Abstract
The genus Helianthus is a diverse taxonomic group with approximately 50 species. Most sunflower genomic investigations are devoted to economically valuable species, e.g., H. annuus, while other Helianthus species, especially perennial, are predominantly a blind spot. In the current study, we have assembled the complete mitogenomes of two perennial species: H. grosseserratus (273,543 bp) and H. strumosus (281,055 bp). We analyzed their sequences and gene profiles in comparison to the available complete mitogenomes of H. annuus. Except for sdh4 and trnA-UGC, both perennial sunflower species had the same gene content and almost identical protein-coding sequences when compared with each other and with annual sunflowers (H. annuus). Common mitochondrial open reading frames (ORFs) (orf117, orf139, and orf334) in sunflowers and unique ORFs for H. grosseserratus (orf633) and H. strumosus (orf126, orf184, orf207) were identified. The maintenance of plastid-derived coding sequences in the mitogenomes of both annual and perennial sunflowers and the low frequency of nonsynonymous mutations point at an extremely low variability of mitochondrial DNA (mtDNA) coding sequences in the Helianthus genus.
1. Introduction
From a genomic point of view, plants are unique organisms, since their cells have three genetic systems—nuclear, plastid, and mitochondrial—which function independently, but are simultaneously intimately interconnected. Nuclear genomes are of primary interest, while cytoplasmic genomes, especially mitochondrial, are often underestimated [1]. Mitochondrial DNA (mtDNA) in higher plants usually has complex multipartite structures, frequent insertions, and rapid rearrangements [2]. Mitochondrial genomes show significant variability not only between species but sometimes even within the same species [3]. Thus, even in species whose nuclear genomes have been investigated, the mitogenomic information is often lacking or incomplete [4].
The genus Helianthus is a diverse taxonomic group with approximately 50 species [5], divided into four sections: two annual sections, Helianthus and Agrestis, and two perennial sections, Ciliares and Divaricati [6]. Most sunflower genomic investigations are devoted to the economically valuable species, such as the annual species H. annuus, and in the case of the perennial species, H. tuberosus, while the other Helianthus species, especially perennial, are predominantly a blind spot. To the best of our knowledge, very few studies of perennial sunflower mtDNA are available [7]. Moreover, to date, the complete mitochondrial genomes of only one species—H. annuus—are deposited in GenBank. Mitochondrial DNA of cytoplasmic male sterility (CMS) sources, obtained from wild progenitors, may be considered as exceptions—for instance, the MAX1 CMS mitogenome [8] was gained through hybridization with Helianthus maximiliani Schrad as a maternal form. Since CMS systems are essential for the hybrid seed production of crops [9], particularly of sunflowers [10], the investigations of mitochondrial genomes of related wild species are also of interest for agricultural purposes.
As with H. maximiliani, the two species H. grosseserratus and H. strumosus represent perennial species and belong to the section Divaricati. H. maximiliani and H. grosseserratus are diploid, whereas H. strumosus can be either tetraploid (2n = 4x = 68) or hexaploid (2n = 6x = 102) [6]. The crossability between diploid species in the genus Helianthus is relatively good. However, while crossing tetraploid or hexaploid species is possible, there are considerable problems which require the application of biotechnological methods [11]. Many sunflower species, mostly diploid ones, have been used to develop new CMS systems for hybrid breeding in sunflowers, but very little is known about the general effects of different cytoplasms on sunflower performance. For instance, in field trials of Jan et al. [12] annual and perennial sunflower species were used to gain 20 cytoplasmic substitution lines, which were compared with the HA89 line. Although alloplasmic lines with cytoplasms of wild species showed predominantly neutral or negative effects on agronomic traits, such as an elevated level of lodging or reduced yield, the beneficial effects were also mentioned [12]. The characteristics of the achene walls are important for the industrial processing of sunflower seeds. H. grosseserratus, H. maximiliani and H. strumosus are characterized by a thin pericarp with a reduced sclerenchymatic layer, which represents an interesting trait in breeding for improved hullability [13].
In the current study, we assembled the complete mitogenomes of two perennial species: H. grosseserratus and H. strumosus. We analyzed their sequences and gene profiles in comparison to the available complete mitogenomes of H. annuus (MN175741.1, MN171345.1, MH704580.1, MG770607.2, MG735191.1, NC_023337.1, CM007908.1).
