RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application
Abstract
1. Introduction
2. Theory
2.1. Backtracking from External Memory
2.2. Increased Memory Efficiency by Reduced Redundancy
2.3. Performance Analysis and Implementation
2.4. New Options in RNALfold
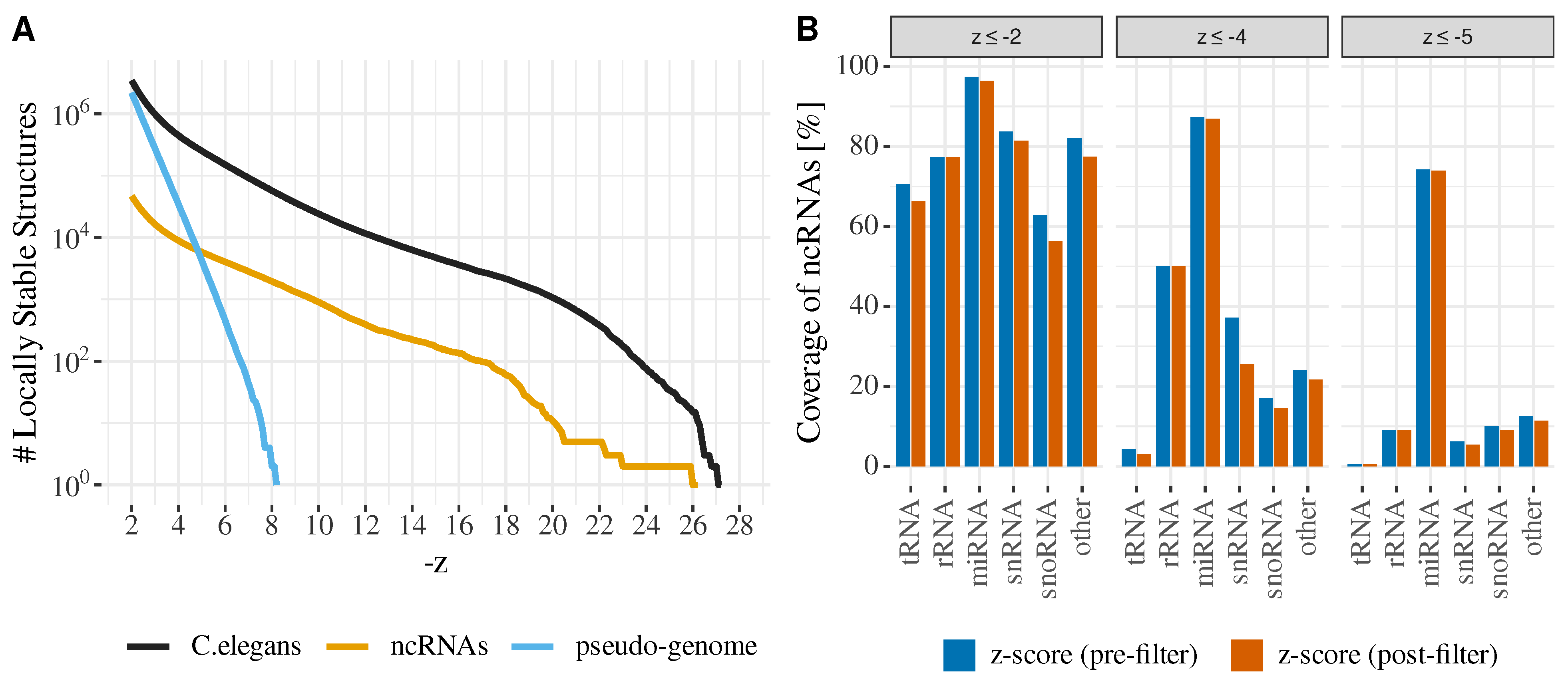
3. Application: Scanning Genomes for “Hyper-Stable” RNA Structures
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MFE | Minimum free energy (secondary structure) |
| miRNA | microRNA |
| ncRNA | Non-coding RNA |
| MEA | Maximum expected accuracy (secondary structure) |
| TLA | Three letter acronym |
References
- Doshi, K.; Cannone, J.; Cobaugh, C.; Gutell, R. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinform. 2004, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Proctor, J.R.P.; Meyer, I.M. CoFold: An RNA secondary structure prediction method that takes co-transcriptional folding into account. Nucleic Acids Res. 2013, 41, e102. [Google Scholar] [CrossRef] [PubMed]
- Amman, F.; Bernhart, S.H.; Doose, G.; Hofacker, I.L.; Qin, J.; Stadler, P.F.; The Students of the Bioinformatics II Lab Class 2013; Will, S. The Trouble with Long-Range Base Pairs in RNA Folding. In Advances in Bioinformatics and Computational Biology, 8th BSB; Setubal, J.C., Almeida, N.F., Eds.; Lect. Notes Comp. Sci.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 8213, pp. 1–11. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Priwitzer, B.; Stadler, P.F. Prediction of Locally Stable RNA Secondary Structures for Genome-Wide Surveys. Bioinformatics 2004, 20, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Kiryu, H.; Kin, T.; Asai, K. Rfold: An exact algorithm for computing local base pairing probabilities. Bioinformatics 2008, 24, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.J.; Daniel, M.; Möhl, M.; Joshua, J.N.; Brown, C.M.; Backofen, R. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012, 40, 5215–5226. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast Folding and Comparison of RNA Secondary Structures. Monatsh. Chem. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Turner, D.H.; Mathews, D.H. NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucl. Acids Res. 2010, 38, D280–D282. [Google Scholar] [CrossRef]
- Do, C.; Woods, D.; Batzoglou, S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 2006, 22, e90–e98. [Google Scholar] [CrossRef]
- Lu, Z.J.; Gloor, J.W.; Mathews, D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA 2009, 15, 1805–1813. [Google Scholar] [CrossRef]
