chemotherapy. Int. J. Oncol. 2013, 42, 1437–1442. [Google Scholar] [CrossRef] [Green Version]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, K.; Hiraki, A.; Hirose, K.; Ito, H.; Suzuki, T.; Wakai, K.; Tajima, K. Impact of theAlcohol-Dehydrogenase(ADH)1CandADH1Bpolymorphisms on drinking behavior in nonalcoholic Japanese. Hum. Mutat. 2007, 28, 506–510. [Google Scholar] [CrossRef]
- Wang, W.; Wang, C.; Xu, H.; Gao, Y. Aldehyde Dehydrogenase, Liver Disease and Cancer. Int. J. Biol. Sci. 2020, 16, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.Y.; Ning, N. Association of ADH1B Arg47His polymorphism with the risk of cancer: A meta-analysis. Biosci. Rep. 2019, 39, BSR20181915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-J.; Yokoyama, A.; Yokoyama, T.; Huang, Y.-C.; Wu, S.-Y.; Shao, Y.; Niu, J.; Wang, J.; Liu, Y.; Zhou, X.-Q.; et al. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: A meta-analysis. World J. Gastroenterol. 2010, 16, 4210–4220. [Google Scholar] [CrossRef]
- Park, B.; Kim, J.-H.; Lee, E.-G.; Jung, S.-Y.; Lee, S.Y.; Kang, H.-S.; Han, J.H. Role of aldehyde dehydrogenases, alcohol dehydrogenase 1B genotype, alcohol consumption, and their combination in breast cancer in East-Asian women. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Krupenko, S.A.; Krupenko, N.I. ALDH1L1 and ALDH1L2 Folate Regulatory Enzymes in Cancer. Adv. Exp. Med. Biol. 2018, 1032, 127–143. [Google Scholar] [CrossRef]
- Krupenko, S.A.; Horita, D.A. The Role of Single-Nucleotide Polymorphisms in the Function of Candidate Tumor Suppressor ALDH1L1. Front. Genet. 2019, 10, 1013. [Google Scholar] [CrossRef]
- Lourembam, D.S.; Singh, A.R.; Sharma, T.D.; Singh, T.S.; Singh, T.R.; Singh, L.S. Evaluation of Risk Factors for Nasopharyngeal Carcinoma in a High-risk Area of India, the Northeastern Region. Asian Pac. J. Cancer Prev. 2015, 16, 4927–4935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.K.; Singh, A.S.; Mondal, R.; Kapfo, W.; Khamo, V.; Singh, Y.I. Dysfunction of mitochondria due to environmental carcinogens in nasopharyngeal carcinoma in the ethnic group of Northeast Indian population. Tumor Biol. 2014, 35, 6715–6724. [Google Scholar] [CrossRef] [PubMed]
- Lakhanpal, M.; Singh, L.C.; Rahman, T.; Sharma, J.; Singh, M.M.; Kataki, A.C.; Verma, S.; Chauhan, P.S.; Singh, Y.M.; Wajid, S.; et al. Contribution of susceptibility locus at HLA class I region and environmental factors to occurrence of nasopharyngeal cancer in Northeast India. Tumor Biol. 2014, 36, 3061–3073. [Google Scholar] [CrossRef]
- Yong, S.K.; Ha, T.C.; Yeo, M.C.R.; Gaborieau, V.; McKay, J.D.; Wee, J. Associations of lifestyle and diet with the risk of nasopharyngeal carcinoma in Singapore: A case–control study. Chin. J. Cancer 2017, 36, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.W.; Chang, W.S.; Gong, C.L.; Shih, L.C.; Chen, L.Y.; Lin, E.Y.; Li, H.T.; Yen, S.T.; Wu, C.N.; Bau, D.T. Contribution of Matrix Metallopeptidase-1 Genotypes, Smoking, Alcohol Drinking and Areca Chewing to Nasopharyngeal Carcinoma Sus-ceptibility. Anticancer Res. 2016, 36, 3335–3340. [Google Scholar] [PubMed]
- Chang, E.T.; Ye, W.; Zeng, Y.-X.; Adami, H.-O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Diehl, S.; Pfeiffer, R.; Chen, C.-J.; Hsu, W.-L.; Dosemeci, M.; Cheng, Y.-J.; Sun, B.; Goldstein, A.M.; Hildesheim, A. Evaluation of Risk Factors for Nasopharyngeal Carcinoma in High-Risk Nasopharyngeal Carcinoma Families in Taiwan. Cancer Epidemiol. Biomark. Prev. 2005, 14, 900–905. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.T.; Liu, Z.; Hildesheim, A.; Liu, Q.; Cai, Y.; Zhang, Z.; Chen, G.; Xie, S.-H.; Cao, S.-M.; Shao, J.-Y.; et al. Active and Passive Smoking and Risk of Nasopharyngeal Carcinoma: A Population-Based Case-Control Study in Southern China. Am. J. Epidemiol. 2017, 185, 1272–1280. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).