2. Materials and Methods
2.1. Plant Material and Mitochondrial DNA Extraction
The perennial species of Helianthus: H. grosseserratus Martens and H. strumosus L. (Figure S1) were obtained from the collection of the N. I. Vavilov All-Russian Institute of Plant Genetic Resources. Plant leaves were used for mitochondrial DNA isolation. For H. strumosus, the previously described method was used [14]. But for H. grosseserratus, we used a slightly different technique of multi-step centrifugations. but for H. grosseserratus, Leaves (5 g, without petiole and midrib) were homogenized by mortar and pestle in 20 ml of STE buffer (0.4 M sucrose, 50 mM Tris pH 7.8, 4 mM EDTA-Na2, 0.2% bovine serum albumin, 0.2% 2-mercaptoethanol) and then centrifuged using several steps: (1) 500 g for 5 min, picking the supernatant, (2) 4000 g for 5 min, picking the supernatant, (3) 10,000 g for 15 min, discarding the supernatant. The pellet was treated using 10 units of DNAse (Syntol, Moscow, Russia) for 7 min and then used for DNA isolation. For both samples, the DNA extraction was performed with a PhytoSorb kit (Syntol, Moscow, Russia), according to the manufacturer’s protocol.
2.2. Next-Generation Sequencing
Next-generation sequencing (NGS) libraries were made with a Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA), following the guidelines of Illumina. The quality and quantity of the libraries were evaluated with the Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and with a Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on different runs of NextSeq 500 (Illumina, San Diego, CA, USA) with the NextSeq 500/550 High Output Kit v2.5: 150 cycles for H. grosseserratus and 300 cycles for H. strumosus. A total number of 1,349,630 75-bp paired reads and 3,014,579 150-bp paired reads were generated.
2.3. Mitochondrial Genome Assembly and Annotation
For quality control of the reads, we used FastQC v 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic v 0.39 software [15]. The generation of contigs was done with SPAdes Genome Assembler v 3.13.1 [16] using different k-mer values. The whole mitochondrial genome assembly was based on high coverage (depths > 50) contigs, using Bandage v 0.8.1 [17] program for visualizing de novo assembly graphs. The genome assembly was validated by remapping reads by Bowtie 2 v 2.3.5.1 [18] with visual revision of coverage uniformity (especially in the junctions of contigs) using Tablet v 1.19.09.03 [19]. Low coverage (depths < 15) regions of the H. strumosus mitochondrial genome were also verified by Sanger sequencing. The mitochondrial genomes were annotated with MITOFY [20], BLAST tool [21] and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder). Graphical genome maps were generated using the OGDRAW tool [22]. The annotated mitochondrial genomes of H. grosseserratus and H. strumosus, were submitted to the NCBI GenBank database (assigned accessions MT588180 and MT588181, respectively).
3. Results
First of all, it should be mentioned that different mitochondrial fraction isolation techniques showed significant variations in the amount of mtDNA derived sequences for the two perennial Helianthus species. In H. strumosus, only about 3.9% of the reads mapped to the mitogenome, whereas the NGS library of H. grosseserratus predominantly contained mitochondrial reads (67.3%). Such difference significantly influenced the complexity of mitogenome assembly. In the case of H. grosseserratus, a relatively small amount (1.3 mln) of short (75 bp) reads were obtained, and the assembly with a k-mer value equal to 65 generated only 14 high coverage (depths > 50) large contigs (N50 = 55,431) which allowed a quick scaffolding. In the case of H. strumous, twice as many elongated reads (3 mln, 150 bp) were observed, but many smaller contigs (N50 = 4,632), containing nuclear and chloroplast sequences, were present, which in turn increased the complexity of scaffolding. Additional filtering of mitochondrial contigs had to be performed in the case of H. strumosus mitochondrial genome assembly. Thus, for mitogenome assembly, the proportion of mtDNA in a sample is more crucial than the number of reads.
For both of the studied sunflower species, master circles of the mitochondrial chromosomes were gained (Figure 1), with lengths of 273,543 bp and 281,055 bp for H. grosseserratus and H. strumosus, respectively. Mitochondrial genomes often have a complex architecture, including sub-genomes or multichromosomal organization [23,24]. However, in the current study, we did not analyze the mtDNA stoichiometry and only focused on the master circle of the mitogenome type.