- Bernhart, S.; Hofacker, I.L.; Stadler, P.F. Local RNA Base Pairing Probabilities in Large Sequences. Bioinformatics 2006, 22, 614–615. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Piecznik, G.; Griggs, J.R.; Kleitman, D.J. Algorithms for Loop Matching. SIAM J. Appl. Math. 1978, 35, 68–82. [Google Scholar] [CrossRef]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 2005, 11, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Kiryu, H.; Sato, K.; Mituyama, T.; Asai, K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 2009, 25, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Sato, K.; Asai, K. Prediction of RNA secondary structure by maximizing pseudo-expected accuracy. BMC Bioinform. 2010, 11, 586. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Hofacker, I.L.; Stadler, P.F. RNA Folding with Hard and Soft Constraints. Algorithms Mol. Biol. 2016, 11, 8. [Google Scholar] [CrossRef]
- Le, S.Y.; Maizel, J.V., Jr. A method for assessing the statistical significance of RNA folding. J. Theor. Biol. 1989, 138, 495–510. [Google Scholar] [CrossRef]
- Rivas, E.; Eddy, S.R. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics 2000, 16, 583–605. [Google Scholar] [CrossRef]
- Clote, P.; Ferré, F.; Kranakis, E.; Krizanc, D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA 2005, 11, 578–591. [Google Scholar] [CrossRef]
- Freyhult, E.; Gardner, P.P.; Moulton, V. A comparison of RNA folding measures. BMC Bioinform. 2005, 6, 241. [Google Scholar] [CrossRef]
- Washietl, S.; Hofacker, I.L.; Stadler, P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 2454–2459. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Findeiß, S.; Washietl, S.; Hofacker, I.L.; Stadler, P.F. RNAz 2.0: Improved noncoding RNA detection. Pac. Symp. Biocomput. 2010, 15, 69–79. [Google Scholar] [CrossRef]
- Klein, R.J.; Misulovin, Z.; Eddy, S.R. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl. Acad. Sci. USA 2002, 99, 7542–7547. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Anderson, J.; Gillespie, J.; Mayne, M. uShuffle: A useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinform. 2008, 9, 192. [Google Scholar] [CrossRef]
- Ciesiolka, A.; Jazurek, M.; Drazkowska, K.; Krzyzosiak, W.J. Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front. Cell. Neurosci. 2017, 11, 97. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenz, R.; Stadler, P.F. RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application. Genes 2021, 12, 14. https://doi.org/10.3390/genes12010014
Lorenz R, Stadler PF. RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application. Genes. 2021; 12(1):14. https://doi.org/10.3390/genes12010014
Chicago/Turabian StyleLorenz, Ronny, and Peter F. Stadler. 2021. "RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application" Genes 12, no. 1: 14. https://doi.org/10.3390/genes12010014
APA StyleLorenz, R., & Stadler, P. F. (2021). RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application. Genes, 12(1), 14. https://doi.org/10.3390/genes12010014