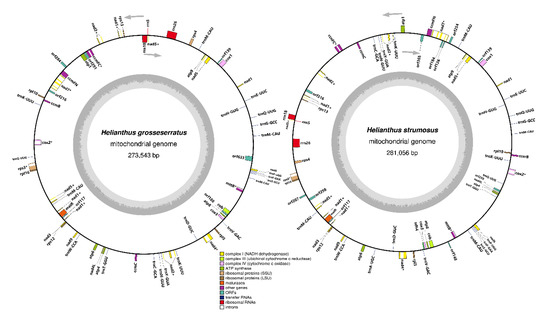
Figure 1.
Mitochondrial genome maps of H. grosseserratus and H. strumosus. Arrows denote the gene’s transcription orientation. Intron-containing genes are marked by an asterisk (*) symbol and trans-spliced genes—by plus (+) symbol.
The mtDNA alignment of perennial sunflower species with H. annuus allowed the identification of 12 syntenic blocks (>1 kbp, >98% similarity), as seen in Figure 2. Thus, the majority (>80%) of mitogenomes is similar for all three species, but there are also unique sequences. The orders of syntenic blocks vary significantly among species and may be considered as rearrangements. Nevertheless, there are common changes for perennial species, e.g., the adjunction of the 3rd and 8th blocks. Among other mtDNA structural differences, the variations in large repeat regions are of interest. In the H. annuus mitogenome, there are two large (>12.5 kbp) repeat regions, but in H. grosseserratus, these repeats are reduced to about 4.5 kbp, and in the case of H. strumosus—to about 1.1 kbp. The most prolonged repeat in H. grosseserratus comprises about 9.4 kbp, whereas in H. strumosus it counts only 7.1 kbp (Supplemental File 2).
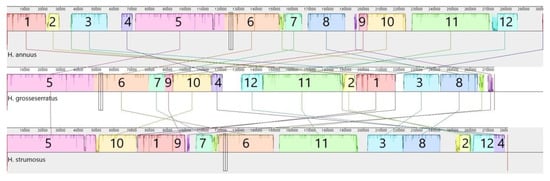
Figure 2.
The progressive MAUVE alignment of H. annuus, H. grosseserratus and H. strumosus complete mitogenomes. According to sequence similarity, 12 syntenic blocks were identified, with a different order for each species.
The mitochondrial gene content was almost the same in the two perennial sunflower species, counting 31–32 protein-coding, three rRNA, 21–22 tRNA genes, and some open reading frames (ORFs). The mtDNA holds most genes that are typical for plant mitogenomes: NADH:ubiquinone oxidoreductase (Complex I) genes (nad1, -2, -3, -4, -4L, -5, -6, -7, and -9), cob (Complex III) genes, cytochrome c oxidase (Complex IV) genes (cox1, -2, and -3), ATP synthase (Complex V) genes (atp1, -4, -8, and -9), cytochrome c biogenesis genes (ccmB, -C, -FC, and -FN), matR, mttB and ribosomal protein genes (rps3, -4, -12, and -13, rpl-5, -10, and -16). In the mitogenomes of the two perennial sunflowers, succinate dehydrogenase (Complex II) genes are only represented by sdh4 in H. strumosus, but in the case of H. grosseserratus, sdh4 is a pseudogene, which has an internal stop codon. In the studied mitogenomes, nine intron-containing genes were identified. Five of the nine genes (nad1, nad2, nad4, nad5, and nad7) belong to mitochondrial complex I, and the remaining three genes were ccmFC, cox2, mttB, and rps3. The trans-spliced introns were observed in nad1, nad2, and nad5, which is consistent with angiosperm plants [25]. Other genes had a single cis-spliced intron, except nad7 which counted four introns. The introns’ locations and splicing were similar to those observed in the mitochondrial genes of other flowering plants [26,27].
While comparing the amino acid sequences of the mitochondrial proteins of perennial sunflowers with annual sunflowers (NCBI accession MN171345.1), a limited number of differences could be detected (Table 1). The most severe changes concerned atp6 and nad6 in the mtDNA of H. grosseserratus. In atp6 two nucleotide changes in the 37th codon resulted in the terminal leading to the N-terminal shortening of ATPase subunit 6, whereas in nad6, a frameshift led to a premature stop codon, resulting in C-terminal truncation of the encoded protein.

Table 1.
Single nucleotide polymorphisms localized in protein-coding genes of perennial sunflower species.
H. grosseserratus mitogenome includes 21 tRNA genes corresponding to 16 amino acids. The same tRNA genes are present in H. strumosus mtDNA, but an additional one (trnA-UGC) allows, in summary, to carry 17 amino acids by the mitogenome of H. strumosus. The in silico prediction of new open reading frames (ORFs) provided a large number of long (more than 300 bp) ORFs in both species. Meanwhile, we annotated only those (Table 2) with significant similarity to proteins and ORFs (according to the protein blast search) in sunflowers or other Magnoliophyta.

Table 2.
ORFs annotated in H. grosseserratus and H. strumosus mitogenomes. The ORFs are named by their amino acid length. orf148—present in H. annuus with PET2 CMS type, orf188—similar ORF presented in H. annuus with ANN2 CMS type.
Among the discovered ORFs, we observed several interesting patterns. For instance, we discovered the presence of an ORF encoding the chloroplast-like ribosomal protein S11, in both currently studied species and all up-to-date, complete mitochondrial genomes of Helianthus in GenBank (MN175741.1, MN171345.1, MH704580.1, MG770607.2, MG735191.1, NC_023337.1, CM007908.1). Different ORFs—orf284 (H. grosseserratus), orf334 (H. annuus), and orf365 (H. strumosus)—are encoding psaA-like proteins, which have almost identical amino acid sequences at the N-terminus. Chimeric mitochondrial genes, including cox2 fragments, are often linked to male sterility inducing mtDNA of plants [28]. The orf188 encoded protein contains 58 amino acids identical to the N-terminus of cytochrome oxidase subunit 2. While the orf316, present in H. annuus and H. strumosus, has 67 amino acids similar to the middle part of the COX2 sequence. The protein encoded by orf188 in H. grosseserratus shares almost the same structure as the cox2-chimeric protein (QFS00065.1), which we identified in ANN2, a CMS line of sunflower in a previous study [29]. According to GenBank data, orf316 is an ORF common to sunflower mtDNA. Notably, an uncharacterized protein from the cupredoxin superfamily (XP_022040088.1) annotated in the nuclear genome of H. annuus has a sequence equal to the orf316 encoded protein. The largest ORF (orf633) was observed in the H. grosseserratus mitogenome. Most of the orf633 encoded protein (397 aa) is identical to the N-terminus of ATP ATPase subunit 1.
4. Discussion
The mitochondrial genomes of perennial sunflower species are 7–9% smaller than annual sunflower mitogenomes. Meanwhile, except for the sdh4 gene, annual and perennial sunflowers share the same protein-coding gene content. The sdh3 and shd4 genes are among the most “unstable” mitochondrial genes, with frequent cases of their transfer to the nucleus, and even related species may have different sets of succinate dehydrogenase genes [25]. The mitochondrial gene sequences showed slight differences between H. grosseserratus and H. strumosus, as well as between H. annuus and both perennial species. Only a few SNPs (1–3 per gene) leading to nonsynonymous substitutions occurred in rps4, cob, rpl16, matR, and atp6. Among the detected polymorphisms, only a single transversion (G to C) in the atp6 gene (Table 1) was common for both perennial species in comparison to H. annuus. Notably, the same SNP in atp6 was observed in the sunflower line with MAX1 (MH704580.1) CMS type, but was absent in PET1 (MG735191.1), PET2 (MG770607.2) CMS lines. Since the CMS lines have cytoplasmic genetic information (plastid and mitochondrial) initially obtained from wild species [30], PET1 and PET2 may be considered as H. petiolaris mitogenomes, and MAX1 as a H. maximiliani mitogenome. This SNP can likely be tracked only in perennial sunflowers, or at least those belonging to the Divaricati section. Sunflower species have unusually diverse karyotypes [31] and high rates of karyotypic evolution [32,33]. Within annual and perennial sunflower diploid species, there is a moderate variation in the sequence of plastid genes and quite a high variation in the nuclear genes [34]. Opposite to nuclear and plastid genes, an extremely low polymorphism level was found in sunflower mitochondrial genes. While only a few species were investigated, future research will clarify this feature. Similar conservation of mitogenome sequences can be observed in other species. Recently, a comparison of the domesticated lettuce (Lactuca sativa) mitogenome with two wild lettuce species (L. saligna and L. serriola) revealed identical sequences and rearrangements for L. sativa and L. serriola, but significant differences with the mitogenome of L. saligna [24]. Such data indicate that domestication had little influence on the mitochondrial genome as L. serriola is regarded as a wild ancestor of the domesticated lettuce [35].
Many mitochondrial genomes have abounded with foreign DNA acquired by horizontal transfer, especially from plastid genomes [36]. However, the functional activity of such plastid-originated insertions is of particular interest. The mitochondrial rps11 gene is excluded from mtDNA in a vast number of eudicots [25,37]. The presence of orf139, encoding a protein that is similar to the chloroplast-like ribosomal protein S11, in the same position (close to cox1 gene) of all up-to-date sunflower mitogenomes, does not seem accidental. The non-coding sequences in plant mitogenomes vary greatly and may rearrange [38], while the coding sequences are conservative [39,40], being under strong purifying selection [41]. Since orf139 has an identical coding sequence in both perennial sunflowers and all up-to-date H. annuus mitogenomes (including CMS lines), we can assume that orf139 plays a vital role in sunflower mitochondria. The functions of the ORFs similar to the psaA gene, are more cryptic. Three ORFs (orf284, orf334, and orf365) have a large identical part and have the same position in the mitogenome—between atp1 and ccmFN (much closer to ccmFN). Thus, it is most likely that the chloroplast DNA (cpDNA) insertion entered the mitochondrial genome in a common sunflower ancestor, and then a limited number of mutations resulted in sequence divergence between these ORFs. Notably, cpDNA-derived sequences with partial homology to the psaA gene have been maintained over long periods in the mtDNA of different Brassica species [42].
Because of high mitochondrial DNA recombination rates, ORFs with partial mitochondrial gene sequences are common in mtDNA [40]. Such chimeric genes often cause the CMS phenotype in plants [28,43]. The orf188 that we annotated in the H. grosseserratus mitogenome is similar to the previously described cox2-chimeric ORF (QFS00065.1), potentially playing a role in the formation of ANN2 CMS type in sunflower [29]. Nevertheless, the coding sequences of these two ORFs are not identical (about 76% similarity), but they have the same appearance in mtDNA. The other notable chimeric ORF in H. grosseserratus is orf633, which has no analogs, among other described sunflower mitogenomes. The orf633 has a large part of sequence which is identical to the atp1 gene, but its role is unclear. Studies have revealed the association between the CMS phenotype with mutations in atp1 or a decrease in protein abundance [44]. However, to the best of our knowledge, there are no data about new atp1-chimeric ORFs involved in CMS. Since the subunit α (F1 sector) of ATPase (atp1 encoded protein) lacks transmembrane domains, the association between additional rearranged atp1 copy (orf633) and CMS phenotype is equivocal. Mitogenomes of higher plants are enriched with ORFs with an unknown function [45,46], while the CMS phenotype is often associated with unusual ORFs present in the mtDNA [47]. Thus, the current study may help to understand whether the ORFs discovered in future research will be new or standard for the sunflower mitogenomes.
5. Conclusions
The complete master circles of mitogenomes were obtained for H. grosseserratus (273,543 bp) and H. strumosus (281,055 bp). Except for sdh4 and trnA-UGC, both perennial sunflower species had the same gene content and almost identical protein-coding sequences when were compared with each other or with the annual sunflower (H. annuus). Mitochondrial ORFs (orf117, orf139, orf334) common to sunflowers and unique ORFs for H. grosseserratus (orf633) and H. strumosus (orf126, orf184, orf207) were determined. The observed maintenance of plastid-derived DNA coding sequences in the mitogenomes of both annual and perennial sunflowers and the low frequency of nonsynonymous mutations point at an extremely low variability of mtDNA coding sequences in Helianthus genus. The current investigation may be useful for future studies in mitochondrial genomics.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/9/982/s1, Figure S1: the appearance of perennial sunflowers, Table S1: large (>200 bp) repeats in mitogenomes of perennial sunflowers.
Author Contributions
Conceptualization, A.U.; Data curation, T.T., K.A. and V.G.; Formal analysis, M.M. and A.K.; Funding acquisition, M.M. and A.U.; Investigation, M.M., K.A. and A.K.; Project administration, A.U.; Resources, V.G.; Software, T.T.; Supervision, A.U., V.G. and R.H.; Validation, M.M. and A.K.; Writing—original draft, M.M., K.A. and R.H.; Writing—review and editing, M.M., T.T. and R.H. All authors have read and agreed to the published version of the manuscript.
Funding
The reported study was funded by RFBR, project number 19-34-60006; the experimental work (nucleic acid extraction, NGS library preparation) was supported by the Ministry of Education and Science of Russia, project No. 6.929.2017/4.6. The NGS sequencing was provided with the support of budgetary subsidy to IITP RAS (Laboratory of Plant Genomics 0053-2019-0005).
Acknowledgments
We thank reviewers and the editor for their insightful suggestions and comments on the paper, which has helped us to improve the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Flood, P.J.; Theeuwen, T.P.J.M.; Schneeberger, K.; Keizer, P.; Kruijer, W.; Severing, E.; Kouklas, E.; Hageman, J.A.; Wijfjes, R.; Calvo-Baltanas, V.; et al. Reciprocal cybrids reveal how organellar genomes affect plant phenotypes. Nat. Plants 2020, 6, 13–21. [Google Scholar] [CrossRef]
- Liu, H.; Cui, P.; Zhan, K.; Lin, Q.; Zhuo, G.; Guo, X.; Ding, F.; Yang, W.; Liu, D.; Hu, S.; et al. Comparative analysis of mitochondrial genomes between a wheat K-type cytoplasmic male sterility (CMS) line and its maintainer line. BMC Genom. 2011, 12, 163. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Logacheva, M.D.; Schelkunov, M.I.; Fesenko, A.N.; Kasianov, A.S.; Penin, A.A. Mitochondrial genome of Fagopyrum esculentum and the genetic diversity of extranuclear genomes in buckwheat. Plants 2020, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Seiler, G.J.; Rieseberg, L.H. Systematics, origin, and germplasm resources of the wild and domesticated sunflower. In Sunflower Technology and Production; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 21–65. ISBN 978-0-89118-227-6. [Google Scholar]
- Timme, R.E.; Simpson, B.B.; Linder, C.R. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S–26S ribosomal DNA external transcribed spacer. Am. J. Bot. 2007, 94, 1837–1852. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Kane, N.C.; Ebert, D.P.; Rieseberg, L.H. Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: Neither from Jerusalem nor an artichoke. New Phytol. 2014, 201, 1021–1030. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Horn, R. Characterization of the mitochondrial genome of the MAX1 type of cytoplasmic male-sterile sunflower. BMC Plant Biol. 2019, 19, 51. [Google Scholar] [CrossRef]
- Garcia, L.; Edera, A.; Marfil, C.; Sanchez-Puerta, M. Male sterility and somatic hybridization in plant breeding. Preprints 2019. [Google Scholar] [CrossRef]
- Tyagi, V.; Dhillon, S.K.; Kaur, G.; Kaushik, P. Heterotic effect of different cytoplasmic combinations in sunflower hybrids cultivated under diverse irrigation regimes. Plants 2020, 9, 465. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Jan, C.-C.; Seiler, G.J.; Hammond, J.J. Effect of wild Helianthus cytoplasms on agronomic and oil characteristics of cultivated sunflower (Helianthus annuus L.). Plant Breed. 2014, 133, 262–267. [Google Scholar] [CrossRef]
- Jocković, J.; Rajčević, N.; Terzić, S.; Zorić, L.; Jocković, M.; Miladinović, D.; Luković, J. Pericarp features of wild perennial Helianthus L. species as a potential source for improvement of technical and technological properties of cultivated sunflower. Ind. Crop. Prod. 2020, 144, 112030. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Markin, N.V.; Azarin, K.V.; Gorbachenko, O.F.; Usatov, N.A. Comparative genomics of domesticated and wild sunflower: Complete chloroplast and mitochondrial genomes. Online J. Biol. Sci. 2016, 16, 71–75. [Google Scholar] [CrossRef][Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In Lecture Notes in Computer Science, Proceedings of the Research in Computational Molecular Biology, Beijing, China, 7–10 April 2013; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Varré, J.-S.; D′Agostino, N.; Touzet, P.; Gallina, S.; Tamburino, R.; Cantarella, C.; Ubrig, E.; Cardi, T.; Drouard, L.; Gualberto, J.M.; et al. Complete sequence, multichromosomal architecture and transcriptome analysis of the Solanum tuberosum mitochondrial genome. Int. J. Mol. Sci. 2019, 20, 4788. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018, 8, 12714. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.G.; Francs-Small, C.D.C.; Ostersetzer, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Kornienko, I.V.; Horn, R. Organization features of the mitochondrial genome of sunflower (Helianthus annuus L.) with ANN2-type male-sterile cytoplasm. Plants 2019, 8, 439. [Google Scholar] [CrossRef]
- Horn, R. Molecular diversity of male sterility inducing and male-fertile cytoplasms in the genus Helianthus. Appl. Genet. 2002, 104, 562–570. [Google Scholar] [CrossRef]
- Qiu, F.; Baack, E.J.; Whitney, K.D.; Bock, D.G.; Tetreault, H.M.; Rieseberg, L.H.; Ungerer, M.C. Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytol. 2019, 221, 1609–1618. [Google Scholar] [CrossRef]
- Kane, N.C.; Burke, J.M.; Marek, L.; Seiler, G.; Vear, F.; Baute, G.; Knapp, S.J.; Vincourt, P.; Rieseberg, L.H. Sunflower genetic, genomic and ecological resources. Mol. Ecol. Resour. 2013, 13, 10–20. [Google Scholar] [CrossRef]
- Ostevik, K.L.; Samuk, K.; Rieseberg, L.H. Ancestral reconstruction of karyotypes reveals an exceptional rate of nonrandom chromosomal evolution in sunflower. Genetics 2020, 214, 1031–1045. [Google Scholar] [CrossRef]
- Stephens, J.D.; Rogers, W.L.; Mason, C.M.; Donovan, L.A.; Malmberg, R.L. Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Am. J. Bot. 2015, 102, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Kesseli, R.; Ochoa, O.; Michelmore, R. Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce (L. sativa). Genome 1991, 34, 430–436. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Gandini, C.L.; Garcia, L.E.; Abbona, C.C.; Sanchez-Puerta, M.V. The complete organelle genomes of Physochlaina orientalis: Insights into short sequence repeats across seed plant mitochondrial genomes. Mol. Phylogenetics Evol. 2019, 137, 274–284. [Google Scholar] [CrossRef]
- Newton, K.J.; Gabay-Laughnan, S.; De Paepe, R. Mitochondrial Mutations in Plants. In Plant Mitochondria: From Genome to Function; Day, D.A., Millar, A.H., Whelan, J., Eds.; Advances in Photosynthesis and, Respiration; Springer: Dordrecht, The Netherlands, 2004; pp. 121–141. ISBN 978-1-4020-2400-9. [Google Scholar]
- Bock, R.; Knoop, V. (Eds.) Genomics of Chloroplasts and Mitochondria; Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2012; ISBN 978-94-007-2919-3. [Google Scholar]
- Bock, D.G.; Andrew, R.L.; Rieseberg, L.H. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 2014, 23, 4899–4911. [Google Scholar] [CrossRef]
- Nugent, J.M.; Palmer, J.D. Location, identity, amount and serial entry of chloroplast DNA sequences in crucifer mitochondrial DNAs. Curr. Genet. 1988, 14, 501–509. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.-G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Zancani, M.; Braidot, E.; Filippi, A.; Lippe, G. Structural and functional properties of plant mitochondrial F-ATP synthase. Mitochondrion 2020, 53, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Grimes, B.T.; Sisay, A.K.; Carroll, H.D.; Cahoon, A.B. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genom. 2014, 15, 31. [Google Scholar] [CrossRef]
- Tang, H.; Zheng, X.; Li, C.; Xie, X.; Chen, Y.; Chen, L.; Zhao, X.; Zheng, H.; Zhou, J.; Ye, S.; et al. Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Res. 2017, 27, 130–146. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).